Venom Ontogeny in the Mexican Lance-Headed Rattlesnake (Crotalus polystictus)
Abstract
:1. Introduction
2. Results
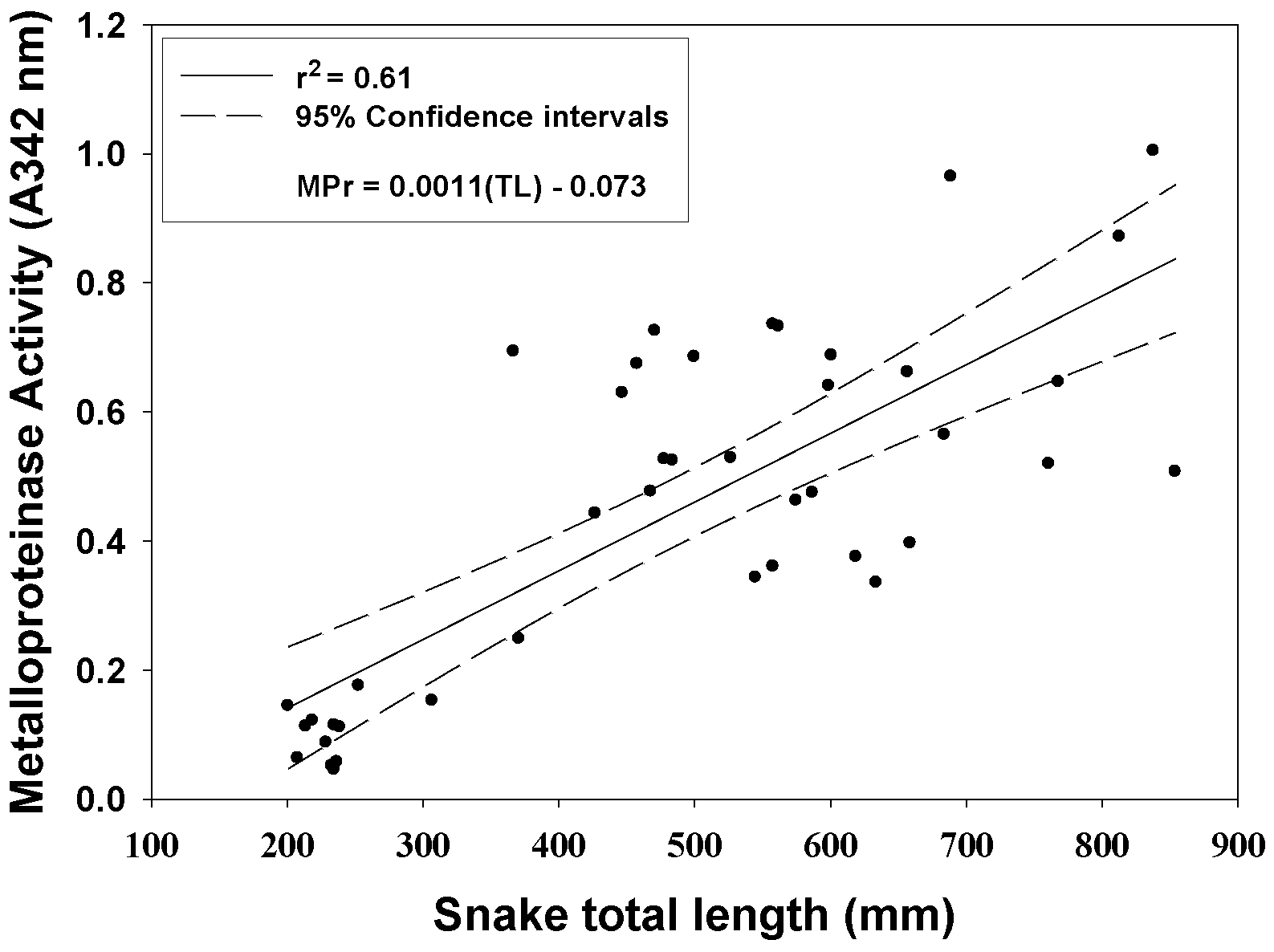
2.1. Enzyme Analyses
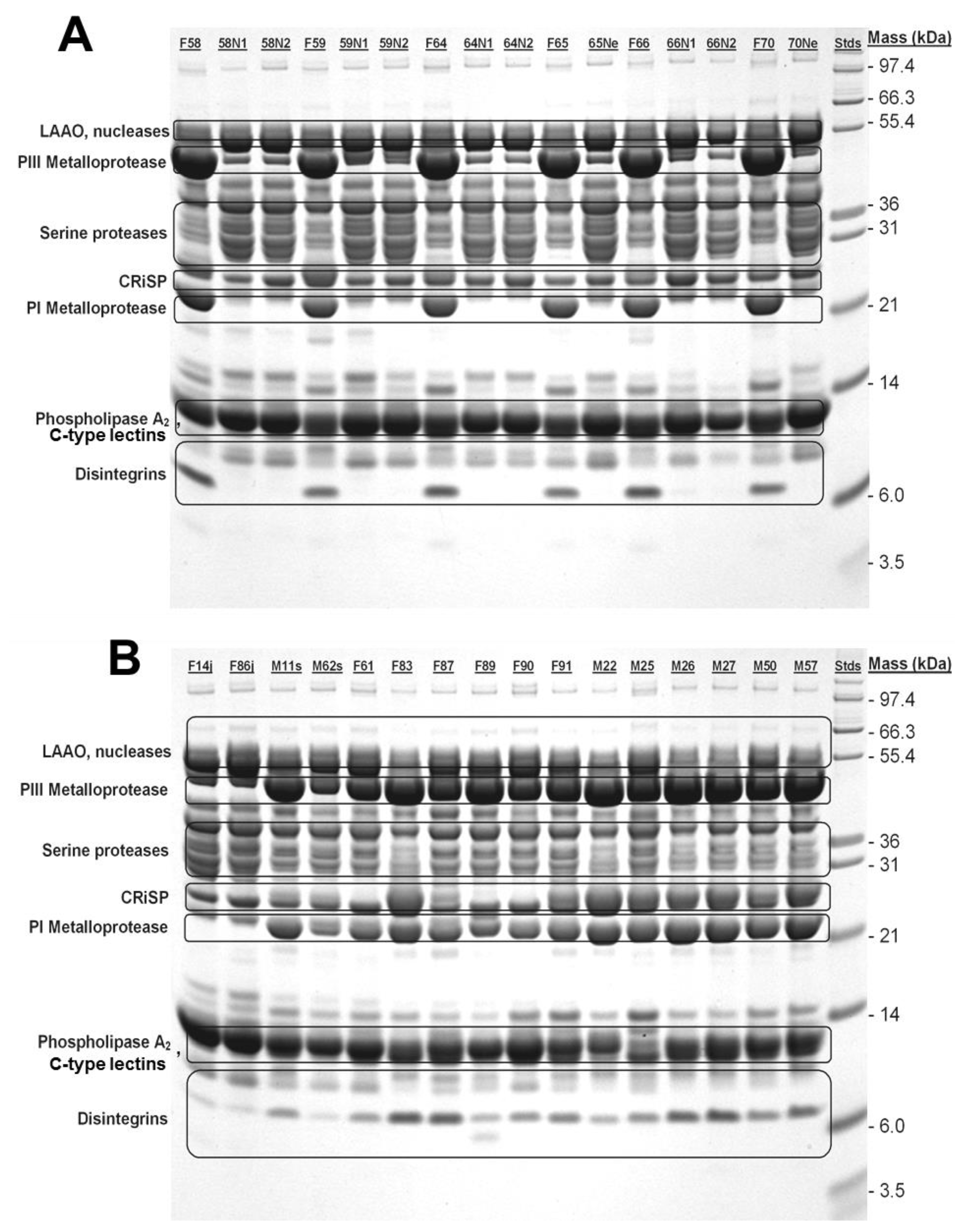
2.2. 1-D SDS-PAGE
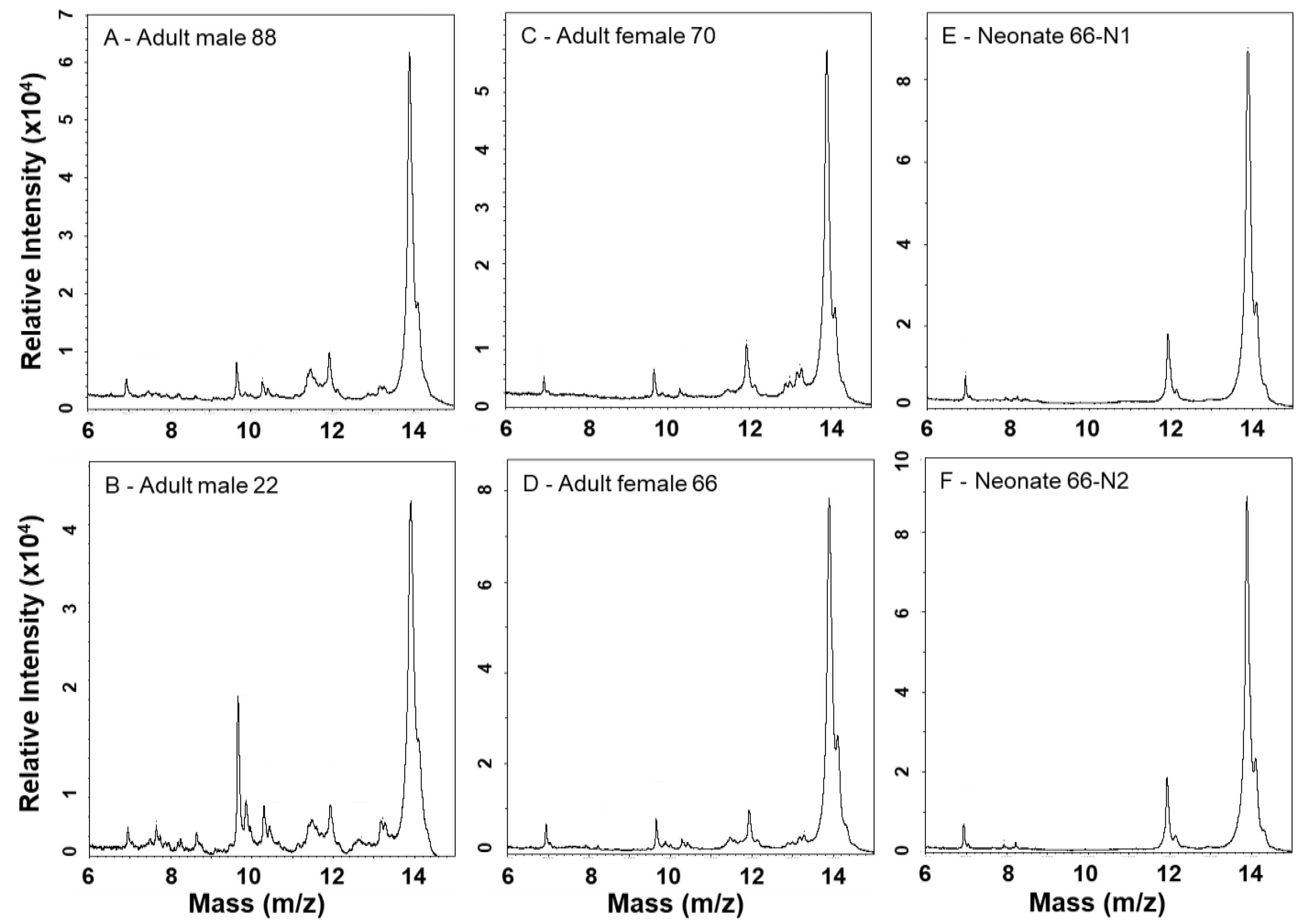
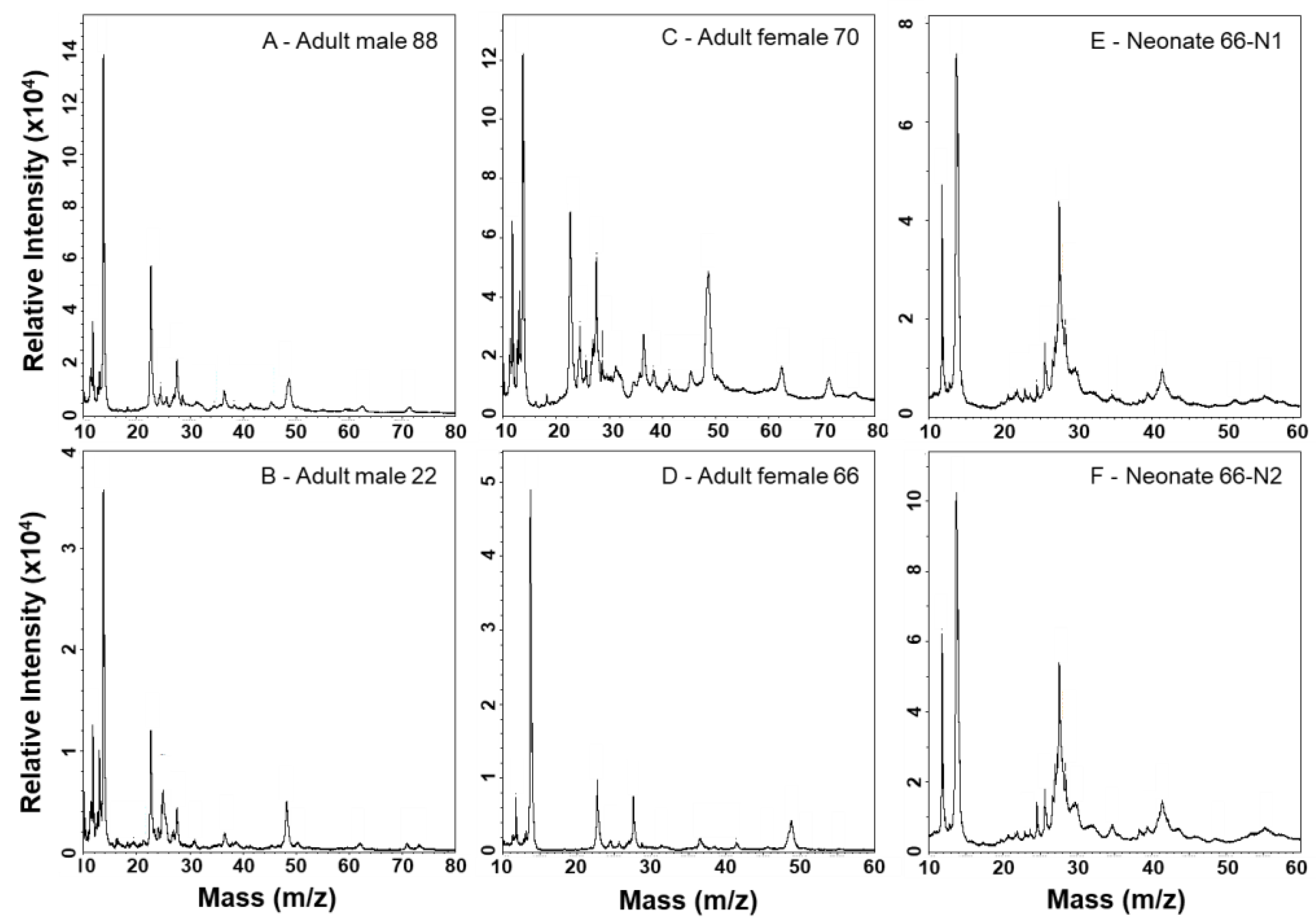
2.3. MALDI-TOF-MS Mass Fingerprinting of Venoms
2.4. MALDI-TOF-MS/MS Identification of Specific Proteins
2.5. Mass Spectroscopy: Orbitrap LC-MS/MS Identification of Venom Proteome
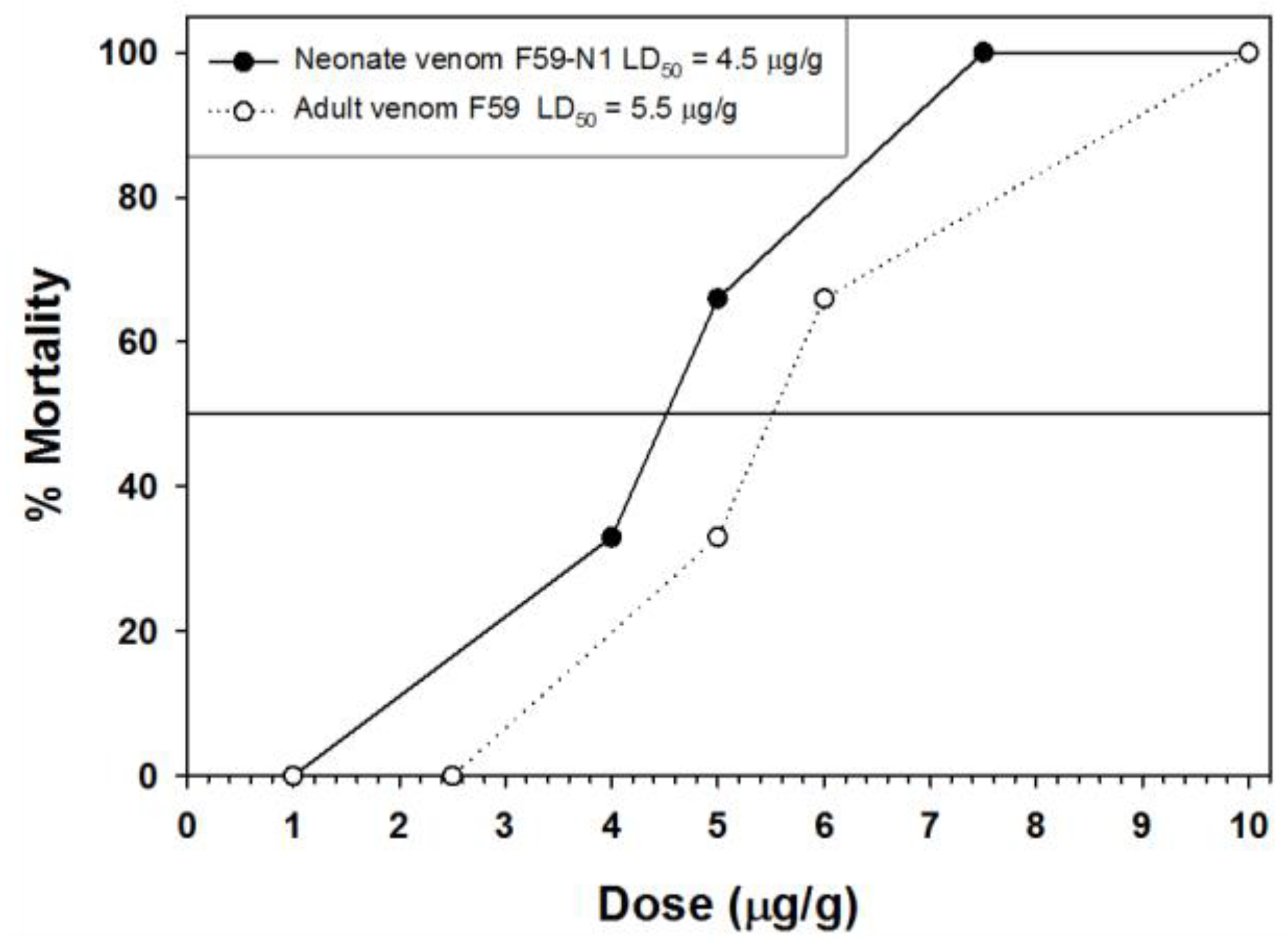
2.6. Toxicity of Neonate and Adult Venom to Mice
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Reagents
5.2. Venoms
5.3. Enzyme Assays
5.4. One-dimensional SDS Gel Electrophoresis
5.5. Mass Spectrometry: MALDI-TOF
5.6. Mass Spectrometry: Orbitrap LC-MS/MS
5.7. Mass Spectrometry: MALDI-TOF-MS/MS
5.8. Venom Lethality
5.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Castoe, T.A.; Parkinson, C.L. Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes). Mol. Phylogenet. Evol. 2006, 39, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Klauber, L.M. Their habits, life histories and influences on Mankind. In Rattlesnakes; University of California Press: Berkeley, CA, USA, 1956; Volume 2, p. 153. [Google Scholar]
- Blair, C.; Sanchez-Ramirez, S. Diversity-dependent cladogenesis throughout western Mexico: Evolutionary biogeography of rattlesnakes (Viperidae: Crotalinae: Crotalus and Sistrurus). Mol. Phylogenet. Evol. 2016, 97, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.A.; Lamar, W.W. The Venomous Reptiles of the Western Hemisphere; Cornell University Press: Ithaca, NY, USA, 2004. [Google Scholar]
- Castro, E.N.; Lomonte, B.; del Carmen Gutiérrez, M.; Alagón, A.; Gutiérrez, J.M. Intraspecies variation in the venom of the rattlesnake Crotalus simus from Mexico: Different expression of crotoxin results in highly variable toxicity in the venoms of three subspecies. J. Proteom. 2013, 87, 103–121. [Google Scholar] [CrossRef] [PubMed]
- Durban, J.; Sanz, L.; Trevisan-Silva, D.; Neri-Castro, E.; Alagón, A.; Calvete, J.J. Integrated venomics and venom gland transcriptome analysis of juvenile and adult Mexican rattlesnakes Crotalus simus, C. tzabcan, and C. culminatus revealed miRNA-modulated ontogenetic shifts. J. Proteome Res. 2017, 16, 3370–3390. [Google Scholar] [CrossRef] [PubMed]
- Borja, M.; Neri-Castro, E.; Castañeda-Gaytán, G.; Strickland, J.L.; Parkinson, C.L.; Castañeda-Gaytán, J.; Ponce-López, R.; Lomonte, B.; Olvera-Rodríguez, A.; Alagón, A.; et al. Biological and Proteolytic variation in the venom of Crotalus scutulatus scutulatus from México. Toxins 2018, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Romero, G.; Rucavado, A.; Lazcano, D.; Gutiérrez, J.M.; Borja, M.; Lomonte, B.; Garza-García, Y.; Zugasti-Cruz, A. Comparison of venom composition and biological activities of the subspecies Crotalus lepidus lepidus, Crotalus lepidus klauberi and Crotalus lepidus morulus from Mexico. Toxicon 2013, 71, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Rivas, E.; Neri-Castro, E.; Bénard-Valle, M.; Hernánez-Dávila, A.I.; Zamudio, F.; Alagón, A. General characterization of the venoms from two species