Gliadin-Specific T-Cells Mobilized in the Peripheral Blood of Coeliac Patients by Short Oral Gluten Challenge: Clinical Applications
Abstract
:1. Introduction
2. Genetic Susceptibility, Clinical Spectrum, and Pathogenesis
3. Current and Emerging Therapies
4. Gluten Oral Challenge as Tool to Monitor Intestinal Gluten-Reactive T-Cells
4.1. Interferon-γ ELISPOT Assay on Peripheral Blood Cells after a Three Day Gluten Challenge
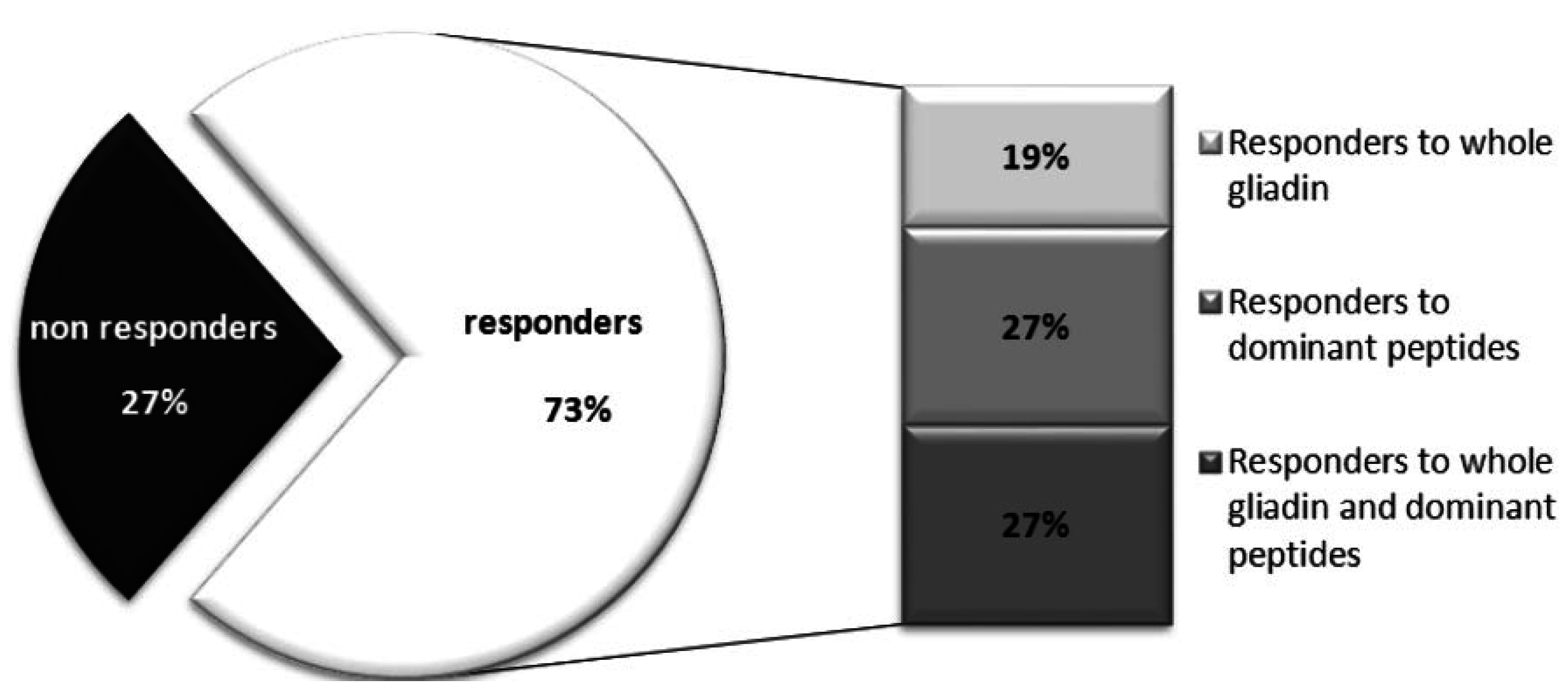
4.2. Interferon-γ ELISA Assay and Multiparametric Masscytometry to Monitor the Anti-Gluten T-Cells Response after a Three Day Gluten Challenge
4.3. HLA-DQ2-Tetramers as Probe to Detect Gliadin-Specific Cells in Peripheral Blood
5. Translational Applications of the Short Gluten Oral Challenge
5.1. Identification of Gluten Immunogenic Peptides
5.2. Validation of Therapeutic Drugs
5.3. Diagnostic Relevance
| Application | Clinical/Research Purpose | References |
|---|---|---|
| Diagnosis | Confirmation of diagnosis in uncertain celiac disease cases | [10,67] |
| Therapies | Validation of new therapeutic drugs | [62] |
| Therapies | Validation of biochemical or enzymatic strategies to detoxify gluten. | [64,65] |
| Therapies | Searching of wheat cultivars with reduced immunotoxic gluten sequences | [68] |
| Pathogenesis | Identification of immunogenic gluten epitopes | [32,60] |
| Pathogenesis | Phenotypic analysis of cell population involved in celiac disease | [53] |
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Lundin, K.E.A.; Qiao, S.-W.; Snir, O.; Sollid, L.M. Coeliac disease—From genetic and immunological studies to clinical applications. Scand. J. Gastroenterol. 2015, 50, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Korponay-Szabó, I.R.; Troncone, R.; Discepolo, V. Adaptive diagnosis of coeliac disease. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 381–398. [Google Scholar] [CrossRef] [PubMed]
- Walker-Smith, J.A.; Guandalini, S.; Schmitz, J.; Shmerling, D.H.; Visakorpi, J.K. Revised criteria for diagnosis of coeliac disease. Arch. Dis. Child. 1990, 65, 909–911. [Google Scholar]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.R.; Mearin, M.L.; Phillips, A.; Shamir, R.; Troncone, R.; Giersiepen, K.; Branski, D.; Catassi, C.; et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition Guidelines for the Diagnosis of Coeliac Disease. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 136–160. [Google Scholar] [CrossRef] [PubMed]
- Currier, J.R.; Kuta, E.G.; Turk, E.; Earhart, L.B.; Loomis-Price, L.; Janetzki, S.; Ferrari, G.; Birx, D.L.; Cox, J.H. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J. Immunol. Methods 2002, 260, 157–172. [Google Scholar] [CrossRef]
- Anderson, R.P.; Degano, P.; Godkin, A.J.; Jewell, D.P.; Hill, A.V. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat. Med. 2000, 6, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.P.; van Heel, D.A.; Tye-Din, J.A.; Barnardo, M.; Salio, M.; Jewell, D.P.; Hill, A.V.S. T cells in peripheral blood after gluten challenge in coeliac disease. Gut 2005, 54, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Camarca, A.; Radano, G.; di Mase, R.; Terrone, G.; Maurano, F.; Auricchio, S.; Troncone, R.; Greco, L.; Gianfrani, C. Short wheat challenge is a reproducible in vivo assay to detect immune response to gluten. Clin. Exp. Immunol. 2012, 169, 129–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ráki, M.; Fallang, L.-E.; Brottveit, M.; Bergseng, E.; Quarsten, H.; Lundin, K.E.A.; Sollid, L.M. Tetramer visualization of gut-homing gluten-specific T cells in the peripheral blood of celiac disease patients. Proc. Natl. Acad. Sci. USA 2007, 104, 2831–2836. [Google Scholar] [CrossRef] [PubMed]
