The Subcellular Location of Selenoproteins and the Impact on Their Function
Abstract
:1. Selenoproteins
2. Subcellular Location of Proteins Affects Their Biological Functions
3. GPx-4
4. GPx-1
5. SBP1
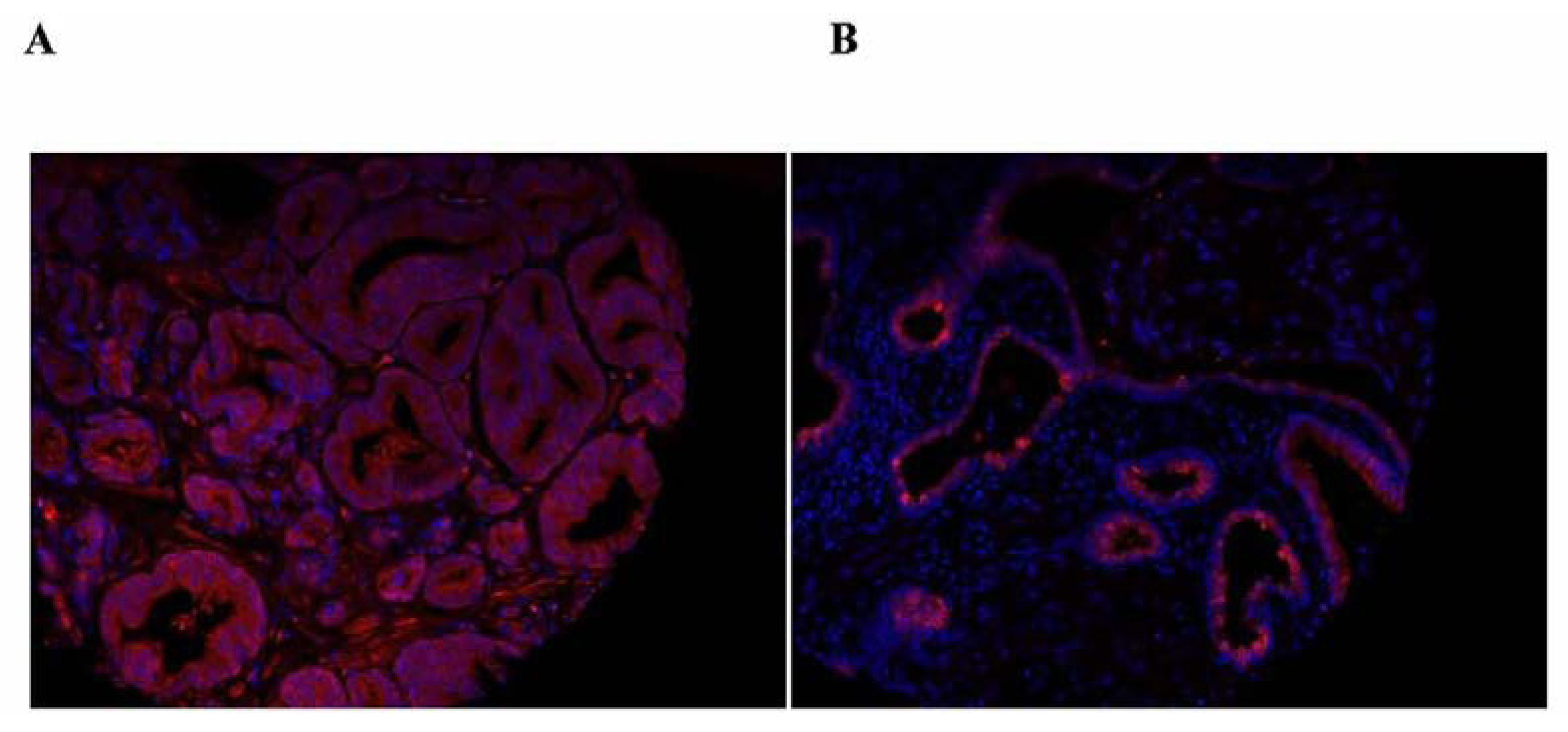
6. Thioredoxin Reductases
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.D.; Tsuji, P.A.; Milner, J.A. Selenoproteins and Cancer Prevention. Annu. Rev. Nutr. 2012, 32, 73–95. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.J.; Banu, L.; Chen, Y.; Mandel, S.J.; Kiefer, J.D.; Harney, J.W.; Larsen, P.R. Recognition of UGA as a selenocysteine codon in Type I deiodinase requires sequences in the 3′ untranslated region. Nature 1991, 353, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, A.V.; Hatfield, D.L.; Gladyshev, V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta 2009, 1790, 1424–1428. [Google Scholar] [CrossRef] [PubMed]
- Bellinger, F.P.; Raman, A.V.; Reeves, M.A.; Berry, M.J. Regulation and function of selenoproteins in human disease. Biochem. J. 2009, 422, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, D.M.; Copeland, P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003, 23, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Ansong, E.; Yang, W.; Diamond, A.M. Molecular cross-talk between members of distinct families of selenium containing proteins. Mol. Nutr. Food Res. 2014, 58, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Flohe, L. The labour pains of biochemical selenology: The history of selenoprotein biosynthesis. Biochim. Biophys. Acta 2009, 1790, 1389–1403. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigo, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D. Introduction to NF-kappaB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef] [PubMed]
- Perkins, N.D. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 2007, 8, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Planchon, S.M.; Waite, K.A.; Eng, C. The nuclear affairs of PTEN. J. Cell Sci. 2008, 121 Pt 3, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Milella, M.; Falcone, I.; Conciatori, F.; Incani, U.C.; del Curatolo, A.; Inzerilli, N.; Nuzzo, C.M.; Vaccaro, V.; Vari, S.; Cognetti, F.; et al. PTEN: Multiple Functions in Human Malignant Tumors. Front. Oncol. 2015, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Eggler, A.L.; Gay, K.A.; Mesecar, A.D. Molecular mechanisms of natural products in chemoprevention: Induction of cytoprotective enzymes by Nrf2. Mol. Nutr. Food Res. 2008, 52 (Suppl. 1), S84–S94. [Google Scholar] [PubMed]
- Brocato, J.; Chervona, Y.; Costa, M. Molecular responses to hypoxia-inducible factor 1alpha and beyond. Mol. Pharmacol. 2014, 85, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Ursini, F.; Maiorino, M.; Gregolin, C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim. Biophys. Acta 1985, 839, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Ursini, F.; Maiorino, M.; Valente, M.; Ferri, L.; Gregolin, C. Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim. Biophys. Acta 1982, 710, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Imai, H.Y. Nakagawa, Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic. Biol. Med. 2003, 34, 145–169. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schneider, M.; Seiler, A.; Bornkamm, G.W. Physiological role of phospholipid hydroperoxide glutathione peroxidase in mammals. Biol. Chem. 2007, 388, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohe, R. Tissue-specific functions of individual glutathione peroxidases. Free Radic. Biol. Med. 1999, 27, 951–965. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Imai, H.; Sumi, D.; Imanaka, T.; Takano, T.; Chiba, N.; Nakagawa, Y. Import into mitochondria of phospholipid hydroperoxide glutathione peroxidase requires a leader sequence. Biochem. Biophys. Res. Commun. 1996, 227, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Pushpa-Rekha, T.R.; Burdsall, A.L.; Oleksa, L.M.; Chisolm, G.M.; Driscoll, D.M. Rat phospholipid-hydroperoxide glutathione peroxidase. cDNA cloning and identification of multiple transcription and translation start sites. J. Biol. Chem. 1995, 270, 26993–26999. [Google Scholar] [CrossRef] [PubMed]
- Ursini, F.; Heim, S.; Kiess, M.; Maiorino, M.; Roveri, A.; Wissing, J.; Flohe, L. Dual function of the selenoprotein PHGPx during sperm maturation. Science 1999, 285, 1393–1396. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Forster, H.; Boersma, A.; Seiler, A.; Wehnes, H.; Sinowatz, F.; Neumuller, C.; Deutsch, M.J.; Walch, A.; de Angelis, M.H.; et al. Mitochondrial glutathione peroxidase 4 disruption causes male infertility. FASEB J. 2009, 23, 3233–3242. [Google Scholar] [CrossRef] [PubMed]
- Baek, I.J.; Seo, D.S.; Yon, J.M.; Lee, S.R.; Jin, Y.; Nahm, S.S.; Jeong, J.H.; Choo, Y.K.; Kang, J.K.; Lee, B.J.; et al. Tissue expression and cellular localization of phospholipid hydroperoxide glutathione peroxidase (PHGPx) mRNA in male mice. J. Mol. Histol. 2007, 38, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Yoo, S.E.; Na, R.; Walter, C.A.; Richardson, A.; Ran, Q. Short form glutathione peroxidase 4 is the essential isoform required for survival and somatic mitochondrial functions. J. Biol. Chem. 2009, 284, 30836–30844. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.G.; Laux, G.; Brielmeier, M.; Bornkamm, G.W.; Conrad, M. Testis-specific expression of the nuclear form of phospholipid hydroperoxide glutathione peroxidase (PHGPx). Biol. Chem. 2003, 384, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Maiorino, M.; Scapin, M.; Ursini, F.; Biasolo, M.; Bosello, V.; Flohe, L. Distinct promoters determine alternative transcription of gpx-4 into phospholipid-hydroperoxide glutathione peroxidase variants. J. Biol. Chem. 2003, 278, 34286–34290. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, R.; Maccari, I.; Pipolo, S.; Conrad, M.; Mangia, F.; Boitani, C. The nuclear form of glutathione peroxidase 4 is associated with sperm nuclear matrix and is required for proper paternal chromatin decondensation at fertilization. J. Cell Physiol. 2012, 227, 1420–1427. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, R.; Maccari, I.; Pipolo, S.; Mangia, F.; Boitani, C. The nuclear form of glutathione peroxidase 4 colocalizes and directly interacts with protamines in the nuclear matrix during mouse sperm chromatin assembly. Spermatogenesis 2014, 4, e28460. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Moreno, S.G.; Sinowatz, F.; Ursini, F.; Kolle, S.; Roveri, A.; Brielmeier, M.; Wurst, W.; Maiorino, M.; Bornkamm, G.W. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol. Cell. Biol. 2005, 25, 7637–7644. [Google Scholar] [CrossRef] [PubMed]
