N-Terminal α-Helices in Domain I of Bacillus thuringiensis Vip3Aa Play Crucial Roles in Disruption of Liposomal Membrane
Abstract
:1. Introduction
2. Results
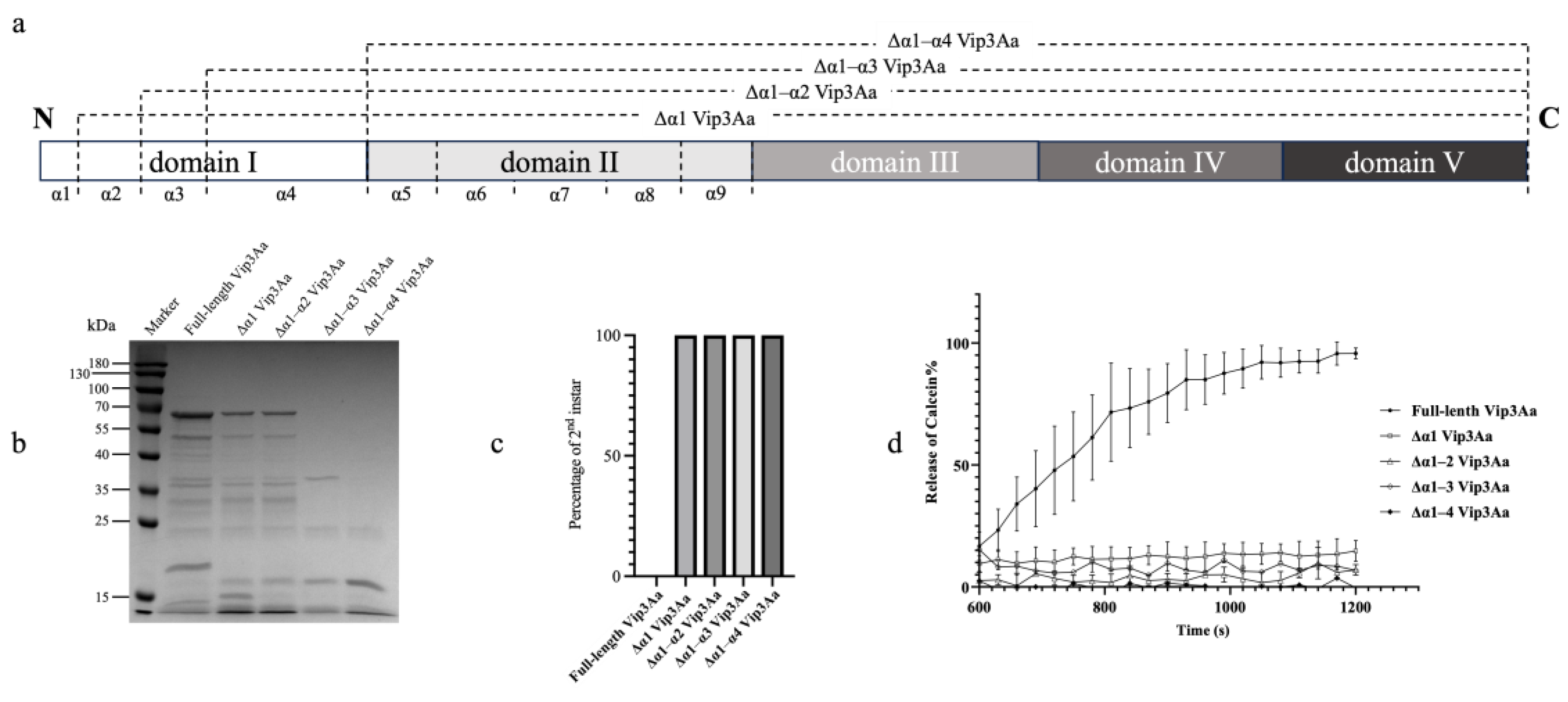
2.1. Proteolytic Processing and Bioactivity of N-Terminus Truncated Vip3Aa Proteins
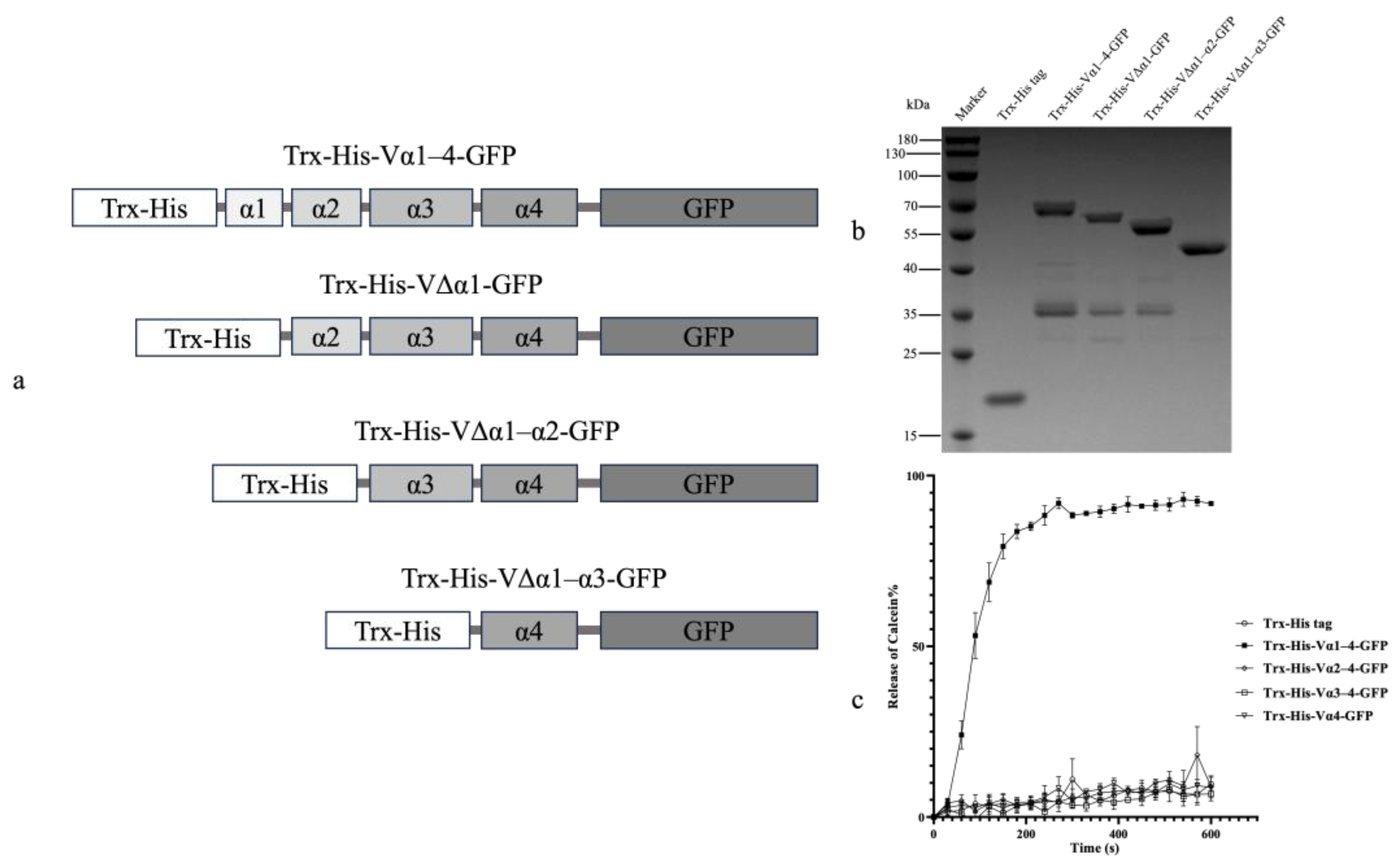
2.2. Liposome Permeability of Vip3Aa Domain I and GFP Fusion Proteins
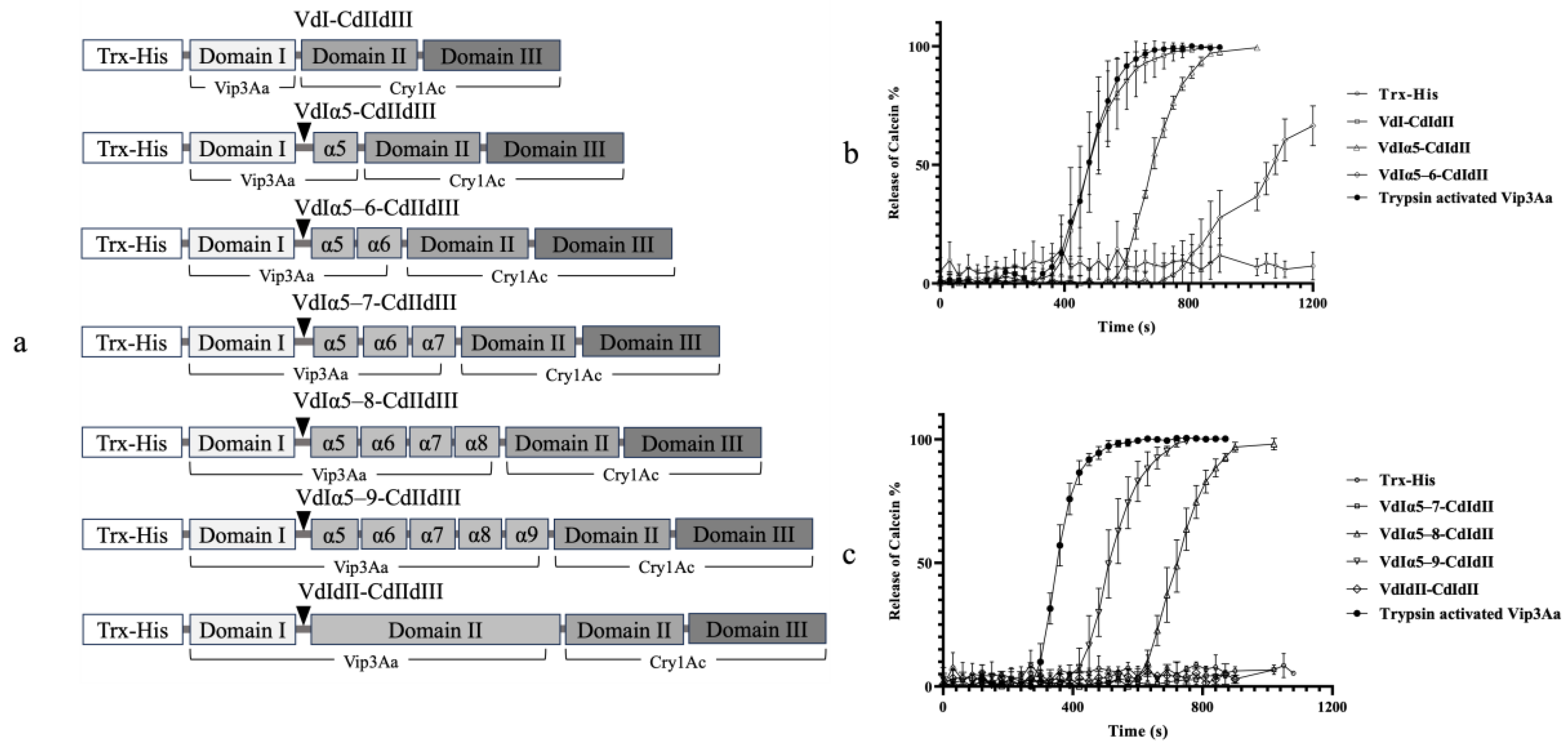
2.3. Bioactivity and Proteolytic Stability of Vip3Aa-Cry1Ac Recombinant Proteins
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Construction of Expression Vectors
5.2. Expression and Purification of Recombinant Proteins
5.3. Insect Rearing and Bioassays
5.4. In Vitro Proteolytic Processing of Recombinant Proteins
5.5. Liposome Fluorescence Permeability Assay of Vip3Aa Complex Activities
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tabashnik, B.E.; Carrière, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 2017, 35, 926–935. [Google Scholar] [CrossRef]
- Bravo, A.; Likitvivatanavong, S.; Gill, S.S.; Soberón, M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423–431. [Google Scholar] [CrossRef]
- Kurtz, R.W. A review of Vip3A mode of action and effects on Bt Cry protein-resistant colonies of lepidopteran larvae. Southwest. Entomol. 2010, 35, 391–394. [Google Scholar] [CrossRef]
- Soares Figueiredo, C.; Nunes Lemes, A.R.; Sebastião, I.; Desidério, J.A. Synergism of the Bacillus thuringiensis Cry1, Cry2, and Vip3 Proteins in Spodoptera frugiperda Control. Appl. Biochem. Biotechnol. 2019, 188, 798–809. [Google Scholar] [CrossRef]
- Yang, F.; González, J.C.S.; Williams, J.; Cook, D.C.; Gilreath, R.T.; Kerns, D.L. Occurrence and ear damage of Helicoverpa zea on transgenic Bacillus thuringiensis maize in the field in Texas, US and its susceptibility to Vip3A protein. Toxins 2019, 11, 102. [Google Scholar] [CrossRef]
- Quan, Y. Studies on the Insecticidal Mechanism of Bacillus thuringiensis Vip3A and Cry Proteins. 2022. Available online: https://dialnet.unirioja.es/servlet/dctes?codigo=305161 (accessed on 6 January 2024).
