Modulation of Saliva Microbiota through Prebiotic Intervention in HIV-Infected Individuals
Abstract
:1. Introduction
2. Material and Methods
2.1. Subjects and Sample Collection
2.2. Bacterial DNA Extraction and Sequencing
2.3. Analysis of 16S rRNA Gene Amplicons
2.4. Quantitative PCR
2.5. Statistical Analysis
2.6. Co-Occurrence of Bacterial Taxa in Salivary and Gut Microbiota
3. Results
3.1. General Features of the Patients and Samples
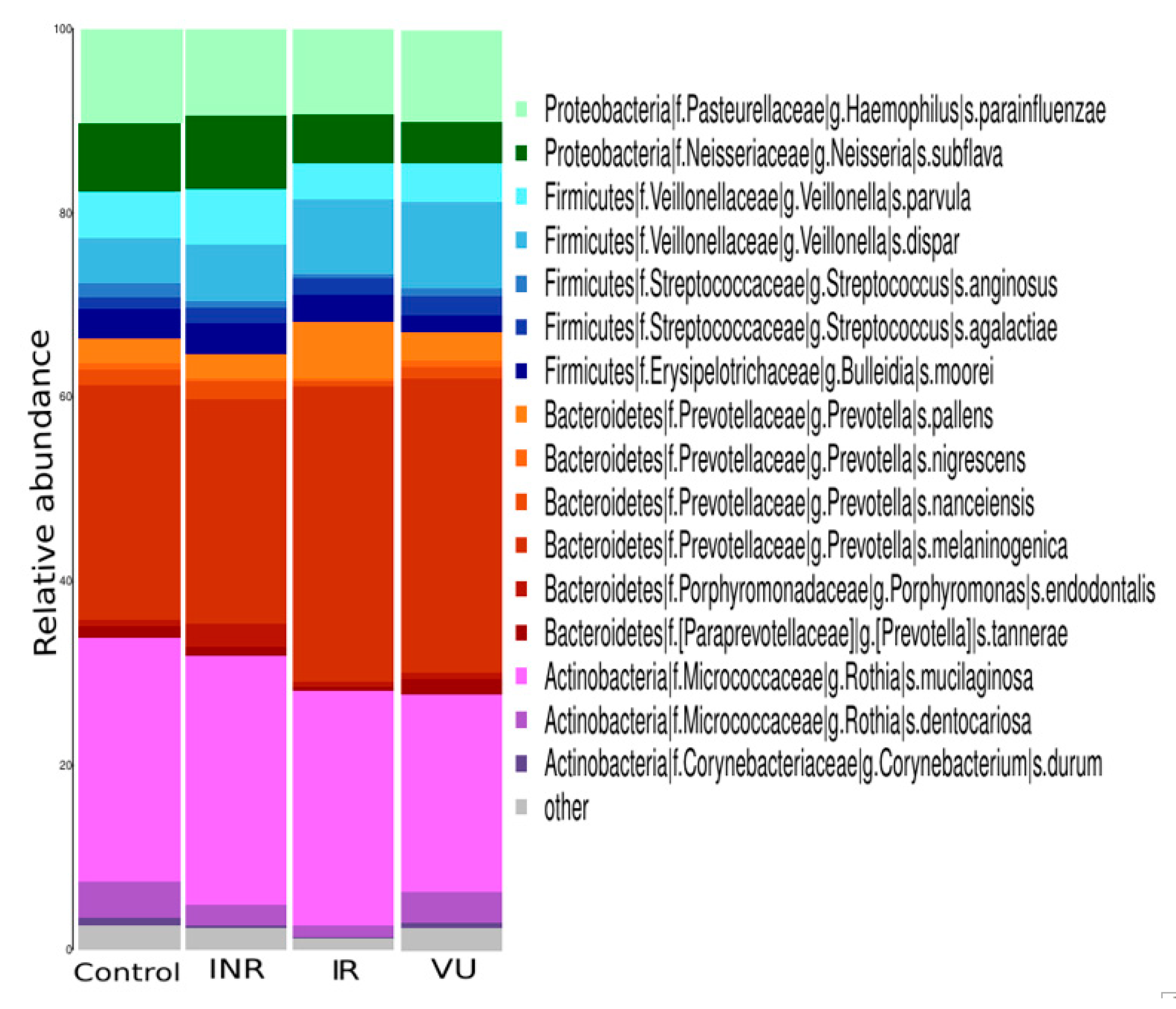
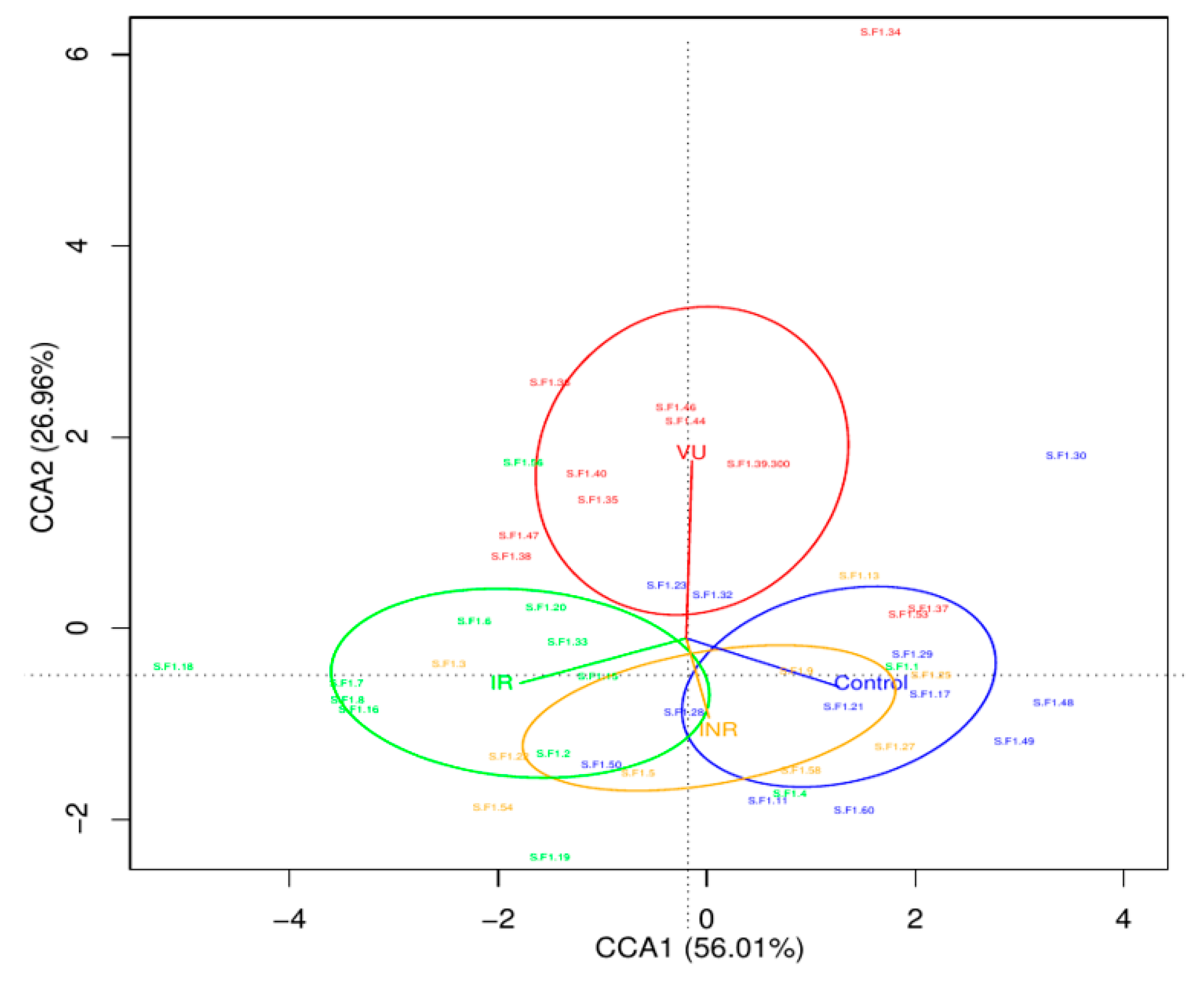
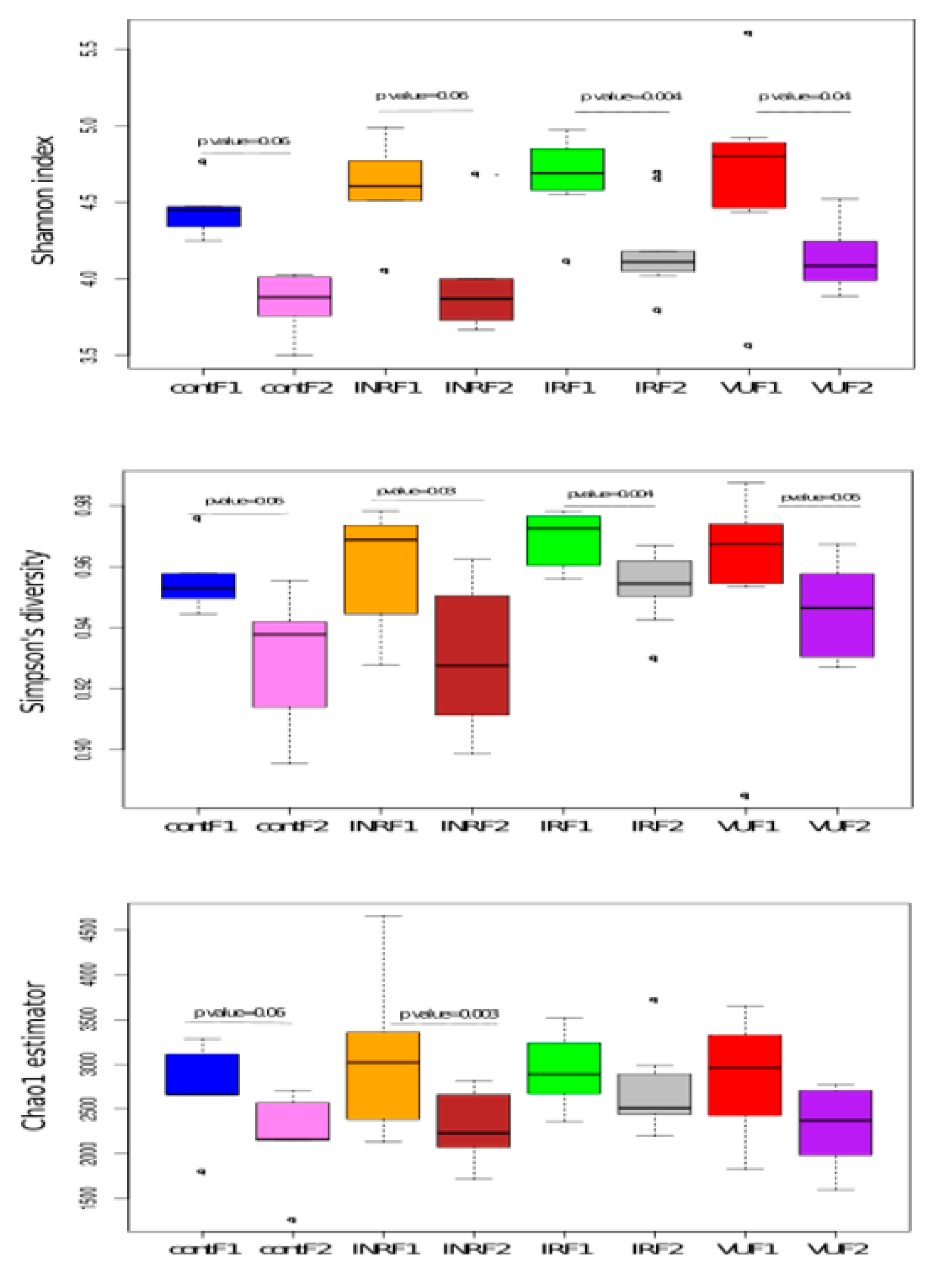
3.2. Salivary Microbiota Analysis in HIV-Infected Individuals
3.3. Effect of Prebiotics on Salivary Microbiota
3.4. Association between Salivary Microbiota and Clinical Markers before and after Prebiotic Intervention
3.5. Comparison between Salivary and Gut Microbiota: Bacterial Co-Occurrence
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marsh, P.D. In Sickness and in Health—What Does the Oral Microbiome Mean to Us? An Ecological Perspective. Adv. Dent. Res. 2018, 29, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Devine, D.A.; Marsh, P.D. Prospects for the development of probiotics and prebiotics for oral applications. J. Oral Microbiol. 2009, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Laleman, I.; Detailleur, V.; Slot, D.E.; Slomka, V.; Quirynen, M.; Teughels, W. Probiotics reduce mutans streptococci counts in humans: A systematic review and meta-analysis. Clin. Oral Investig. 2014, 18, 1539–1552. [Google Scholar] [CrossRef] [PubMed]
- Gruner, D.; Paris, S.; Schwendicke, F. Probiotics for managing caries and periodontitis: Systematic review and meta-analysis. J. Dent. 2016, 48, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Palmer, S.R.; Ahn, S.J.; Richards, V.P.; Williams, M.L.; Nascimento, M.M.; Burne, R.A. A highly arginolytic Streptococcus species that potently antagonizes Streptococcus mutans. Appl. Environ. Microbiol. 2016, 82, 2187–2201. [Google Scholar] [CrossRef]
- Saxena, D.; Li, Y.; Devota, A.; Pushalkar, S.; Abrams, W.; Barber, C.; Corby, P.; Poles, M.; Phelan, J.; Malamud, D. Modulation of the orodigestive tract microbiome in HIV-infected patients. Oral Dis. 2016, 22, 73–78. [Google Scholar] [CrossRef] [Green Version]
- López-López, A.; Camelo-Castillo, A.; Ferrer, M.D.; Simon-Soro, A.; Mira, A. Health-associated niche inhabitants as oral probiotics: The case of Streptococcus dentisani. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Slomka, V.; Hernandez-Sanabria, E.; Herrero, E.R.; Zaidel, L.; Bernaerts, K.; Boon, N.; Quirynena, M.; Teughelsa, W. Nutritional stimulation of commensal oral bacteria suppresses pathogens: The prebiotic concept. J. Clin. Periodontol. 2017, 44, 344–352. [Google Scholar] [CrossRef]