- Brottveit, M.; Ráki, M.; Bergseng, E.; Fallang, L.-E.; Simonsen, B.; Løvik, A.; Larsen, S.; Løberg, E.M.; Jahnsen, F.L.; Sollid, L.M.; Lundin, K.E.A. Assessing possible celiac disease by an HLA-DQ2-gliadin Tetramer Test. Am. J. Gastroenterol. 2011, 106, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Abadie, V.; Sollid, L.M.; Barreiro, L.B.; Jabri, B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu. Rev. Immunol. 2011, 29, 493–525. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, D.A.; Hunt, K.; Greco, L.; Wijmenga, C. Genetics in coeliac disease. Best Pract. Res. Clin. Gastroenterol. 2005, 19, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Thorsby, E. HLA susceptibility genes in celiac disease: Genetic mapping and role in pathogenesis. Gastroenterology 1993, 105, 910–922. [Google Scholar] [PubMed]
- Karell, K.; Louka, A.S.; Moodie, S.J.; Ascher, H.; Clot, F.; Greco, L.; Ciclitira, P.J.; Sollid, L.M.; Partanen, J. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: Results from the European Genetics Cluster on Celiac Disease. Hum. Immunol. 2003, 64, 469–477. [Google Scholar] [CrossRef]
- Kumar, V.; Gutierrez-Achury, J.; Kanduri, K.; Almeida, R.; Hrdlickova, B.; Zhernakova, D.V.; Westra, H.J.; Karjalainen, J.; Ricaño-Ponce, I.; Li, Y.; et al. Systematic annotation of celiac disease loci refines pathological pathways and suggests a genetic explanation for increased interferon-gamma levels. Hum. Mol. Genet. 2015, 24, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Bodd, M.; Ráki, M.; Bergseng, E.; Jahnsen, J.; Lundin, K.E.A.; Sollid, L.M. Direct cloning and tetramer staining to measure the frequency of intestinal gluten-reactive T cells in celiac disease. Eur. J. Immunol. 2013, 43, 2605–2612. [Google Scholar] [CrossRef] [PubMed]
- Oberhuber, G.; Granditsch, G.; Vogelsang, H. The histopathology of coeliac disease: Time for a standardized report scheme for pathologists. Eur. J. Gastroenterol. Hepatol. 1999, 11, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F.; Leffler, D.A.; Bai, J.C.; Biagi, F.; Fasano, A.; Green, P.H.R.; Hadjivassiliou, M.; Kaukinen, K.; Kelly, C.P.; Leonard, J.N.; et al. The Oslo definitions for coeliac disease and related terms. Gut 2013, 62, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Auricchio, R.; Tosco, A.; Piccolo, E.; Galatola, M.; Izzo, V.; Maglio, M.; Paparo, F.; Troncone, R.; Greco, L. Potential celiac children: 9-year follow-up on a gluten-containing diet. Am. J. Gastroenterol. 2014, 109, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Christophersen, A.; Ráki, M.; Bergseng, E.; Lundin, K.E.; Jahnsen, J.; Sollid, L.M.; Qiao, S.