- Quan, Y.; Lázaro-Berenguer, M.; Hernández-Martínez, P.; Ferré, J.; Rudi, K. Critical Domains in the Specific Binding of Radiolabeled Vip3Af Insecticidal Protein to Brush Border Membrane Vesicles from Spodoptera spp. and Cultured Insect Cells. Appl. Environ. Microbiol. 2021, 87, e01787-21. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; Jin, M.; Wu, C.; Chakraborty, P.; Xiao, Y. Bacillus thuringiensis vegetative insecticidal protein family Vip3A and mode of action against pest Lepidoptera. Pest Manag. Sci. 2020, 76, 1612–1617. [Google Scholar] [CrossRef]
- Chakroun, M.; Banyuls, N.; Bel, Y.; Escriche, B.; Ferré, J. Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 329–350. [Google Scholar] [CrossRef] [PubMed]
- Bel, Y.; Banyuls, N.; Chakroun, M.; Escriche, B.; Ferré, J. Insights into the structure of the Vip3Aa insecticidal protein by protease digestion analysis. Toxins 2017, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Ferré, J. Structural Domains of the Bacillus thuringiensis Vip3Af Protein Unraveled by Tryptic Digestion of Alanine Mutants. Toxins 2019, 11, 368. [Google Scholar] [CrossRef]
- Shao, E.; Zhang, A.; Yan, Y.; Wang, Y.; Jia, X.; Sha, L.; Guan, X.; Wang, P.; Huang, Z. Oligomer Formation and Insecticidal Activity of Bacillus thuringiensis Vip3Aa Toxin. Toxins 2020, 12, 274. [Google Scholar] [CrossRef]
- Banyuls, N.; Hernández-Martínez, P.; Quan, Y.; Ferré, J. Artefactual band patterns by SDS-PAGE of the Vip3Af protein in the presence of proteases mask the extremely high stability of this protein. Int. J. Biol. Macromol. 2018, 120, 59–65. [Google Scholar] [CrossRef]
- Núñez-Ramírez, R.; Huesa, J.; Bel, Y.; Ferré, J.; Casino, P.; Arias-Palomo, E. Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis. Nat. Commun. 2020, 11, 3974. [Google Scholar] [CrossRef]