- Slomka, V.; Herrero, E.R.; Boon, N.; Bernaerts, K.; Trivedi, H.M.; Daep, C.; Quirynen, M.; Teughels, W. Oral prebiotics and the influence of environmental conditions in vitro. J. Periodontol. 2018, 89, 708–717. [Google Scholar] [CrossRef]
- Rosier, B.T.; Marsh, P.D.; Mira, A. Resilience of the oral microbiota in health: Mechanisms that prevent dysbiosis. J. Dent. Res. 2018, 97, 371–380. [Google Scholar] [CrossRef]
- Dillon, S.M.; Frank, D.N.; Cara, C.W. The gut microbiome and HIV-1 pathogenesis: A two way street. AIDS 2016, 30, 2737–2751. [Google Scholar] [CrossRef] [PubMed]
- Gori, A.; Rizzardini, G.; Van’T Land, B.; Amor, K.B.; Van Schaik, J.; Torti, C.; Quirino, T.; Tincati, C.; Bandera, A.; Knol, J.; et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: Results of the “COPA” pilot randomized trial. Mucosal Immunol. 2011, 4, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Castellanos, J.F.; Serrano-Villar, S.; Latorre, A.; Artacho, A.; Ferrús, M.L.; Madrid, N.; Vallejo, A.; Sainz, T.; Martínez-Botas, J.; Ferrando-Martínez, S.; et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 2015, 8, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Villar, S.; Vázquez-Castellanos, J.F.; Vallejo, A.; Latorre, A.; Sainz, T.; Ferrando-Martínez, S.; Rojo, D.; Martínez-Botas, J.; del Romero, J.; Madrid, N.; et al. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol. 2017, 10, 1279–1293. [Google Scholar] [CrossRef] [PubMed]
- Villar-García, J.; Güerri-Fernández, R.; Moya, A.; Gonzalez, A.; Hernandez, J.J.; Lerma, E.; Guelar, A.; Sorli, L.; Horcajada, J.P.; Artacho, A.; et al. Impact of probiotic Saccharomyces boulardii on the gut microbiome composition in HIV-treated patients: A double-blind, randomised, placebo-controlled trial. PLoS ONE 2017, 12, e0173802. [Google Scholar] [CrossRef] [PubMed]
- Asmuth, D.M.; Ma, Z.M.; Albanese, A.; Sandler, N.G.; Devaraj, S.; Knight, T.H.; Flynn, N.M.; Yotter, T.; Garcia, J.C.; Tsuchida, E.; et al. Oral serum-derived bovine immunoglobulin improves duodenal immune reconstitution and absorption function in patients with HIV enteropathy. AIDS 2013, 27, 2207–2217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahn, P.; Ruxrungtham, K.; Gazzard, B.; Diaz, R.S.; Gori, A.; Kotler, D.P.; Vriesema, A.; Georgiou, N.A.; Garssen, J.; Clerici, M.; et al. The immunomodulatory nutritional intervention NR100157 reduced CD4+ T-cell decline and immune activation: A 1-year multicenter randomized controlled double-blind trial in HIV-infected persons not receiving antiretroviral therapy (The BITE Study). Clin. Infect. Dis. 2013, 57, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Saxena, D.; Chen, Z.; Liu, G.; Abrams, W.R.; Phelan, J.A.; Norman, R.G.; Fisch, G.S.; Corby, P.M.; Dewhirst, F.; et al. HIV infection and microbial diversity in saliva. J. Clin. Microbiol. 