-W. Tetramer-visualized gluten-specific CD4+ T cells in blood as a potential diagnostic marker for coeliac disease without oral gluten challenge. United Eur. Gastroenterol. J. 2014, 2, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Gianfrani, C.; Auricchio, S.; Troncone, R. Adaptive and innate immune responses in celiac disease. Immunol. Lett. 2005, 99, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Matysiak-Budnik, T.; Moura, I.C.; Arcos-Fajardo, M.; Lebreton, C.; Ménard, S.; Candalh, C.; ben-Khalifa, K.; Dugave, C.; Tamouza, H.; van Niel, G.; et al. Secretory IgA mediates retrotranscytosis of intact gliadin peptides via the transferrin receptor in celiac disease. J. Exp. Med. 2008, 205, 143–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, A.; Lammers, K.M.; Goldblum, S.; Shea-Donohue, T.; Netzel-Arnett, S.; Buzza, M.S.; Antalis, T.M.; Vogel, S.N.; Zhao, A.; Yang, S.; et al. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proc. Natl. Acad. Sci. USA 2009, 106, 16799–16804. [Google Scholar] [CrossRef] [PubMed]
- Lammers, K.M.; Lu, R.; Brownley, J.; Lu, B.; Gerard, C.; Thomas, K.; Rallabhandi, P.; Shea-Donohue, T.; Tamiz, A.; Alkan, S.; et al. Gliadin induces an increase in intestinal permeability and zonulin release by binding to the chemokine receptor CXCR3. Gastroenterology 2008, 135, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A.; Not, T.; Wang, W.; Uzzau, S.; Berti, I.; Tommasini, A.; Goldblum, S.E. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet 2000, 355, 1518–1519. [Google Scholar] [CrossRef]
- Molberg, O.; Mcadam, S.N.; Körner, R.; Quarsten, H.; Kristiansen, C.; Madsen, L.; Fugger, L.; Scott, H.; Norén, O.; Roepstorff, P.; et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 1998, 4, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Pinkas, D.M.; Strop, P.; Brunger, A.T.; Khosla, C. Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Jabri, B. Celiac disease and transglutaminase 2: A model for posttranslational modification of antigens and HLA association in the pathogenesis of autoimmune disorders. Curr. Opin. Immunol. 2011, 23, 732–873. [Google Scholar] [CrossRef] [PubMed]
- Camarca, A.; del Mastro, A.; Gianfrani, C. Repertoire of gluten peptides active in celiac disease patients: Perspectives for translational therapeutic applications. Endocr. Metab. Immune Disord. Drug Targets 2012, 12, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Molberg, Ø.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef] [PubMed]
- Tye-Din, J.A.; Stewart, J.A.; Dromey, J.A.; Beissbarth, T.; van Heel, D.A.; Tatham, A.; Henderson, K.; Mannering, S.I.; Gianfrani, C.; Jewell, D.P.; et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci. Transl. Med. 2010. [Google Scholar] [CrossRef] [PubMed]
- Camarca, A.; Anderson, R.P.; Mamone, G.; Fierro, O.; Facchiano, A.; Costantini, S.; Zanzi, D.; Sidney, J.; Auricchio, S.; Sette, A.; et al. Intestinal T cell responses to gluten peptides are largely heterogeneous: Implications for a peptide-based therapy in celiac disease. J. Immunol. 2009, 182, 4158–4166. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 2012, 64, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Lundin, K.E.A. Diagnosis and treatment of celiac disease. Mucosal Immunol. 2009, 2, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Greco, L.; Mayer, M.; Ciccarelli, G.; Troncone, R.; Auricchio, S. Compliance to a gluten-free diet in adolescents, or “what do 300 coeliac adolescents eat every day?”. Ital. J. Gastroenterol. Hepatol. 1997, 29, 305–310. [Google Scholar] [PubMed]