- Byrne, M.J.; Iadanza, M.G.; Perez, M.A.; Maskell, D.P.; George, R.M.; Hesketh, E.L.; Beales, P.A.; Zack, M.D.; Berry, C.; Thompson, R.F. Cryo-EM structures of an insecticidal Bt toxin reveal its mechanism of action on the membrane. Nat. Commun. 2021, 12, 2791. [Google Scholar] [CrossRef]
- Zheng, M.; Evdokimov, A.G.; Moshiri, F.; Lowder, C.; Haas, J. Crystal structure of a Vip3B family insecticidal protein reveals a new fold and a unique tetrameric assembly. Protein Sci. 2020, 29, 824–829. [Google Scholar] [CrossRef]
- Lázaro-Berenguer, M.; Paredes-Martínez, F.; Bel, Y.; Núñez-Ramírez, R.; Arias-Palomo, E.; Casino, P.; Ferré, J. Structural and functional role of Domain I for the insecticidal activity of the Vip3Aa protein from Bacillus thuringiensis. Microb. Biotechnol. 2022, 15, 2607–2618. [Google Scholar] [CrossRef]
- Jiang, K.; Zhang, Y.; Chen, Z.; Wu, D.; Cai, J.; Gao, X. Structural and functional insights into the C-terminal fragment of insecticidal Vip3A toxin of Bacillus thuringiensis. Toxins. 2020, 12, 438. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Carroll, J.; Ellar, D.J. Crystal structure of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature 1991, 353, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Grochulski, P.; Masson, L.; Borisova, S.; Pusztai-Carey, M.; Schwartz, J.L.; Brousseau, R.; Cygler, M. Bacillus thuringiensis CrylA (a) Insecticidal Toxin: Crystal Structure and Channel Formation. J. Mol. Biol. 1995, 254, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Pardo-López, L.; Soberón, M.; Bravo, A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 2013, 37, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Palma, L.; Muñoz, D.; Berry, C.; Murillo, J.; Caballero, P. Bacillus thuringiensis toxins: An overview of their biocidal activity. Toxins 2014, 6, 3296–3325. [Google Scholar] [CrossRef]
- Pacheco, S.; Gómez, I.; Peláez-Aguilar, A.E.; Verduzco-Rosas, L.A.; García-Suárez, R.; do Nascimento, N.A.; Rivera-Nájera, L.Y.; Cantón, P.E.; Soberón, M.; Bravo, A. Structural changes upon membrane insertion of the insecticidal pore-forming toxins produced by Bacillus thuringiensis. Front. Insect Sci. 2023, 3, 1188891. [Google Scholar] [CrossRef]