2014, 52, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Kistler, J.O.; Arirachakaran, P.; Poovorawan, Y.; Dahlén, G.; Wade, W.G. The oral microbiome in human immunodeficiency virus (HIV)-positive individuals. J. Med. Microbiol. 2015, 64, 1094–1101. [Google Scholar] [CrossRef]
- Lewy, T.; Hong, B.Y.; Weiser, B.; Burger, H.; Tremain, A.; Weinstock, G.; Anastos, K.; George, M.D. Oral Microbiome in HIV-infected women: Shifts in the abundance of pathogenic and beneficial bacteria are associated with aging, HIV load, CD4 count, and antiretroviral therapy. AIDS Res. Hum. Retrovir. 2019, 35, 276–286. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Chandra, J.; Retuerto, M.; Tatsuoka, C.; Ghannoum, M.A.; McComsey, G.A. Dysbiosis in the oral bacterial and fungal microbiome of HIV-infected subjects is associated with clinical and immunologic variables of HIV infection. PLoS ONE 2018, 13, e0200285. [Google Scholar] [CrossRef] [PubMed]
- Presti, R.M.; Handley, S.A.; Droit, L.; Ghannoum, M.; Jacobson, M.; Shiboski, C.H.; Webster-Cyriaque, J.; Brown, T.; Yin, M.T.; Overton, E.T. Alterations in the oral microbiome in HIV-infected participants after antiretroviral therapy administration are influenced by immune status. AIDS 2018, 32, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Boix-Amorós, A.; Collado, M.C.; Mira, A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsk, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Zaura, E.; Mira, A. The oral microbiome in an ecological perspective. Front. Cell. Infect. Microbiol. 2015, 5, 2014–2015. [Google Scholar] [CrossRef]
- Saito, T.; Shimazaki, Y.; Sakamoto, M. Obesity and Periodontitis. N. Engl. J. Med. 1998, 339, 482–483. [Google Scholar] [CrossRef]
- Koren, O.; Spora, A.; Felinb, J.; Fåkb, F.; Stombaughd, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4592–4598. [Google Scholar] [CrossRef]
- Spear, G.T.; Sikaroodi, M.; Zariffard, M.R.; Landay, A.L.; French, A.L.; Gillevet, P.M. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J. Infect. Dis. 2008, 198, 1131–1140. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Said, H.S.; Suda, W.; Nakagome, S.; Chinen, H.; Oshima, K.; Kim, S.; Kimura, R.; Iraha, A.; Ishida, H.; Fujita, J.; et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014, 21, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wu, X.; Li, J.; Wang, D.; Liang, D.