Characterization of Sinorhizobium sp. LM21 Prophages and Virus-Encoded DNA Methyltransferases in the Light of Comparative Genomic Analyses of the Sinorhizobial Virome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Media, and Growth Conditions
2.2. DNA Sequencing
2.3. Bioinformatics
2.4. Standard Molecular Biology Procedures
2.5. Cloning, Overexpression, Purification, and Testing of Putative DNA MTases Activities
2.6. Cloning, Overexpression and Testing of Φ2LM21-Encoded Lytic Enzyme Activity
2.7. Nucleotide Sequence Accession Number
3. Results and Discussion
3.1. Identification and Classification of the Sinorhizobium sp. LM21 Prophages
3.2. Characterization of the Φ2LM21 and Φ3LM21 Prophages
3.3. Characterization of the Φ4LM21 Prophage Remnant
3.4. Functional Analyses of DNA Methyltransferases Encoded by the Sinorhizobium sp. LM21 Prophages
3.5. Comparative Genomics and Networking of the Sinorhizobial (Pro)phages
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Denison, R.F.; Toby Kiers, E. Why are most rhizobia beneficial to their plant hosts, rather than parasitic? Microbes Infect. 2004, 6, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Dziewit, L.; Czarnecki, J.; Prochwicz, E.; Wibberg, D.; Schluter, A.; Puhler, A.; Bartosik, D. Genome-guided insight into the methylotrophy of Paracoccus aminophilus JCM 7686. Front. Microbiol. 2015, 6, 852. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.K.; Narra, H.P.; Sahni, A.; Walker, D.H. Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiol. 2013, 8, 1265–1288. [Google Scholar] [CrossRef] [PubMed]
- Dicenzo, G.C.; Checcucci, A.; Bazzicalupo, M.; Mengoni, A.; Viti, C.; Dziewit, L.; Finan, T.M.; Galardini, M.; Fondi, M. Metabolic modelling reveals the specialization of secondary replicons for niche adaptation in Sinorhizobium meliloti. Nat. Commun. 2016, 7, 12219. [Google Scholar] [CrossRef] [PubMed]
- Dziewit, L.; Czarnecki, J.; Wibberg, D.; Radlinska, M.; Mrozek, P.; Szymczak, M.; Schlüter, A.; Pühler, A.; Bartosik, D. Architecture and functions of a multipartite genome of the methylotrophic bacterium Paracoccus aminophilus JCM 7686, containing primary and secondary chromids. BMC Genom. 2014, 15, 124. [Google Scholar] [CrossRef] [PubMed]
- Galardini, M.; Pini, F.; Bazzicalupo, M.; Biondi, E.G.; Mengoni, A. Replicon-dependent bacterial genome evolution: The case of Sinorhizobium meliloti. Genome Biol. Evol. 2013, 5, 542–558. [Google Scholar] [CrossRef] [PubMed]
- Schneiker-Bekel, S.; Wibberg, D.; Bekel, T.; Blom, J.; Linke, B.; Neuweger, H.; Stiens, M.; Vorholter, F.J.; Weidner, S.; Goesmann, A.; et al. The complete genome sequence of the dominant Sinorhizobium meliloti field isolate SM11 extends the S. meliloti pan-genome. J. Biotechnol. 2011, 155, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Lagares, A.; Sanjuan, J.; Pistorio, M. The plasmid mobilome of the model plant-symbiont Sinorhizobium meliloti: Coming up with new questions and answers. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Weidner, S.; Puhler, A.; Kuster, H. Genomics insights into symbiotic nitrogen fixation. Curr. Opin. Biotechnol. 2003, 14, 200–205. [Google Scholar] [CrossRef]