- Zack, M.D.; Sopko, M.S.; Frey, M.L.; Wang, X.; Tan, S.Y.; Arruda, J.M.; Letherer, T.T.; Narva, K.E. Functional characterization of Vip3Ab1 and Vip3Bc1: Two novel insecticidal proteins with differential activity against lepidopteran pests. Sci. Rep. 2017, 7, 11112. [Google Scholar] [CrossRef]
- Selvapandiyan, A.; Arora, N.; Rajagopal, R.; Jalali, S.; Venkatesan, T.; Singh, S.; Bhatnagar, R.K. Toxicity analysis of N-and C-terminus-deleted vegetative insecticidal protein from Bacillus thuringiensis. Appl. Environ. Microbiol. 2001, 67, 5855–5858. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.B. Soluble expression of archaeal proteins in Escherichia coli by using fusion-partners. Protein Expr. Purif. 2008, 62, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Pardo-López, L.; Gómez, I.; Muñoz-Garay, C.; Jiménez-Juarez, N.; Soberón, M.; Bravo, A. Structural and functional analysis of the pre-pore and membrane-inserted pore of Cry1Ab toxin. J. Invertebr. Pathol. 2006, 92, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Lebel, G.; Vachon, V.; Préfontaine, G.; Girard, F.; Masson, L.; Juteau, M.; Bah, A.; Larouche, G.; Vincent, C.; Laprade, R. Mutations in domain I interhelical loops affect the rate of pore formation by the Bacillus thuringiensis Cry1Aa toxin in insect midgut brush border membrane vesicles. Appl. Environ. Microbiol. 2009, 75, 3842–3850. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Z.; Geng, L.; Chi, B.; Liu, R.; Li, H.; Gao, J.; Zhang, J. Vip3Aa domain IV and V mutants confer higher insecticidal activity against Spodoptera frugiperda and Helicoverpa armigera. Pest Manag. Sci. 2022, 78, 2324–2331. [Google Scholar] [CrossRef] [PubMed]