- Errichiello, S.; Esposito, O.; di Mase, R.; Camarca, M.E.; Natale, C.; Limongelli, M.G.; Marano, C.; Coruzzo, A.; Lombardo, M.; Strisciuglio, P.; et al. Celiac disease: Predictors of compliance with a gluten-free diet in adolescents and young adults. J. Pediatr. Gastroenterol. Nutr. 2010, 50, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Lamacchia, C.; Camarca, A.; Picascia, S.; Di Luccia, A.; Gianfrani, C. Cereal-based gluten-free food: How to reconcile nutritional and technological properties of wheat proteins with safety for celiac disease patients. Nutrients 2014, 6, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Diamanti, A.; Capriati, T.; Basso, M.S.; Panetta, F.; di Ciommo Laurora, V.M.; Bellucci, F.; Cristofori, F.; Francavilla, R. Celiac disease and overweight in children: An update. Nutrients 2014, 6, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.M.; Kelleher, D. Refractory celiac disease. Gastroenterology 2000, 119, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Malamut, G.; Cellier, C. Complications of coeliac disease. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Kaukinen, K.; Lindfors, K.; Mäki, M. Advances in the treatment of coeliac disease: An immunopathogenic perspective. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F.; Card, T.; Ciclitira, P.J.; Swift, G.L.; Nasr, I.; Sanders, D.S.; Ciacci, C. Support for patients with celiac disease: A literature review. United Eur. Gastroenterol. J. 2015, 3, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Leffler, D.; Schuppan, D.; Pallav, K.; Najarian, R.; Goldsmith, J.D.; Hansen, J.; Kabbani, T.; Dennis, M.; Kelly, C.P. Kinetics of the histological, serological and symptomatic responses to gluten challenge in adults with coeliac disease. Gut 2013, 62, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Tack, G.J.; van de Water, J.M.W.; Bruins, M.J.; Kooy-Winkelaar, E.M.C.; van Bergen, J.; Bonnet, P.; Vreugdenhil, A.C.E.; Korponay-Szabo, I.; Edens, L.; von Blomberg, B.M.E.; et al. Consumption of gluten with gluten-degrading enzyme by celiac patients: A pilot-study. World J. Gastroenterol. 2013, 19, 5837–5847. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Greco, L.; Troncone, R.; Grimaldi, M.; Pansa, G. Early prediction of relapse during gluten challenge in childhood celiac disease. J. Pediatr. Gastroenterol. Nutr. 1989, 8, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Gjertsen, H.A.; Sollid, L.M.; Ek, J.; Thorsby, E.; Lundin, K.E. T cells from the peripheral blood of coeliac disease patients recognize gluten antigens when presented by HLA-DR, -DQ, or -DP molecules. Scand. J. Immunol. 1994, 39, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.; Sollid, L.M.; Scott, H.; Paulsen, G.; Kett, K.; Thorsby, E.; Lundin, K.E. Gliadin-specific T cell responses in peripheral blood of healthy individuals involve T cells restricted by the coeliac disease associated DQ2 heterodimer. Scand. J. Immunol. 1995, 42, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Quiding-Järbrink, M.; Ahlstedt, I.; Lindholm, C.; Johansson, E.L.; Lönroth, H. Homing commitment of lymphocytes activated in the human gastric and intestinal mucosa. Gut 2001, 49, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Berlin, C.; Berg, E.L.; Briskin, M.J.; Andrew, D.P.; Kilshaw, P.J.; Holzmann, B.; Weissman, I.L.; Hamann, A.; Butcher, E.C. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell 1993, 74, 185–195. [Google Scholar] [CrossRef]