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Moyes, D.L.; Saxena, D.; John, M.D.; Malamud, D. The gut and oral microbiome in HIV disease: A workshop report. Oral Dis. 2016, 22, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Lira-Junior, R.; Boström, E.A. Oral-gut connection: One step closer to an integrated view of the gastrointestinal tract? Mucosal Immunol. 2018, 11, 316–318. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Sathirapongsasuti, J.F.; Izard, J.; Segata, N.; Gevers, D.; Raes, J.; Huttenhower, C. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 2012, 8, e1002606. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Morgan, X.C.; Segata, N.; Waldron, L.; Reyes, J.; Earl, A.M.; Giannoukos, G.; Boylanc, M.R.; Ciulla, D.; Gevers, D.; et al. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. USA 2014, 111, E2329–E2338. [Google Scholar] [CrossRef] [Green Version]




| Saliva F1 a | Saliva F2 b | Feces F1 a | Feces F2 b | HIV Group c |
|---|---|---|---|---|
| S-F1-11 | F1-11 | Control | ||
| S-F1-17 | F1-17 | Control | ||
| S-F1-21 | F1-21 | Control | ||
| S-F1-23 | S-F2-23 | F1-23 | F2-23 | Control |
| S-F1-28 | S-F2-28 | F1-28 | F2-28 | Control |
| S-F1-29 | S-F2-29 | F1-29 | F2-29 | Control |
| S-F1-30 | F1-30 | Control | ||
| S-F1-31 | F1-31 | Control | ||
| S-F1-32 | F1-32 | Control | ||
| S-F1-45 | S-F2-45 | F1-45 | F2-45 | Control |
| S-F1-48 | F1-48 | Control | ||
| S-F1-49 | S-F2-49 | F1-49 | F2-49 | Control |
| S-F1-50 | S-F2-50 | F1-50 | F2-50 | Control |
| S-F1-60 | S-F2-60 | F1-60 | F2-60 | Control |
| S-F1-13 | S-F2-13 | F1-13 | F2-13 | INR |
| S-F1-22 | S-F2-22 | F1-22 | F2-22 | INR |
| S-F1-25 | S-F2-25 | F1-25 | F2-25 | INR |
| S-F1-27 | S-F2-27 | F1-27 | F2-27 | INR |
| S-F1-3 | F1-3 | INR | ||
| S-F1-5 | S-F2-5 | F1-5 | F2-5 | INR |
| S-F1-54 | F1-54 | INR | ||
| S-F1-58 | F1-58 | F2-58 | INR | |
| S-F1-9 | S-F2-9 | F1-9 | F2-9 | INR |
| S-F1-1 | S-F2-1 | F1-1 | IR | |
| S-F1-10 | F1-10 | IR | ||
| S-F1-12 | F1-12 | IR | ||
| S-F1-14 | S-F2-14 | F1-14 | F2-14 | IR |
| S-F1-15 | F1-15 | F2-15 | IR | |
| S-F1-16 | S-F2-16 | F1-16 | F2-16 | IR |
| S-F1-18 | S-F2-18 | F1-18 | F2-18 | IR |
| S-F1-19 | F1-19 | F2-19 | IR | |
| S-F1-2 | S-F2-2 | F1-2 | F2-2 | IR |
| S-F1-20 | S-F2-20 | F1-20 | F2-20 | IR |
| S-F1-24 | F1-24 | IR | ||
| S-F1-26 | F1-26 | IR | ||
| S-F1-33 | S-F2-33 | F1-33 | F2-33 | IR |
| S-F1-4 | S-F2-4 | F1-4 | F2-4 | IR |
| S-F1-56 | F1-56 | IR | ||
| S-F1-6 | F1-6 | IR | ||
| S-F1-7 | S-F2-7 | F1-7 | F2-7 | IR |
| S-F1-8 | S-F2-8 | F1-8 | F2-8 | IR |
| S-F1-34 | VU | |||
| S-F1-35 | S-F2-35 | F1-35 | F2-35 | VU |