- Gomez, I.; Arenas, I.; Benitez, I.; Miranda-Rios, J.; Becerril, B.; Grande, R.; Almagro, J.C.; Bravo, A.; Soberon, M. Specific epitopes of domains II and III of Bacillus thuringiensis Cry1Ab toxin involved in the sequential interaction with cadherin and aminopeptidase-N receptors in Manduca sexta. J. Biol. Chem. 2006, 281, 34032–34039. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Solis, M.; Pinos, D.; Endo, H.; Portugal, L.; Sato, R.; Ferre, J.; Herrero, S.; Hernández-Martínez, P. Role of Bacillus thuringiensis Cry1A toxins domains in the binding to the ABCC2 receptor from Spodoptera exigua. Insect Biochem. Mol. Biol. 2018, 101, 47–56. [Google Scholar] [CrossRef]
- Herrero, S.; Bel, Y.; Hernández-Martínez, P.; Ferré, J. Susceptibility, mechanisms of response and resistance to Bacillus thuringiensis toxins in Spodoptera spp. Curr. Opin. Insect Sci. 2016, 15, 89–96. [Google Scholar] [CrossRef]
- Dong, F.; Shi, R.; Zhang, S.; Zhan, T.; Wu, G.; Shen, J.; Liu, Z. Fusing the vegetative insecticidal protein Vip3Aa7 and the N terminus of Cry9Ca improves toxicity against Plutella xylostella larvae. Appl. Microbiol. Biotechnol. 2012, 96, 921–929. [Google Scholar] [CrossRef]
- Gulzar, A.; Wright, D.J. Sub-lethal effects of Vip3A toxin on survival, development and fecundity of Heliothis virescens and Plutella xylostella. Ecotoxicology 2015, 24, 1815–1822. [Google Scholar] [CrossRef]
- Mosbahi, K.; Walker, D.; Lea, E.; Moore, G.R.; James, R.; Kleanthous, C. Destabilization of the colicin E9 Endonuclease domain by interaction with negatively charged phospholipids: Implications for colicin translocation into bacteria. J. Biol. Chem. 2004, 279, 22145–22151. [Google Scholar] [CrossRef]
- Zakharov, S.D.; Cramer, W.A. Colicin crystal structures: Pathways and mechanisms for colicin insertion into membranes. Biochim. Biophys. Acta-Biomembr. 2002, 1565, 333–346. [Google Scholar] [CrossRef]