- Donnarumma, F.; Bazzicalupo, M.; Blazinkov, M.; Mengoni, A.; Sikora, S.; Babic, K.H. Biogeography of Sinorhizobium meliloti nodulating alfalfa in different Croatian regions. Res. Microbiol. 2014, 165, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Marek-Kozaczuk, M.; Wielbo, J.; Pawlik, A.; Skorupska, A. Nodulation competitiveness of Ensifer meliloti alfalfa nodule isolates and their potential for application as inoculants. Pol. J. Microbiol. 2014, 63, 375–386. [Google Scholar] [PubMed]
- Johnson, M.C.; Tatum, K.B.; Lynn, J.S.; Brewer, T.E.; Lu, S.; Washburn, B.K.; Stroupe, M.E.; Jones, K.M. Sinorhizobium meliloti phage ΦM9 defines a new group of T4 superfamily phages with unusual genomic features but a common T=16 capsid. J. Virol. 2015, 89, 10945–10958. [Google Scholar] [CrossRef] [PubMed]
- Brewer, T.E.; Stroupe, M.E.; Jones, K.M. The genome, proteome and phylogenetic analysis of Sinorhizobium meliloti phage ΦM12, the founder of a new group of T4-superfamily phages. Virology 2014, 450–451, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Dziewit, L.; Oscik, K.; Bartosik, D.; Radlinska, M. Molecular characterization of a novel temperate Sinorhizobium bacteriophage, ФLM21, encoding DNA methyltransferase with CcrM-like specificity. J. Virol. 2014, 88, 13111–13124. [Google Scholar] [CrossRef] [PubMed]
- Dorgai, L.; Papp, I.; Papp, P.; Kalman, M.; Orosz, L. Nucleotide sequences of the sites involved in the integration of phage 16–3 of Rhizobium meliloti 41. Nucleic Acids Res. 1993, 21, 1671. [Google Scholar] [CrossRef] [PubMed]
- Dziewit, L.; Pyzik, A.; Szuplewska, M.; Matlakowska, R.; Mielnicki, S.; Wibberg, D.; Schlüter, A.; Pühler, A.; Bartosik, D. Diversity and role of plasmids in adaptation of bacteria inhabiting the Lubin copper mine in Poland, an environment rich in heavy metals. Front. Microbiol. 2015, 6, 152. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.; Kozdon, J.B.; McAdams, H.H.; Shapiro, L.; Collier, J. The functions of DNA methylation by CcrM in Caulobacter crescentus: A global approach. Nucleic Acids Res. 2014, 42, 3720–3735. [Google Scholar] [CrossRef] [PubMed]
- Kahng, L.S.; Shapiro, L. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 2001, 183, 3065–3075. [Google Scholar] [CrossRef] [PubMed]
- Siwek, W.; Czapinska, H.; Bochtler, M.; Bujnicki, J.M.; Skowronek, K. Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI. Nucleic Acids Res. 2012, 40, 7563–7572. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Berriman, M.; Tivey, A.; Patel, C.; Bohme, U.; Barrell, B.G.; Parkhill, J.; Rajandream, M.A. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008, 24, 2672–2676. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- MOTIF Search. Available online: http://www.genome.jp/tools/motif/ (accessed on 15 April 2017).
- Soding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2010, 38, D234–D236. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.; Tavares, P.; Petit, M.A.; Guerois, R.; Zinn-Justin, S. Automated classification of tailed bacteriophages according to their neck organization. BMC Genom. 2014, 15, 1027. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV). Available online: ictvonline.org (accessed on 10 April 2017).