of rattlesnakes and an intergrade population (C. lepidus x aquilus) from Aguascalientes and Zacatecas, Mexico. Toxicon 2017, 138, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Saviola, A.J.; Gandara, A.J.; Bryson, R.W., Jr.; Mackessy, S.P. Venom phenotypes of the Rock Rattlesnake (Crotalus lepidus) and the Ridge-nosed Rattlesnake (Crotalus willardi) from México and the United States. Toxicon 2017, 138, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P. Morphology and ultrastructure of the venom glands of the northern Pacific rattlesnake Crotalus viridis oreganus. J. Morphol. 1991, 208, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Baxter, L.M. Bioweapons synthesis and storage: The venom gland of front-fanged snakes. Zool. Anz. 2006, 245, 147–159. [Google Scholar] [CrossRef]
- Sanz, L.; Gibbs, H.L.; Mackessy, S.P.; Calvete, J.J. Venom proteomes of closely related Sistrurus rattlesnakes with divergent diets. J. Proteome Res. 2006, 5, 2098–2112. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Juárez, P.; Sanz, L. Snake venomics. Strategy and applications. J. Mass Spectrom. 2007, 42, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P. Venom composition in rattlesnakes: Trends and biological significance. In The Biology of Rattlesnakes; Hayes, W.K., Beaman, K.R., Cardwell, M.D., Bush, S.P., Eds.; Loma Linda University Press: Loma Linda, CA, USA, 2008; pp. 495–510. [Google Scholar]
- Mackessy, S.P. The field of reptile toxinology: Snakes, lizards and their venoms. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2010; pp. 3–23. [Google Scholar]
- Tu, A.T. Rattlesnake venoms: Their actions and treatments. Can. Vet. J. 1983, 24, 203–204. [Google Scholar]
- Minton, S.A. Observations on toxicity and antigenetic makeup of venoms from juvenile snakes. In Animal Toxins; Russell, F.E., Saunders, P.R., Eds.; Pergamon Press: Oxford, UK, 1967; pp. 211–222. [Google Scholar]
- Reid, H.A.; Theakston, R.D.G. Changes in coagulation effects by venoms of Crotalus atrox as snakes age. Am. J. Trop. Med. Hyg. 1978, 27, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Gené, J.A.; Gutiérrez, J.M.; Cerdas, L. Estudio comparativo de los venenos de serpiente Cascabel (Crotalus durissus durissus) de ejemplares adultos y recien nacidos. Toxicon 1983, 21, 379–384. [Google Scholar] [CrossRef]
- Mackessy, S.P. Venom ontogeny in the Pacific rattlesnakes Crotalus viridis helleri and C. v. oreganus. Copeia 1988, 1988, 92–101. [Google Scholar] [CrossRef]
- Mackessy, S.P. Kallikrein-like and thrombin-like proteases from the venoms of juvenile and adult northern Pacific rattlesnakes (Crotalus viridis oreganus). J. Nat. Toxins 1993, 2, 223–239. [Google Scholar]
- Mackessy, S.P. Fibrinogenolytic proteases from the venoms of juvenile and adult northern Pacific rattlesnakes (Crotalus viridis oreganus). Comp. Biochem. Physiol. 1993, 106B, 181–189. [Google Scholar] [CrossRef]
- Mackessy, S.P.; Williams, K.; Ashton, K.G. Ontogenetic variation in venom composition and diet of Crotalus oreganus concolor: A case of venom paedomorphosis? Copeia 2003, 2003, 769–782. [Google Scholar] [CrossRef]