- Kunthic, T.; Watanabe, H.; Kawano, R.; Tanaka, Y.; Promdonkoy, B.; Yao, M.; Boonserm, P. pH regulates pore formation of a protease activated Vip3Aa from Bacillus thuringiensis. Biochim. Biophys. Acta-Biomembr. 2017, 1859, 2234–2241. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Higuchi, M.; Endo, R.; Maruyama, T.; Haginoya, K.; Shitomi, Y.; Hayakawa, T.; Mitsui, T.; Sato, R.; Hori, H. Bacillus thuringiensis Cry1Ab, but not Cry1Aa or Cry1Ac, disrupts liposomes. Pestic. Biochem. Physiol. 2006, 84, 1–9. [Google Scholar] [CrossRef]
- Fortea, E.; Lemieux, V.; Potvin, L.; Chikwana, V.; Griffin, S.; Hey, T.; McCaskill, D.; Narva, K.; Tan, S.Y.; Xu, X. Cry6Aa1, a Bacillus thuringiensis nematocidal and insecticidal toxin, forms pores in planar lipid bilayers at extremely low concentrations and without the need of proteolytic processing. J. Biol. Chem. 2017, 292, 13122–13132. [Google Scholar] [CrossRef] [PubMed]
- Coux, F.; Vachon, V.; Rang, C.; Moozar, K.; Masson, L.; Royer, M.; Bes, M.; Rivest, S.; Brousseau, R.; Schwartz, J.-L. Role of interdomain salt bridges in the pore-forming ability of the Bacillus thuringiensis toxins Cry1Aa and Cry1Ac. J. Biol. Chem. 2001, 276, 35546–35551. [Google Scholar] [CrossRef] [PubMed]
- Sellami, S.; Jemli, S.; Abdelmalek, N.; Cherif, M.; Abdelkefi-Mesrati, L.; Tounsi, S.; Jamoussi, K. A novel Vip3Aa16-Cry1Ac chimera toxin: Enhancement of toxicity against Ephestia kuehniella, structural study and molecular docking. Int. J. Biol. Macromol. 2018, 117, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Chen, C.; Wu, S.; Shao, E.; Li, M.; Guan, X.; Huang, Z. Transcriptional profiling analysis of Spodoptera litura larvae challenged with Vip3Aa toxin and possible involvement of trypsin in the toxin activation. Sci. Rep. 2016, 6, 23861. [Google Scholar] [CrossRef]
- Pogulis, R.J.; Vallejo, A.N.; Pease, L.R. In vitro recombination and mutagenesis by overlap extension PCR. Vitr. Mutagen. Protoc. 1996, 57, 167–176. [Google Scholar]