- Imai, H.; Hakkaku, N.; Iwamoto, R.; Suzuki, J.; Suzuki, T.; Tajima, Y.; Konishi, K.; Minami, S.; Ichinose, S.; Ishizaka, K.; et al. Depletion of selenoprotein GPx4 in spermatocytes causes male infertility in mice. J. Biol. Chem. 2009, 284, 32522–32532. [Google Scholar] [CrossRef] [PubMed]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997. [Google Scholar] [CrossRef] [PubMed]
- Utsunomiya, H.; Komatsu, N.; Yoshimura, S.; Tsutsumi, Y.; Watanabe, K. Exact ultrastructural localization of glutathione peroxidase in normal rat hepatocytes: Advantages of microwave fixation. J. Histochem. Cytochem. 1991, 39, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Asayama, K.; Yokota, S.; Dobashi, K.; Hayashibe, H.; Kawaoi, A.; Nakazawa, S. Purification and immunoelectron microscopic localization of cellular glutathione peroxidase in rat hepatocytes: Quantitative analysis by postembedding method. Histochemistry 1994, 102, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Esworthy, R.S.; Ho, Y.S.; Chu, F.F. The Gpx1 gene encodes mitochondrial glutathione peroxidase in the mouse liver. Arch. Biochem. Biophys. 1997, 340, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Bera, S.; Weinberg, F.; Ekoue, D.N.; Ansenberger-Fricano, K.; Mao, M.; Bonini, M.G.; Diamond, A.M. Natural allelic variations in glutathione peroxidase-1 affect its subcellular localization and function. Cancer Res. 2014, 74, 5118–5126. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, P.; Goldberg, M.; Herman, L.; Lee, B.S.; Wang, H.; Brown, R.L.; Foster, C.B.; Peters, U.; Diamond, A.M. Molecular consequences of genetic variations in the glutathione peroxidase 1 selenoenzyme. Cancer Res. 2009, 69, 8183–8190. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.J.; Diamond, A.M. Role of glutathione peroxidase 1 in breast cancer: Loss of heterozygosity and allelic differences in the response to selenium. Cancer Res. 2003, 63, 3347–3351. [Google Scholar] [PubMed]
- Shepard, B.D.; Tuma, D.J.; Tuma, P.L. Chronic ethanol consumption induces global hepatic protein hyperacetylation. Alcohol. Clin. Exp. Res. 2010, 34, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, K.S.; Galligan, J.J.; Hirschey, M.D.; Verdin, E.; Petersen, D.R. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J. Proteome Res. 2012, 11, 1633–1643. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Leng, Y.; Huang, W.; Liu, X.; Kufe, D. Glutathione peroxidase 1 is regulated by the c-Abl and Arg tyrosine kinases. J. Biol. Chem. 2003, 278, 39609–39614. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.C.; Mao, M.; de Abreu, A.L.P.; Ansenberger-Fricano, K.; Ekoue, D.N.; Ganini, D.; Kajdacsy-Balla, A.; Diamond, A.M.; Minshall, R.D.; Consolaro, M.E.L.; et al. MnSOD upregulation sustains the Warburg effect via mitochondrial ROS and AMPK-dependent signalling in cancer. Nat. Commun. 