- Calvete, J.J.; Sanz, L.; Cid, P.; de la Torre, P.; Flores-Díaz, M.; Dos Santos, M.C.; Borges, A.; Bremo, A.; Angulo, Y.; Lomonte, B.; et al. Snake venomics of the Central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res. 2010, 9, 528–544. [Google Scholar] [CrossRef] [PubMed]
- Saviola, A.J.; Pla, D.; Sanz, L.; Castoe, T.A.; Calvete, J.J.; Mackessy, S.P. Comparative venomics of the Prairie Rattlesnake (Crotalus viridis viridis) from Colorado: Identification of a novel pattern of ontogenetic changes in venom composition and assessment of the immunoreactivity of the commercial antivenom CroFab®. J. Proteom. 2015, 121, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Wray, K.P.; Margres, M.J.; Seavy, M.; Rokyta, D.R. Early significant ontogenetic changes in snake venoms. Toxicon 2015, 96, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Glenn, J.L.; Straight, R. Mojave rattlesnake Crotalus scutulatus venom: Variation in toxicity with geographical origin. Toxicon 1978, 16, 81–84. [Google Scholar] [CrossRef]
- Núñez, V.; Cid, P.; Sanz, L.; De La Torre, P.; Angulo, Y.; Lomonte, B.; Gutiérrez, J.M.; Calvete, J.J. Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J. Proteom. 2009, 73, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Boldrini-França, J.; Corrêa-Netto, C.; Silva, M.M.; Rodrigues, R.S.; De La Torre, P.; Pérez, A.; Soares, A.M.; Zingali, R.B.; Nogueira, R.A.; Rodrigues, V.M.; et al. Snake venomics and antivenomics of Crotalus durissus subspecies from Brazil: Assessment of geographic variation and its implication on snakebite management. J. Proteom. 2010, 73, 1758–1776. [Google Scholar] [CrossRef] [PubMed]
- Glaudas, X.; Jezkova, T.; Rodríguez-Robles, J.A. Feeding ecology of the Great Basin Rattlesnake (Crotalus lutosus, Viperidae). Can. J. Zool. 2008, 86, 723–734. [Google Scholar] [CrossRef]
- Durban, J.; Pérez, A.; Sanz, L.; Gómez, A.; Bonilla, F.; Rodríguez, S.; Chacón, D.; Sasa, M.; Angulo, Y.; Gutiérrez, J.M.; et al. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genet. 2013, 14, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mociño-Deloya, E.; Setser, K.; Pleguezuelos, J.M.; Kardon, A.; Lazcano, D. Cannibalism of nonviable offspring by postparturient Mexican lance-headed rattlesnakes, Crotalus polystictus. Anim. Behav. 2009, 77, 145–150. [Google Scholar] [CrossRef]
- Setser, K.; Mociño-Deloya, E.; Pleguezuelos, J.M.; Lazcano, D.; Kardon, A. Reproductive ecology of female Mexican lance-headed rattlesnakes. J. Zool. 2010, 281, 175–182. [Google Scholar] [CrossRef]
- Meik, J.M.; Setser, K.; Mociño-Deloya, E.; Lawing, A.M. Sexual differences in head form and diet in a population of Mexican lance-headed rattlesnakes, Crotalus polystictus. Biol. J. Linn. Soc. 2012, 106, 633–640. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, J.W.; Serrano, S.M. Snake venom metalloproteinases. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2010; pp. 95–113. [Google Scholar]
- Jia, L.G.; Shimokawa, K.I.; Bjarnason, J.B.; Fox, J.W. Snake venom metalloproteinases: Structure, function and relationship to the ADAMs family of proteins. Toxicon 1996, 34, 1269–1276. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M.T. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 2005, 45, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Saviola, A.J. Understanding biological roles of venoms among the Caenophidia: The importance of rear-fanged snakes. Integr. Comp. Biol. 2016, 56, 1004–1021. [Google Scholar] [CrossRef] [PubMed]