| S-F1-36 | S-F2-36 | F1-36 | F2-36 | VU |
| S-F1-37 | F1-37 | VU | ||
| S-F1-38 | F1-38 | F2-38 | VU | |
| S-F1-39 | S-F2-39 | F1-39 | F2-39 | VU |
| S-F1-40 | S-F2-40 | F1-40 | F2-40 | VU |
| S-F1-44 | S-F2-44 | F1-44 | F2-44 | VU |
| S-F1-46 | S-F2-46 | F1-46 | F2-46 | VU |
| S-F1-47 | S-F2-47 | F1-47 | F2-47 | VU |
| S-F1-51 | S-F2-51 | F1-51 | F2-51 | VU |
| S-F1-53 | S-F2-53 | F1-53 | F2-53 | VU |
| Control | INR | IR | VU | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | Prebiotics | Baseline | Prebiotics | Baseline | Prebiotics | Baseline | Prebiotics | p.value Baseline a | Adj.fdr p.value Baseline a | p.value Prebiotics b | Adj.fdr p.value Prebiotics b | |
| Metabolic profile in plama | ||||||||||||
| Glucose (mg/dL) | 91 (88–94) | 90(83–98) | 93(89–96) | 94(88–108) | 93(89–97) | 90(86–99) | 85(82–87) | 92(85–94) | 0.0615 | 0.1285 | 0.6181 | 0.6476 |
| Creatinine (mg/dL) | 0.97 (0.82–1) | 0.91 (0.84–0.99) | 1(0.95–1) | 1.1 (0.96–1.2) | 1(0.96–1.1) | 0.97 (0.93–1.1) | 1(0.94–1.2) | 0.96(0.89–1) | 0.3194 | 0.4081 | 0.1024 | 0.1733 |
| Total cholesterol (mg/dL) | 207 (160–220) | 149 (136–192) | 148 (145–198) | 155 (135–185) | 185 (153–208) | 190 (156–220) | 158 (147–184) | 164 (160–177) | 0.0924 | 0.1771 | 0.2299 | 0.3161 |
| HDL-cholesterol (mg/dL) | 54(49–75) | 55(52–76) | 50(44–57) | 49(43–61) | 53(47–58) | 56(48–60) | 48(41–53) | 49(40–53) | 0.3141 | 0.4081 | 0.3028 | 0.3918 |
| LDL-cholesterol (mg/dL) | 116 (82–134) | 89 (74–126) | 78 (71–116) | 81 (69–99) | 105 (93–119) | 115 (89–132) | 88 (86–100) | 94 (86–103) | 0.1423 | 0.2517 | 0.3867 | 0.4477 |
| Triglycerides (mg/dL) | 80 (66–161) | 67(54–76) | 102(87–147) | 97(75–120) | 102(81–127) | 104(75–152) | 107(63–158) | 110 (103–135) | 0.9266 | 0.9687 | 0.9188 | 0.9188 |
| T cell markers | ||||||||||||
| CD4+T-cell counts (cells/μl) | 716 (601–862) | 742 (629–880) | 271 (204–321) | 289 (227–319) | 561 (426–667) | 534 (432–668) | 521 (385–738) | 549 (458–645) | 0.0001 | 0.0007 | 0.0031 | 0.0115 |
| %HLADR+ CD38+ | 1(0.7–1.1) | 1.2 (0.89–1.4) | 3.4(3–4) | 3.3(2.6–3.8) | 2.3(1.8–2.6) | 1.9(1.5–2) | 3.8(3.6–7.8) | 3.4(2.8–3.6) | 0.0000 | 0.0000 | 0.0000 | 0.0001 |
| %CD25+ | 2.6(2–4) | 2.6(2.1–3.2) | 6.1(5.5–8.3) | 6.6(6–7.5) | 4.7(4.2–6.6) | 4.5(4.2–6.4) | 8.2(5.4–10) | 6.2(4.6–7.3) | 0.0028 | 0.0108 | 0.0013 | 0.0058 |
| %CD57+ | 3.2 (2.2–5.9) | 5.1(2.4–5.9) | 11(3.7–12) | 8.3(4.3–11) | 5.7(4–8.6) | 6.8(3.3–10) | 13(5.7–22) | 9.3(8.7–22) | 0.0211 | 0.0485 | 0.0498 | 0.1095 |
| CD8+ T-cell counts (cells/μl) | 403 (306–552) | 429 (308–541) | 404 (377–695) | 434 (226–566) | 606 (480–730) | 532 (436–670) | 1004 (794–1114) | 1075 (829–1271) | 0.0079 | 0.0228 | 0.0140 | 0.0439 |