- Adang, M.J.; Staver, M.J.; Rocheleau, T.A.; Leighton, J.; Barker, R.F.; Thompson, D.V. Characterized full-length and truncated plasmid clones of the crystal protein of Bacillus thuringiensis subsp. kurstaki HD-73 and their toxicity to Manduca sexta. Gene 1985, 36, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Amir, M.B.; Liao, Y.; Yu, H.; He, W.; Lu, Z. New insights into the Plutella xylostella detoxifying enzymes: Sequence evolution, structural similarity, functional diversity, and application prospects of glucosinolate sulfatases. J. Agric. Food Chem. 2023, 71, 10952–10969. [Google Scholar] [CrossRef] [PubMed]
- Kain, W.C.; Zhao, J.-Z.; Janmaat, A.F.; Myers, J.; Shelton, A.M.; Wang, P. Inheritance of resistance to Bacillus thuringiensis Cry1Ac toxin in a greenhouse-derived strain of cabbage looper (Lepidoptera: Noctuidae). J. Econ. Entomol. 2004, 97, 2073–2078. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, J.-Z.; Rodrigo-Simón, A.; Kain, W.; Janmaat, A.F.; Shelton, A.M.; Ferré, J.; Myers, J. Mechanism of resistance to Bacillus thuringiensis toxin Cry1Ac in a greenhouse population of the cabbage looper, Trichoplusia ni. Appl. Environ. Microbiol. 2007, 73, 1199–1207. [Google Scholar] [CrossRef]
- Pan, Z.-Z.; Xu, L.; Liu, B.; Chen, Q.-X.; Zhu, Y.-J. Key residues of Bacillus thuringiensis Cry2Ab for oligomerization and pore-formation activity. AMB Express 2021, 11, 112. [Google Scholar] [CrossRef]

| Primers | Sequences (5′ to 3′) |
|---|---|
| Vip3Aa-F | acaaggccatggctgatatcGGATCCCTGGAGGTGCTGTTCCAGGGGCCCATGAACAAGAATAATACTAAATTA |
| Δα1 Vip3Aa-F | acaaggccatggctgatatcGGATCCCTGGAAGTTCTGTTCCAGGGGCCCACGGATACAGGTGGTGATCTAA |
| Δα1–α2 Vip3Aa-F | acaaggccatggctgatatcGGATCCCTGGAAGTTCTGTTCCAGGGGCCCGGAAACTTAAATACAGAATTAT |
| Δα1–α3 Vip3Aa-F | acaaggccatggctgatatcGGATCCCTGGAAGTTCTGTTCCAGGGGCCCAAGTTGGATATTATTAATGTAA |
| Δα4 Vip3Aa-F | acaaggccatggctgatatcGGATCCCTGGAAGTTCTGTTCCAGGGGCCCGAAACTAGTTCAAAAGTA |
| Vip3Aa-R | agtggtggtggtggtggtg CTCGAGTTACTTAATAGAGACATCGT |
| Vip3A-DI-GFP-R | agtgaaaagttcttctcctttactcatAGCAAAAGTTAATTCCTCAAATT |
| GFP-F | TTGAGGAATTAACTTTTGCTatgagtaaaggagaagaacttttcact |
| GFP-R | gttagcagccggatctcagtggtggtggtggtggtgCTCGAGttatttgtatagttcatccatgccatgt |
| Primers | Sequences (5′ to 3′) |