- Saravia, P.; Rojas, E.; Arce, V.; Guevara, C.; López, J.C.; Chaves, E.; Velásquez, R.; Rojas, G.; Gutiérrez, J.M. Geographic and ontogenic variability in the venom of the neotropical rattlesnake Crotalus durissus: Pathophysiological and therapeutic implications. Rev. Biol. Trop. 2002, 50, 337–346. [Google Scholar] [PubMed]
- Kini, R.M.; Evans, H.J. Structural domains in venom proteins: Evidence that metalloproteinases and nonenzymatic platelet aggregation inhibitors (disintegrins) from snake venoms are derived by proteolysis from a common precursor. Toxicon 1992, 30, 265–293. [Google Scholar] [CrossRef]
- Calvete, J.J.; Marcinkiewicz, C.; Monleón, D.; Esteve, V.; Celda, B.; Juárez, P.; Sanz, L. Snake venom disintegrins: Evolution of structure and function. Toxicon 2005, 45, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wagstaff, S.C.; Harrison, R.A.; Renjifo, C.; Wüster, W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 2011, 28, 2637–2649. [Google Scholar] [CrossRef] [PubMed]
- Scarborough, R.M.; Rose, J.W.; Naughton, M.A.; Phillips, D.R.; Nannizzi, L.; Arfsten, A.; Campbell, A.M.; Charo, I.F. Characterization of the integrin specificities of disintegrins isolated from American pit viper venoms. J. Biol. Chem. 1993, 268, 1058–1065. [Google Scholar] [PubMed]
- Saviola, A.J.; Chiszar, D.; Busch, C.; Mackessy, S.P. Molecular basis for prey relocation in viperid snakes. BMC Biol. 2013, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Saviola, A.J.; Mackessy, S.P. Observations on the chemosensory responses of the midget faded rattlesnake (Crotalus oreganus concolor): Discrimination of envenomated prey in a type II venom species. J. Ethol. 2017, 35, 245–250. [Google Scholar] [CrossRef]
- Mackessy, S.P. Thrombin-like enzymes in snake venoms. In Toxins and Hemostasis; Kini, R., Clemetson, K., Markland, F., McLane, M., Morita, T., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 519–557. [Google Scholar]
- Swenson, S.; Markland, F.S. Snake venom fibrin(ogen)olytic enzymes. Toxicon 2005, 45, 1021–1039. [Google Scholar] [CrossRef] [PubMed]
- Markland, F.S.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Russell, F.E.; Beuss, F.W.; Woo, M.Y. Zootoxicological properties of venom phosphodiesterase. Toxicon 1963, 1, 99–108. [Google Scholar] [CrossRef]
- Aird, S.D. Ophidian envenomation strategies and the role of purines. Toxicon 2002, 40, 335–393. [Google Scholar] [CrossRef]
- Izidoro, L.F.M.; Sobrinho, J.C.; Mendes, M.M.; Costa, T.R.; Grabner, A.N.; Rodrigues, V.M.; da Silva, S.L.; Zanchi, F.B.; Zuliani, J.P.; Fernandes, C.F.; et al. Snake venom L-amino acid oxidases: Trends in pharmacology and biochemistry. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.S.; da Silva, J.F.; França, J.B.; Fonseca, F.P.; Otaviano, A.R.; Silva, F.H.; Hamaguchi, A.; Magro, A.J.; Braz, A.S.K.; dos Santos, J.I.; et al. Structural and functional properties of Bp-LAAO, a new L-amino acid oxidase isolated from Bothrops pauloensis snake venom. Biochimie 2009, 91, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Samel, M.; Vija, H.; Rönnholm, G.; Siigur, J.; Kalkkinen, N.; Siigur, E. Isolation and characterization of an apoptotic and platelet aggregation inhibiting L-amino acid oxidase from Vipera berus berus (common viper) venom. BBA Proteins Proteom. 2006, 1764, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Souza, D.H.; Eugenio, L.M.; Fletcher, J.E.; Jiang, M.S.; Garratt, R.C.; Oliva, G.; Selistre-de-Araujo, H.S. Isolation and structural characterization of a cytotoxic L-amino acid oxidase from Agkistrodon contortrix laticinctus snake venom: Preliminary crystallographic data. Arch. Biochem. Biophys. 1999, 368, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.L.; Fung, S.Y.; Chung, I.; Pailoor, J.; Cheah, S.H.; Tan, N.H. King cobra (Ophiophagus hannah) venom L-amino acid oxidase induces apoptosis in PC-3 cells and suppresses PC-3 solid tumor growth in a tumor xenograft mouse model. Int. J. Med. Sci. 2014, 11, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Saviola, A.J.; Burns, P.D.; Mackessy, S.P. Apoptosis induction in human breast cancer (MCF-7) cells by a novel venom L-amino acid oxidase (Rusvinoxidase) is independent of its enzymatic activity and is accompanied by caspase-7 activation and reactive oxygen species production. Apoptosis 2015, 20, 1358–1372. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.J.; Graham, C.; McClean, S.; Chen, T.; O’Rourke, M.; Hirst, D.; Theakston, D.; Shaw, C. Identification and functional analysis of a novel bradykinin inhibitory peptide in the venoms of New World Crotalinae pit vipers. Biochem. Biophys. Res. Commun. 2005, 338, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Fasoli, E.; Sanz, L.; Boschetti, E.; Righetti, P.G. Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 2009, 8, 3055–3067. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Lomonte, B.; Sanz, L.; Angulo, Y.; Gutiérrez, J.M.; Calvete, J.J. Snake venomics of Bothriechis nigroviridis reveals extreme variability among palm pitviper venoms: Different evolutionary solutions for the same trophic purpose. J. Proteome Res. 2010, 9, 4234–4241. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Tsai, W.C.; Ureña-Diaz, J.M.; Sanz, L.; Mora-Obando, D.; Sánchez, E.E.; Fry, B.G.; Gutiérrez, J.M.; Gibbs, H.L.; Sovic, M.G.; et al. Venomics of New World pit vipers: Genus-wide comparisons of venom proteomes across Agkistrodon. J. Proteom. 2014, 96, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Pla, D.; Sanz, L.; Molina-Sánchez, P.; Zorita, V.; Madrigal, M.; Flores-Díaz, M.; Alape-Girón, A.; Núñez, V.; Andrés, V.; Gutiérrez, J.M.; Calvete, J.J. Snake venomics of Lachesis muta rhombeata and genus-wide antivenomics assessment of the paraspecific immunoreactivity of two antivenoms evidence the high compositional and immunological conservation across Lachesis. J. Proteom. 