| %HLADR +CD38+ | 2(1.2–2.1) | 1.5(0.6–1.6) | 4.8(4.4–6.3) | 4.9(3.7–8) | 3.3(2.6–8.3) | 4.8(3.8–5.7) | 13(10–17) | 10(8.1–11) | 0.0000 | 0.0000 | 0.0001 | 0.0004 |
| %CD25+ | 0.33 (0.095–0.41) | 0.19 (0.13–0.23) | 0.73 (0.57–1.6) | 0.84 (0.47–1.2) | 0.47 (0.32–0.74) | 0.46 (0.26–0.6) | 0.95 (0.93–1.1) | 0.77 (0.72–0.85) | 0.0022 | 0.0102 | 0.0184 | 0.0507 |
| %CD57+ | 24(16–46) | 18(18–57) | 27(18–34) | 29(19–34) | 26(18–42) | 31(21–47) | 42(39–55) | 46(36–56) | 0.2309 | 0.3540 | 0.3801 | 0.4477 |
| CD4/CD8 ratio | 1.5 (1.2–1.7) | 1.7 (1.3–2) | 0.65 (0.36–0.89) | 0.88 (0.36–2.2) | 1(0.91–1.2) | 1.2(1.1–1.3) | 0.54 (0.45–0.74) | 0.56 (0.51–0.62) | 0.0001 | 0.0006 | 0.0011 | 0.0058 |
| Thymic function | ||||||||||||
| sj/β-TREC c ratio | 9(8.2–9.5) | 8.8(7.1–8.9) | 7.8(7.6–8.4) | 8.2 (7.9–8.8) | 8.2(7.7–9.2) | 9.1(8.3–9.6) | 8.7(8.3–9) | 8.4 (7.4–8.8) | 0.0195 | 0.0485 | 0.5480 | 0.6029 |
| Endotelial function | ||||||||||||
| ADMA d (μM/L) | 1.1 (1.1–1.1) | 1(0.9–1.1) | 0.97 (0.86–1.2) | 1.2(1–1.3) | 0.97 (0.96–1.1) | 0.99(0.95–1) | 0.96 (0.92–1.1) | 1.1(1–1.2) | 0.2506 | 0.3602 | 0.2074 | 0.3042 |
| Bacterial translocation | ||||||||||||
| BPI e (ng/mL) | 35(12–35) | 8.3(2.6–9.1) | 13(4.1–49) | 3(2.3–5.7) | 28(1.3–114) | 6(1.8–8.6) | 17(5.3–60) | 9.8(5.3–17) | 0.9768 | 0.9768 | 0.0000 | 0.0002 |
| SCD14 f (ng/mL) | 1424 (1254–1484) | 1272 (1162–1362) | 1636 (1454–1999) | 1894 (1641–2402) | 1663 (1483–1732) | 1610 (1251–1750) | 1552 (1360–1613) | 1507 (1295–1721) | 0.1587 | 0.2607 | 0.0463 | 0.1095 |
| Inflammation | ||||||||||||
| IL6 g (pg/mL) | 2(2–4.1) | 2(2–2) | 2(2–2.2) | 7.3(5.3–8.1) | 2(2–2) | 2(2–2.7) | 2(2–2) | 2(2–4.3) | 0.3811 | 0.4614 | 0.0665 | 0.1329 |
| Hs-CRP h (mg/L) | 0.095 (0.055–0.3) | 0.07 (0.035–0.22) | 0.11 (0.06–0.15) | 0.57 (0.28–1.3) | 0.18 (0.09–0.29) | 0.19 (0.062–0.34) | 0.09 (0.053–0.18) | 0.15 (0.07–0.26) | 0.6838 | 0.7489 | 0.0835 | 0.1530 |
| Thrombosis | ||||||||||||
| Dimers-D (ng/mL) | 180 (120–274) | 240 (209–304) | 202 (113–306) | 261 (237–290) | 215 (176–293) | 202 (184–232) | 280 (202–322) | 238 (216–397) | 0.4032 | 0.4637 | 0.1428 | 0.2244 |
| Correlations Before Prebiotic Intervention | ||||
| Clinical Variable | Taxa | Spearman Correlation Index | p.value | Adj.fdr_p.value |
| Plasma Metabolite | ||||
| Creatinine | Faecalibacterium prausnitzii | 0.5893 | 0.0004 | 0.0344 |
| Creatinine | Haemophilus parainfluenzae | 0.5670 | 0.0007 | 0.0376 |
| Creatinine | Actinobacillus porcinus | 0.4944 | 0.0040 | 0.0710 |
| HDL-cholesterol | Prevotella tannerae | −0.4664 | 0.0071 | 0.1025 |
| HDL-cholesterol | Lactobacillus coleohominis | −0.4136 | 0.0186 | 0.1346 |