|---|---|
| Vip3Aa-F | acaaggccatggctgatatcGGATCCCTGGAGGTGCTGTTCCAGGGGCCCATGAACAAGAATAATACTAAATTA |
| VdI-Cry-R | aattgggaaactgttcgaattggTTCTGTAGCAAAAGTTAAT |
| VdI-Cry-F | TTCTTGATGAGTTAACTGAGccaattcgaacagtttcccaatt |
| VdIα5-Cry-R | aattgggaaactgttcgaattggTACACTTTTCGCTAGTTCAGTT |
| VdIα5-Cry-F | TTAACTGAACTAGCGAAAAGTGTAccaattcgaacagtttcccaatt |
| VdIα5–6-Cry-R | aattgggaaactgttcgaattggCATTACATCGTGGAATGTATTAA |
| VdIα5–6-Cry-F | TTAATACATTCCACGATGTAATGccaattcgaacagtttcccaatt |
| VdIα5–7-Cry-R | aattgggaaactgttcgaattggATTTTCTTTAGTAATTAATTCC |
| VdIα5–7-Cry-F | GGAATTAATTACTAAAGAAAATccaattcgaacagtttcccaatt |
| VdIα5–8-Cry-R | aattgggaaactgttcgaattggTAATAATTTTCGGCATGTTGTTAA |
| VdIα5–8-Cry-F | TTAACAACATGCCGAAAATTATTAccaattcgaacagtttcccaatt |
| VdIα5–9-Cry-R | aattgggaaactgttcgaattggGTTTACTCTAAATTCCTCTTTTTC |
| VdIα5–9-Cry-F | GAAAAAGAGGAATTTAGAGTAAACccaattcgaacagtttcccaatt |
| VdIdII-Cry-R | aattgggaaactgttcgaattggAGAAAGTGTAGGGAGGATGTTTACTC |
| VdIdII-Cry-F | GAGTAAACATCCTCCCTACACTTTCTccaattcgaacagtttcccaatt |
| Cry1Ac-R | gtggtggtggtggtggtgctcgagttatgcagtaactggaataaattcaaatc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, E.; Huang, H.; Yuan, J.; Yan, Y.; Ou, L.; Chen, X.; Pan, X.; Guan, X.; Sha, L. N-Terminal α-Helices in Domain I of Bacillus thuringiensis Vip3Aa Play Crucial Roles in Disruption of Liposomal Membrane. Toxins 2024, 16, 88. https://doi.org/10.3390/toxins16020088
Shao E, Huang H, Yuan J, Yan Y, Ou L, Chen X, Pan X, Guan X, Sha L. N-Terminal α-Helices in Domain I of Bacillus thuringiensis Vip3Aa Play Crucial Roles in Disruption of Liposomal Membrane. Toxins. 2024; 16(2):88. https://doi.org/10.3390/toxins16020088
Chicago/Turabian StyleShao, Ensi, Hanye Huang, Jin Yuan, Yaqi Yan, Luru Ou, Xiankun Chen, Xiaohong Pan, Xiong Guan, and Li Sha. 2024. "N-Terminal α-Helices in Domain I of Bacillus thuringiensis Vip3Aa Play Crucial Roles in Disruption of Liposomal Membrane" Toxins 16, no. 2: 88. https://doi.org/10.3390/toxins16020088