2013, 89, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, J.; Kini, R.M. Snake venom glutaminyl cyclase. Toxicon 2006, 48, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Huang, K.F.; Tsai, I.H. Snake venom glutaminyl cyclases: Purification, cloning, kinetic study, recombinant expression, and comparison with the human enzyme. Toxicon 2014, 86, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Wermelinger, L.S.; Dutra, D.L.S.; Oliveira-Carvalho, A.L.; Soares, M.R.; Bloch, C., Jr.; Zingali, R.B. Fast analysis of low molecular mass compounds present in snake venom: Identification of ten new pyroglutamate-containing peptides. Rapid Commun. Mass Spectrom. 2005, 19, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, B.; Zhu, S.; Rong, R. Hypotensive peptides from snake venoms: Structure, function and mechanism. Curr. Top. Med. Chem. 2015, 15, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Munekiyo, S.M.; Mackessy, S.P. Presence of peptide inhibitors in rattlesnake venoms and their effects on endogenous metalloproteases. Toxicon 2005, 45, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Bernheimer, A.W.; Weinstein, S.A.; Linder, R. Isoelectric analysis of some Australian elapid snake venoms with special reference to phospholipase B and hemolysis. Toxicon 1986, 24, 841–849. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Linder, R.; Weinstein, S.A.; Kim, K.S. Isolation and characterization of a phospholipase B from venom of Collett’s snake, Pseudechis colletti. Toxicon 1987, 25, 547–554. [Google Scholar] [CrossRef]
- Takasaki, C.; Tamiya, N. Isolation and properties of lysophospholipases from the venom of an Australian elapid snake, Pseudechis australis. Biochem. J. 1982, 203, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Kemparaju, K.; Girish, K.S. Snake venom hyaluronidase: A therapeutic target. Cell Biochem. Funct. 2006, 24, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Guércio, R.A.; Shevchenko, A.; Shevchenko, A.; López-Lozano, J.L.; Paba, J.; Sousa, M.V.; Ricart, C.A. Ontogenetic variations in the venom proteome of the Amazonian snake Bothrops atrox. Proteome Sci. 2006, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Alape-Girón, A.; Sanz, L.; Escolano, J.; Flores-Díaz, M.; Madrigal, M.; Sasa, M.; Calvete, J.J. Snake venomics of the lancehead pitviper Bothrops asper: Geographic, individual, and ontogenetic variations. J. Proteome Res. 2008, 7, 3556–3571. [Google Scholar] [CrossRef] [PubMed]
- Madrigal, M.; Sanz, L.; Flores-Díaz, M.; Sasa, M.; Núñez, V.; Alape-Girón, A.; Calvete, J.J. Snake venomics across genus Lachesis. Ontogenetic changes in the venom composition of Lachesis stenophrys and comparative proteomics of the venoms of adult Lachesis melanocephala and Lachesis acrochorda. J. Proteom. 2012, 77, 280–297. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Margres, M.J.; Ward, M.J.; Sanchez, E.E. The genetics of venom ontogeny in the eastern diamondback rattlesnake (Crotalus adamanteus). Peer J. 2017, 5, 3249. [Google Scholar] [CrossRef] [PubMed]
- Pla, D.; Sanz, L.; Sasa, M.; Acevedo, M.E.; Dwyer, Q.; Durban, J.; Pérez, A.; Rodriguez, Y.; Lomonte, B.; Calvete, J.J. Proteomic analysis of venom variability and ontogeny across the arboreal palm-pitvipers (genus Bothriechis). J. Proteom. 2017, 152, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Modahl, C.M.; Mukherjee, A.K.; Mackessy, S.P. An analysis of venom ontogeny and prey-specific toxicity in the Monocled Cobra (Naja kaouthia). Toxicon 2016, 119, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.N.; Koludarov, I.; Ali, S.A.; Dobson, J.; Zdenek, C.N.; Dashevsky, D.; Masci, P.P.; Nouwens, A.; Josh, P.; Goldenberg, J.; et al. Rapid radiations and the race to redundancy: An investigation of the evolution of Australian elapid snake venoms. Toxins 2016, 8, 309. [Google Scholar] [CrossRef] [PubMed]