2015, 6, 6053. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.G.; Tamimi, R.M.; Hunter, D.J. Gene x Gene interaction between MnSOD and GPX-1 and breast cancer risk: A nested case-control study. BMC Cancer 2006, 6, 217. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Goldberg, M.L.; Pohl, N.M.; Bi, X.; Tong, C.; Xiong, B.; Koh, T.J.; Diamond, A.M.; Yang, W. Functional and physical interaction between the selenium-binding protein 1 (SBP1) and the glutathione peroxidase 1 selenoprotein. Carcinogenesis 2010, 31, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Bansal, M.P.; Mukhopadhyay, T.; Scott, J.; Cook, R.G.; Mukhopadhyay, R.; Medina, D. DNA sequencing of a mouse liver protein that binds selenium: Implications for selenium’s mechanism of action in cancer prevention. Carcinogenesis 1990, 11, 2071–2073. [Google Scholar] [CrossRef] [PubMed]
- Bansal, M.P.; Oborn, C.J.; Danielson, K.G.; Medina, D. Evidence for two selenium-binding proteins distinct from glutathione peroxidase in mouse liver. Carcinogenesis 1989, 10, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Pohl, N.M.; Tong, C.; Fang, W.; Bi, X.; Li, T.; Yang, W. Transcriptional regulation and biological functions of selenium-binding protein 1 in colorectal cancer in vitro and in nude mouse xenografts. PLoS ONE 2009, 4, e7774. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ding, G.; Gu, C.; Zhou, J.; Kuang, M.; Ji, Y.; He, Y.; Kondo, T.; Fan, J. Decreased Selenium-Binding Protein 1 Enhances Glutathione Peroxidase 1 Activity and Downregulates HIF-1alpha to Promote Hepatocellular Carcinoma Invasiveness. Clin. Cancer Res. 2012, 18, 3042–3053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, F.; Younes, M.; Liu, H.; Chen, C.; Yao, Q. Reduced selenium-binding protein 1 in breast cancer correlates with poor survival and resistance to the anti-proliferative effects of selenium. PLoS ONE 2013, 8, e63702. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, W.; Pan, W.; Wang, N.; Li, G.; Fan, X.; Xu, X.; Shen, S.; Das, U.N. Selenium-binding protein 1 may decrease gastric cellular proliferation and migration. Int. J. Oncol. 2013, 42, 1620–1629. [Google Scholar] [PubMed]
- Silvers, A.L.; Lin, L.; Bass, A.J.; Chen, G.; Wang, Z.; Thomas, D.G.; Lin, J.; Giordano, T.J.; Orringer, M.B.; Beer, D.G.; et al. Decreased selenium-binding protein 1 in esophageal adenocarcinoma results from posttranscriptional and epigenetic regulation and affects chemosensitivity. Clin. Cancer Res. 2010, 16, 2009–2021. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Zhou, J.R.; Gao, C.; Feldman, L.; Sytkowski, A.J. Human selenium binding protein-1 (hSP56) is a negative regulator of HIF-1alpha and suppresses the malignant characteristics of prostate cancer cells. BMB Rep. 2014, 47, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Wang, Y.; Sytkowski, A.J. Human selenium binding protein-1 (hSP56) interacts with VDU1 in a selenium-dependent manner. Biochem. Biophys. Res. Commun. 