- Ontiveros, N.; Tye-Din, J.A.; Hardy, M.Y.; Anderson, R.P. Ex-vivo whole blood secretion of interferon (IFN)-γ and IFN-γ-inducible protein-10 measured by enzyme-linked immunosorbent assay are as sensitive as IFN-γ enzyme-linked immunospot for the detection of gluten-reactive T cells in human leucocyte antigen (HLA)-DQ2·5(+) -associated coeliac disease. Clin. Exp. Immunol. 2014, 175, 305–315. [Google Scholar] [PubMed]
- Mazzarella, G.; Stefanile, R.; Camarca, A.; Giliberti, P.; Cosentini, E.; Marano, C.; Iaquinto, G.; Giardullo, N.; Auricchio, S.; Sette, A.; et al. Gliadin activates HLA class I-restricted CD8+ T cells in celiac disease intestinal mucosa and induces the enterocyte apoptosis. Gastroenterology 2008, 134, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Newell, E.W.; Glanville, J.; Fernandez-Becker, N.; Khosla, C.; Chien, Y.-H.; Davis, M.M. Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proc. Natl. Acad. Sci. USA 2013, 110, 13073–13078. [Google Scholar] [CrossRef] [PubMed]
- Altman, J.D.; Moss, P.A.; Goulder, P.J.; Barouch, D.H.; McHeyzer-Williams, M.G.; Bell, J.I.; McMichael, A.J.; Davis, M.M. Phenotypic analysis of antigen-specific T lymphocytes. Science 1996, 274, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Klenerman, P.; Cerundolo, V.; Dunbar, P.R. Tracking T cells with tetramers: New tales from new tools. Nat. Rev. Immunol. 2002, 2, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.-W.; Ráki, M.; Gunnarsen, K.S.; Løset, G.-Å.; Lundin, K.E.A.; Sandlie, I.; Sollid, L.M. Posttranslational modification of gluten shapes TCR usage in celiac disease. J. Immunol. 2011, 187, 3064–3071. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.-W.; Christophersen, A.; Lundin, K.E.A.; Sollid, L.M. Biased usage and preferred pairing of α- and β-chains of TCRs specific for an immunodominant gluten epitope in coeliac disease. Int. Immunol. 2014, 26, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Broughton, S.E.; Petersen, J.; Theodossis, A.; Scally, S.W.; Loh, K.L.; Thompson, A.; van Bergen, J.; Kooy-Winkelaar, Y.; Henderson, K.N.; Beddoe, T.; et al. Biased T-cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 2012, 37, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Tanner, G.J.; Howitt, C.A.; Forrester, R.I.; Campbell, P.M.; Tye-Din, J.A.; Anderson, R.P. Dissecting the T-cell response to hordeins in coeliac disease can develop barley with reduced immunotoxicity. Aliment. Pharmacol. Ther. 2010, 32, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Hardy, M.Y.; Girardin, A.; Pizzey, C.; Cameron, D.J.; Watson, K.A.; Picascia, S.; Auricchio, R.; Greco, L.; Gianfrani, C.; La Gruta, N.L.; et al. Consistency in Polyclonal T-cell Responses to Gluten Between Children and Adults with Celiac Disease. Gastroenterology 2015, 6, 1541–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, H.J. Emerging drugs for celiac disease. Expert Opin. Emerg. Drugs 2015, 20, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Tye-Din, J.A.; Anderson, R.P.; Ffrench, R.A.; Brown, G.J.; Hodsman, P.; Siegel, M.; Botwick, W.; Shreeniwas, R. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin. Immunol. 2010, 134, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Lähdeaho, M.-L.; Kaukinen, K.; Laurila, K.; Vuotikka, P.; Koivurova, O.-P.; Kärjä-Lahdensuu, T.; Marcantonio, A.; Adelman, D.C.; Mäki, M. Glutenase ALV003 attenuates gluten-induced mucosal injury in patients with celiac disease. Gastroenterology 2014, 146, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Greco, L.; Gobbetti, M.; Auricchio, R.; di Mase, R.; Landolfo, F.; Paparo, F.; di Cagno, R.; de Angelis, M.; Rizzello, C.G.; Cassone, A.; et al. Safety for patients with celiac disease of baked goods made of wheat flour hydrolyzed during food processing. Clin. Gastroenterol. Hepatol. 2011, 9, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Gianfrani, C.; Siciliano, R.A.; Facchiano, A.M.; Camarca, A.; Mazzeo, M.F.; Costantini, S.; Salvati, V.M.; Maurano, F.; Mazzarella, G.; Iaquinto, G.; et al. Transamidation of wheat flour inhibits the response to gliadin of intestinal T cells in celiac disease. Gastroenterology 2007, 133, 780–789. [Google Scholar] [CrossRef] [PubMed]
- NPD Group. Dieting Monitor: Eating Patterns in America; NPD Group: Port Washington, NY, USA, 2013. [Google Scholar]
- Brottveit, M.; Beitnes, A.C.; Tollefsen, S.; Bratlie, J.E.; Jahnsen, F.L.; Johansen, F.E.; Sollid, L.M.; Lundin, K.E.A. Mucosal Cytokine Response After Short-Term Gluten Challenge in Celiac Disease and Non-Celiac Gluten Sensitivity. Am. J. Gastroenterol. 2013, 108, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Gianfrani, C.; Camarca, A.; Mazzarella, G.; di Stasio, L.; Giardullo, N.; Ferranti, P.; Picariello, G.; Rotondi Aufiero, V.; Picascia, S.; Troncone, R.; et al. Extensive in vitro gastrointestinal digestion markedly reduces the immune-toxicity of Triticum monococcum wheat: Implication for celiac disease. Mol. Nutr. Food Res. 2015. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picascia, S.; Mandile, R.; Auricchio, R.; Troncone, R.; Gianfrani, C. Gliadin-Specific T-Cells Mobilized in the Peripheral Blood of Coeliac Patients by Short Oral Gluten Challenge: Clinical Applications. Nutrients 2015, 7, 10020-10031. https://doi.org/10.3390/nu7125515
Picascia S, Mandile R, Auricchio R, Troncone R, Gianfrani C. Gliadin-Specific T-Cells Mobilized in the Peripheral Blood of Coeliac Patients by Short Oral Gluten Challenge: Clinical Applications. Nutrients. 2015; 7(12):10020-10031. https://doi.org/10.3390/nu7125515
Chicago/Turabian StylePicascia, Stefania, Roberta Mandile, Renata Auricchio, Riccardo Troncone, and Carmen Gianfrani. 2015. "Gliadin-Specific T-Cells Mobilized in the Peripheral Blood of Coeliac Patients by Short Oral Gluten Challenge: Clinical Applications" Nutrients 7, no. 12: 10020-10031. https://doi.org/10.3390/nu7125515
APA StylePicascia, S., Mandile, R., Auricchio, R., Troncone, R., & Gianfrani, C. (2015). Gliadin-Specific T-Cells Mobilized in the Peripheral Blood of Coeliac Patients by Short Oral Gluten Challenge: Clinical Applications. Nutrients, 7(12), 10020-10031. https://doi.org/10.3390/nu7125515