- Darzentas, N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef] [PubMed]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Drozdz, M.; Piekarowicz, A.; Bujnicki, J.M.; Radlinska, M. Novel non-specific DNA adenine methyltransferases. Nucleic Acids Res. 2012, 40, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Cevallos, M.A.; Cervantes-Rivera, R.; Gutierrez-Rios, R.M. The repABC plasmid family. Plasmid 2008, 60, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, R.I.; Bustos, P.; Sepulveda-Robles, O.; Lozano, L.; Rodriguez, C.; Fernandez, J.L.; Juarez, S.; Kameyama, L.; Guarneros, G.; Davila, G.; et al. Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl. Environ. Microbiol. 2014, 80, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Duffy, C.; Feiss, M. The large subunit of bacteriophage lambda’s terminase plays a role in DNA translocation and packaging termination. J. Mol. Biol. 2002, 316, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Velleman, M.; Arber, W. Three functions of bacteriophage P1 involved in cell lysis. J. Bacteriol. 1996, 178, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Muller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Munch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Anand, S.; Iyer, L.M. Novel autoproteolytic and DNA-damage sensing components in the bacterial SOS response and oxidized methylcytosine-induced eukaryotic DNA demethylation systems. Biol. Direct 2013, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Motamedi, H.; Shafiee, A.; Cai, S.J.; Streicher, S.L.; Arison, B.H.; Miller, R.R. Characterization of methyltransferase and hydroxylase genes involved in the biosynthesis of the immunosuppressants FK506 and FK520. J. Bacteriol. 1996, 178, 5243–5248. [Google Scholar] [CrossRef] [PubMed]
- Gilch, S.; Meyer, O.; Schmidt, I. A soluble form of ammonia monooxygenase in Nitrosomonas europaea. Biol. Chem. 2009, 390, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Hove-Jensen, B.; Nygaard, P. Phosphoribosylpyrophosphate synthetase of Escherichia coli, Identification of a mutant enzyme. Eur. J. Biochem. 1982, 126, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Kovalyova, I.V.; Billington, S.J.; Patrick, A.N.; Butts, B.D.; Guichard, J.A.; Pitcher, T.J.; Guthrie, C.C.; Sydlaske, A.D.; Barnhill, L.M.; et al. The genome of ε15, a serotype-converting, Group E1 Salmonella enterica-specific bacteriophage. Virology 2007, 369, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Losick, R.; Robbins, P.W. Mechanism of epsilon-15 conversion studies with a bacterial mutant. J. Mol. Biol. 1967, 30, 445–455. [Google Scholar] [CrossRef]
- Bujnicki, J.M.; Radlinska, M.; Zaleski, P.; Piekarowicz, A. Cloning of the Haemophilus influenzae Dam methyltransferase and analysis of its relationship to the Dam methyltransferase encoded by the HP1 phage. Acta Biochim. Pol. 2001, 48, 969–983. [Google Scholar] [PubMed]
- Dziewit, L.; Radlinska, M. Two novel temperate bacteriophages co-existing in Aeromonas sp. ARM81—Characterization of their genomes, proteomes and DNA methyltransferases. J. Gen. Virol. 2016, 97, 2008–2022. [Google Scholar] [CrossRef] [PubMed]
- Galibert, F.; Finan, T.M.; Long, S.R.; Puhler, A.; Abola, P.; Ampe, F.; Barloy-Hubler, F.; Barnett, M.J.; Becker, A.; Boistard, P.; et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 2001, 293, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.M.; Stroupe, M.E.; Sousa, D.R.; Kropinski, A.M.; Adriaenssens, E.M.; Kuhn, J.H.; Grose, J.H. ICTV Taxonomic Proposal 2016.025a-dB.A.v1.M12virus. Create Genus M12virus in the Family Myoviridae, Order Caudovirales. International Committee on Taxonomy in Viruses (ICTV), 2016. Available online: http://www.ictv.global/proposals-16/2016.025a-dB.A.v1.M12virus.pdf (accessed on 26 June 2017).