| HDL-cholesterol | Pyramidobacter piscolens | −0.3985 | 0.0239 | 0.1356 |
| HDL-cholesterol | Lactobacillus reuteri | −0.3955 | 0.0250 | 0.1356 |
| HDL-cholesterol | Pasteurella multocida | −0.3851 | 0.0295 | 0.1356 |
| T cell markers | ||||
| CD4+T cell | Streptococcus anginosus | −0.3965 | 0.0247 | 0.1356 |
| CD4+T cell | Pyramidobacter piscolens | −0.3682 | 0.0381 | 0.1356 |
| CD4+ CD25+T cell | Veillonella parvula | 0.4170 | 0.0176 | 0.1346 |
| CD4+ CD25+T cell | Prevotella pallens | −0.3736 | 0.0352 | 0.1356 |
| CD4+ CD57+T cell | Prevotella copri | 0.3864 | 0.0289 | 0.1356 |
| CD4+ CD57+T cell | Pasteurella multocida | 0.3587 | 0.0438 | 0.1356 |
| CD4+ HLADR+ CD38+T cell | Prevotella nigrescens | 0.3592 | 0.0442 | 0.1356 |
| Thymic function | ||||
| sj/β-TREC ratio a | Actinomyces hyovaginalis | 0.5852 | 0.0004 | 0.0344 |
| sj/β-TREC ratio | Streptococcus sobrinus | −0.5077 | 0.0030 | 0.0680 |
| sj/β-TREC ratio | Streptococcus agalactiae | 0.4780 | 0.0062 | 0.0975 |
| sj/β-TREC ratio | Pyramidobacter piscolens | −0.3710 | 0.0366 | 0.1356 |
| Bacterial translocation | ||||
| BPI b | Streptobacillus moniliformis | 0.3550 | 0.0462 | 0.1356 |
| Correlations After Prebiotic Intervention | ||||
| Clinical Variable | Taxa | Spearman Correlation Index | p.value | Adj.fdr p.value |
| Plasma Metabolite | ||||
| Creatinine | Campylobacter rectus | 0.3741 | 0.0349 | 0.0689 |
| Glucose | Actinomyces hyovaginalis | −0.4022 | 0.0225 | 0.0689 |
| Glucose | Collinsella aerofaciens | 0.3575 | 0.0446 | 0.0689 |
| HDL-cholesterol | Lactobacillus coleohominis | −0.3839 | 0.0301 | 0.0689 |
| HDL-cholesterol | Prevotella melaninogenica | 0.3761 | 0.0339 | 0.0689 |
| LDL-cholesterol | Pasteurella multocida | −0.3893 | 0.0276 | 0.0689 |
| Triglycerides | Pasteurella multocida | −0.4501 | 0.0098 | 0.0689 |
| Triglycerides | Lactobacillus coleohominis | 0.4211 | 0.0164 | 0.0689 |
| Triglycerides | Lactobacillus reuteri | 0.3714 | 0.0364 | 0.0689 |
| T cell markers | ||||
| CD4+T cell | Ruminococcus gnavus | 0.5321 | 0.0017 | 0.0283 |
| CD4+ HLADR+ CD38+T cell | Bifidobacterium adolescentis | 0.3928 | 0.0262 | 0.0689 |
| CD4+ HLADR+ CD38+T cell | Ruminococcus gnavus | −0.3618 | 0.0419 | 0.0689 |
| CD4+ HLADR+ CD38+T cell | Capnocytophaga ochracea | 0.3590 | 0.0436 | 0.0689 |
| CD4+ CD25+T cell | Ruminococcus gnavus | −0.5393 | 0.0014 | 0.0283 |
| CD4+ CD25+T cell | Actinobacillus porcinus | 0.3916 | 0.0267 | 0.0689 |
| CD4+ CD25+T cell | Prevotella_intermedia | 0.3901 | 0.0273 | 0.0689 |
| CD4+ CD25+T cell | Selenomonas_noxia | −0.3821 | 0.0309 | 0.0689 |
| CD4+ CD57+T cell | Neisseria_subflava | −0.4621 | 0.0078 | 0.0689 |