- Cipriani, V.; Debono, J.; Goldenberg, J.; Jackson, T.N.; Arbuckle, K.; Dobson, J.; Koludarov, I.; Li, B.; Hay, C.; Dunstan, N.; et al. Correlation between ontogenetic dietary shifts and venom variation in Australian brown snakes (Pseudonaja). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2017, 197, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Sixberry, N.M.; Heyborne, W.H.; Fritts, T. Venom of the Brown Treesnake, Boiga irregularis: Ontogenetic shifts and taxa-specific toxicity. Toxicon 2006, 47, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Pla, D.; Petras, D.; Saviola, A.J.; Modahl, C.M.; Sanz, L.; Pérez, A.; Juárez, E.; Frietze, S.; Dorrestein, P.C.; Mackessy, S.P.; et al. Transcriptomics-guided bottom-up and top-down venomics of neonate and adult specimens of the arboreal rear-fanged Brown Treesnake, Boiga irregularis, from Guam. J. Proteom. 2018, 174, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wüster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. R. Soc. Lond. B. Biol. Sci. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–251. [Google Scholar] [CrossRef]
- Aird, S.D.; da Silva, N.J., Jr. Comparative enzymatic composition of Brazilian coral snake (Micrurus) venoms. Comp. Biochem. Physiol. 1991, 99B, 287–294. [Google Scholar] [CrossRef]
- Björk, W. Purification of phosphodiesterase from Bothrops atrox venom, with special consideration of the elimination of monophosphatases. J. Biol. Chem. 1963, 238, 2487–2490. [Google Scholar] [PubMed]
- Laskowski, M. Purification and properties of venom phosphodiesterase. Meths. Enzymol. 1980, 65, 276–284. [Google Scholar]
- Smith, C.F.; Mackessy, S.P. The effects of hybridization on divergent venom phenotypes: Characterization of venom from Crotalus scutulatus scutulatus × Crotalus oreganus helleri hybrids. Toxicon 2016, 120, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Smith, J.W.; Huang, C.M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- McIlwain, S.; Mathews, M.; Bereman, M.S.; Rubel, E.W.; MacCoss, M.J.; Noble, W.S. Estimating relative abundances of proteins from shotgun proteomics data. BMC Bioinform. 2012, 13, 308. [Google Scholar] [CrossRef] [PubMed]
- MASCOT Ver. 2.2 Search Engine. Available online: http://www.matrixscience.com (accessed on 10 September 2017).
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- NCBI BLAST program. Available online: http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 10 September 2017).
- U.S. Environmental Protection Agency. Trimmed Spearman–Karber (TSK) Program Version 1.5. Ecological Monitoring Research Division, Environmental Monitoring Systems Laboratory, Cincinnati, OH. 1990. Available online: http://www.scirp.org/%28S%28i43dyn45teexjx455qlt3d2q%29%29/reference/ReferencesPapers.aspx?ReferenceID=1130679 (accessed on 15 September 2017).