2009, 379, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Diamond, A.M. Selenium-binding protein 1 as a tumor suppressor and a prognostic indicator of clinical outcome. Biomark. Res. 2013, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, H.; Miller, C.T.; Thomas, D.G.; Gharib, T.G.; Misek, D.E.; Giordano, T.J.; Orringer, M.B.; Hanash, S.M.; Beer, D.G. Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. J. Pathol. 2004, 202, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dong, W.G.; Lin, J. Reduced selenium-binding protein 1 is associated with poor survival rate in gastric carcinoma. Med. Oncol. 2011, 28, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhan, N.; Dong, W.G. Altered expression of selenium-binding protein 1 in gastric carcinoma and precursor lesions. Med. Oncol. 2011, 28, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Ansong, E.; Ying, Q.; Ekoue, D.N.; Deaton, R.; Hall, A.R.; Kajdacsy-Balla, A.; Yang, W.; Gann, P.H.; Diamond, A.M. Evidence that Selenium Binding Protein 1 is a Tumor Suppressor in Prostate Cancer. PLoS ONE 2015, in press. [Google Scholar]
- Jerome-Morais, A.; Wright, M.E.; Liu, R.; Yang, W.; Jackson, M.I.; Combs, G.F., Jr.; Diamond, A.M. Inverse association between glutathione peroxidase activity and both selenium-binding protein 1 levels and Gleason score in human prostate tissue. Prostate 2012, 72, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Stadtman, T.C. A new selenoprotein from human lung adenocarcinoma cells: Purification, properties, and thioredoxin reductase activity. Proc. Natl. Acad. Sci. USA 1996, 93, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.A. Holmgren, Selenoproteins. J. Biol. Chem. 2009, 284, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Kim, J.R.; Kwon, K.S.; Yoon, H.W.; Levine, R.L.; Ginsburg, A.; Rhee, S.G. Molecular cloning and characterization of a mitochondrial selenocysteine-containing thioredoxin reductase from rat liver. J. Biol. Chem. 1999, 274, 4722–4734. [Google Scholar] [CrossRef] [PubMed]
- Rozell, B.; Hansson, H.A.; Luthman, M.; Holmgren, A. Immunohistochemical localization of thioredoxin and thioredoxin reductase in adult rats. Eur. J. Cell Biol. 1985, 38, 79–86. [Google Scholar] [PubMed]
- Su, D.; Novoselov, S.V.; Sun, Q.A.; Moustafa, M.E.; Zhou, Y.; Oko, R.; Hatfield, D.L.; Gladyshev, V.N. Mammalian selenoprotein thioredoxin-glutathione reductase. Roles in disulfide bond formation and sperm maturation. J. Biol. Chem. 2005, 280, 26491–26498. [Google Scholar]
- Arner, E.S. Focus on mammalian thioredoxin reductases—Important selenoproteins with versatile functions. Biochim. Biophys. Acta 2009, 1790, 495–526. [Google Scholar] [CrossRef] [PubMed]
- Mustacich, D.; Powis, G. Thioredoxin reductase. Biochem. J. 2000, 15, 1–8. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diamond, A.M. The Subcellular Location of Selenoproteins and the Impact on Their Function. Nutrients 2015, 7, 3938-3948. https://doi.org/10.3390/nu7053938
Diamond AM. The Subcellular Location of Selenoproteins and the Impact on Their Function. Nutrients. 2015; 7(5):3938-3948. https://doi.org/10.3390/nu7053938
Chicago/Turabian StyleDiamond, Alan M. 2015. "The Subcellular Location of Selenoproteins and the Impact on Their Function" Nutrients 7, no. 5: 3938-3948. https://doi.org/10.3390/nu7053938