| CD4+ CD57+T cell | Atopobium_rimae | 0.4522 | 0.0094 | 0.0689 |
| CD4+ CD57+T cell | Haemophilus_parainfluenzae | −0.4359 | 0.0133 | 0.0689 |
| CD4+ CD57+T cell | Veillonella dispar | 0.4245 | 0.0162 | 0.0689 |
| CD4+ CD57+T cell | Pasteurella multocida | −0.4036 | 0.0220 | 0.0689 |
| CD4+ CD57+T cell | Prevotella intermedia | 0.3979 | 0.0241 | 0.0689 |
| CD4+ CD57+T cell | Bifidobacterium adolescentis | 0.3612 | 0.0423 | 0.0689 |
| CD8+T cell | Capnocytophaga ochracea | 0.3983 | 0.0240 | 0.0689 |
| CD8+ CD25+T cell | Pseudomonas veronii | −0.4773 | 0.0057 | 0.0672 |
| CD8+ CD25+T cell | Bulleidia moorei | −0.3796 | 0.0321 | 0.0689 |
| CD8+ CD25+T cell | Bifidobacterium longum | −0.3699 | 0.0372 | 0.0689 |
| CD8+ CD57+T cell | Pasteurella multocida | −0.5469 | 0.0012 | 0.0283 |
| CD8+ CD57+T cell | Pasteurella multocida | −0.5469 | 0.0012 | 0.0283 |
| CD8+ CD57+T cell | Flexispira rappini | −0.3800 | 0.0319 | 0.0689 |
| CD8+ CD57+T cell | Flexispira rappini | −0.3800 | 0.0319 | 0.0689 |
| CD8+ CD57+T cell | Neisseria subflava | −0.3598 | 0.0431 | 0.0689 |
| CD8+ CD57+T cell | Streptobacillus moniliformis | −0.3577 | 0.0444 | 0.0689 |
| CD4/CD8 ratio | Ruminococcus gnavus | 0.4574 | 0.0085 | 0.0689 |
| Endotelial function | ||||
| ADMA c | Faecalibacterium prausnitzii | 0.4219 | 0.0162 | 0.0689 |
| ADMA | Prevotella tannerae | 0.3859 | 0.0292 | 0.0689 |
| ADMA | Lactobacillus coleohominis | 0.3617 | 0.0420 | 0.0689 |
| Bacterial translocation | ||||
| BPI | Lactobacillus helveticus | 0.3642 | 0.0405 | 0.0689 |
| SCD14 d | Lactobacillus vaginalis | −0.4307 | 0.0139 | 0.0689 |
| sCD14 | Streptococcus sobrinus | −0.3978 | 0.0242 | 0.0689 |
| sCD14 | Lactobacillus salivarius | −0.3584 | 0.0440 | 0.0689 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Hernández, N.; Serrano-Villar, S.; Domingo, A.; Pons, X.; Artacho, A.; Estrada, V.; Moya, A.; Gosalbes, M.J. Modulation of Saliva Microbiota through Prebiotic Intervention in HIV-Infected Individuals. Nutrients 2019, 11, 1346. https://doi.org/10.3390/nu11061346
Jiménez-Hernández N, Serrano-Villar S, Domingo A, Pons X, Artacho A, Estrada V, Moya A, Gosalbes MJ. Modulation of Saliva Microbiota through Prebiotic Intervention in HIV-Infected Individuals. Nutrients. 2019; 11(6):1346. https://doi.org/10.3390/nu11061346
Chicago/Turabian StyleJiménez-Hernández, Nuria, Sergio Serrano-Villar, Alba Domingo, Xavier Pons, Alejandro Artacho, Vicente Estrada, Andrés Moya, and María José Gosalbes. 2019. "Modulation of Saliva Microbiota through Prebiotic Intervention in HIV-Infected Individuals" Nutrients 11, no. 6: 1346. https://doi.org/10.3390/nu11061346