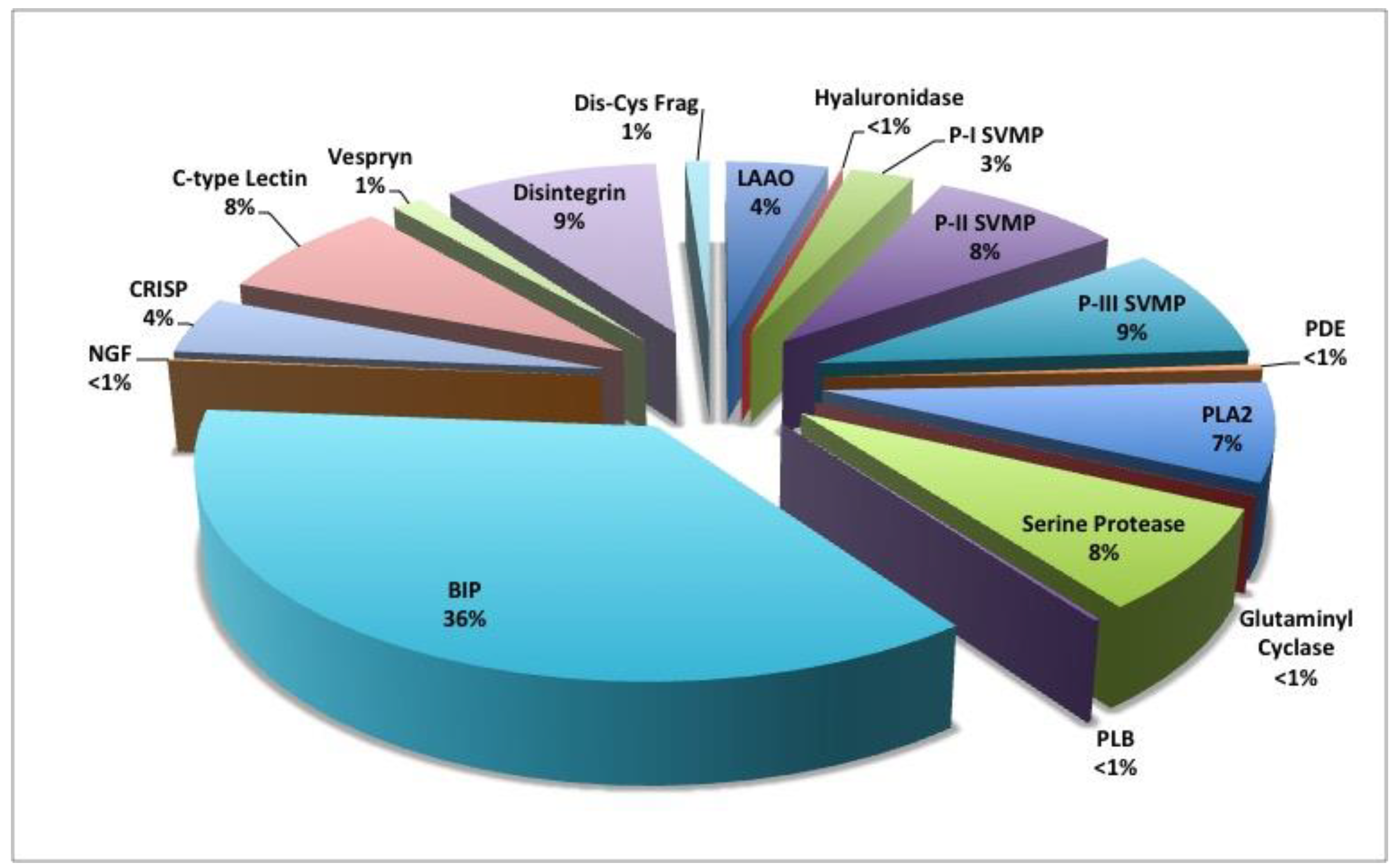
| Protein Family | % of Total Venom Proteins |
|---|---|
| L-Amino Acid Oxidase | 4.35 |
| Snake Venom Metalloproteinase (SVMP) | 19.53 |
| • P-I SVMP | 2.72 |
| • P-II SVMP | 8.23 |
| • P-III SVMP | 8.58 |
| Phosphodiesterase (Exonuclease) | <1.0 |
| Phospholipase A2 | 7.49 |
| • Acidic PLA2 | 1.47 |
| • Basic PLA2 | 6.02 |
| Serine Protease | 7.84 |
| • Thrombin-like | 5.69 |
| • Kallikrein-like | 1.90 |
| • Plasminogen Activator | <1.0 |
| Glutaminyl Cyclase | <1.0 |
| Hyaluronidase | <1.0 |
| Phospholipase B | <1.0 |
| Bradykinin-Inhibitory Peptide | 36.32 |
| Cysteine-rich Secretory Protein | 4.41 |
| C-type Lectin | 7.80 |
| Vespryn | 1.31 |
| Disintegrin | 9.07 |
| Dis-Cys Fragments | 1.04 |
| Nerve Growth Factor | <1.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mackessy, S.P.; Leroy, J.; Mociño-Deloya, E.; Setser, K.; Bryson, R.W.; Saviola, A.J. Venom Ontogeny in the Mexican Lance-Headed Rattlesnake (Crotalus polystictus). Toxins 2018, 10, 271. https://doi.org/10.3390/toxins10070271
Mackessy SP, Leroy J, Mociño-Deloya E, Setser K, Bryson RW, Saviola AJ. Venom Ontogeny in the Mexican Lance-Headed Rattlesnake (Crotalus polystictus). Toxins. 2018; 10(7):271. https://doi.org/10.3390/toxins10070271
Chicago/Turabian StyleMackessy, Stephen P., Jamie Leroy, Estrella Mociño-Deloya, Kirk Setser, Robert W. Bryson, and Anthony J. Saviola. 2018. "Venom Ontogeny in the Mexican Lance-Headed Rattlesnake (Crotalus polystictus)" Toxins 10, no. 7: 271. https://doi.org/10.3390/toxins10070271