- Casjens, S.R.; Gilcrease, E.B.; Winn-Stapley, D.A.; Schicklmaier, P.; Schmieger, H.; Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Hatfull, G.F.; Hendrix, R.W. The generalized transducing Salmonella bacteriophage ES18: Complete genome sequence and DNA packaging strategy. J. Bacteriol. 2005, 187, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef] [PubMed]
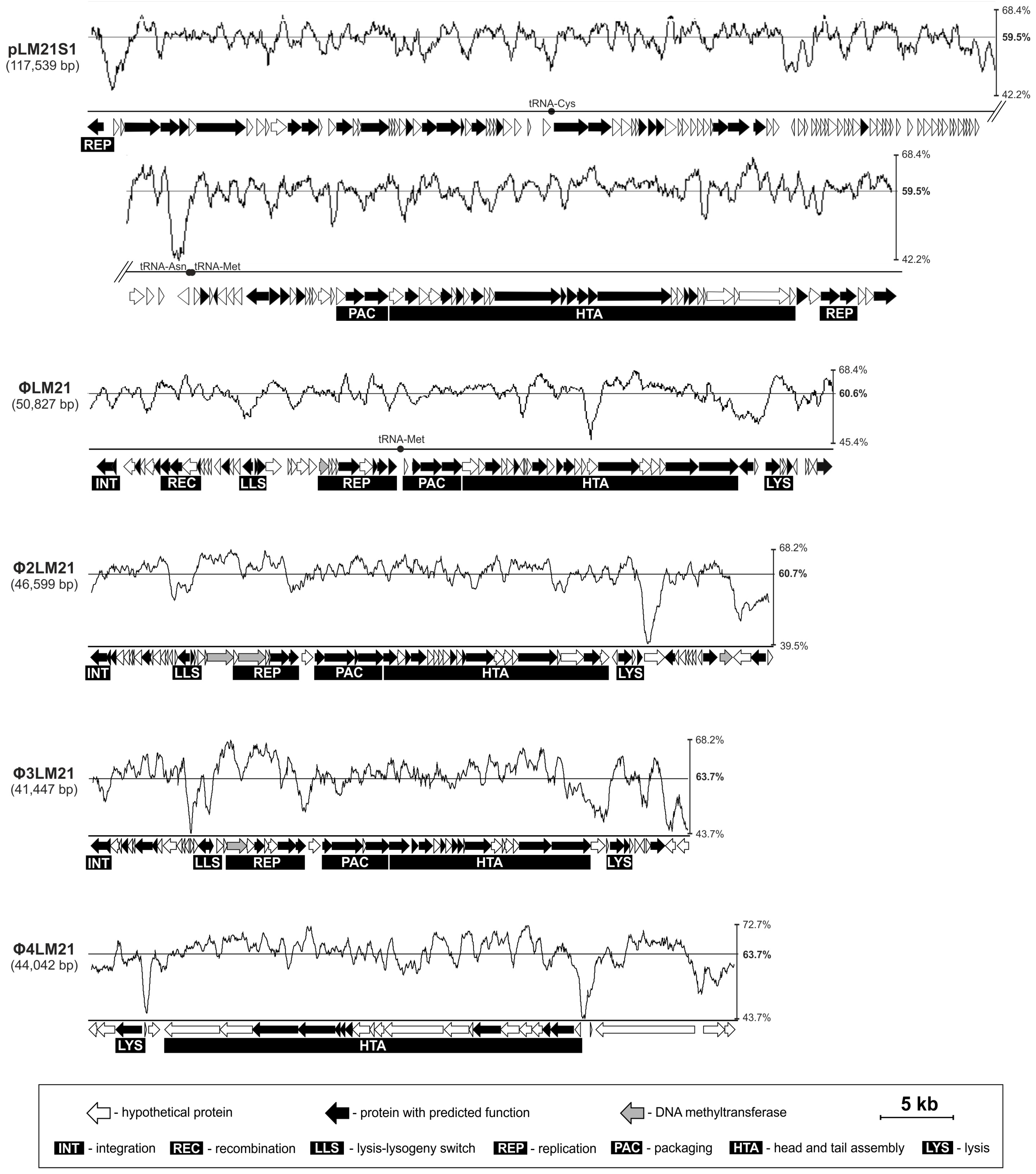

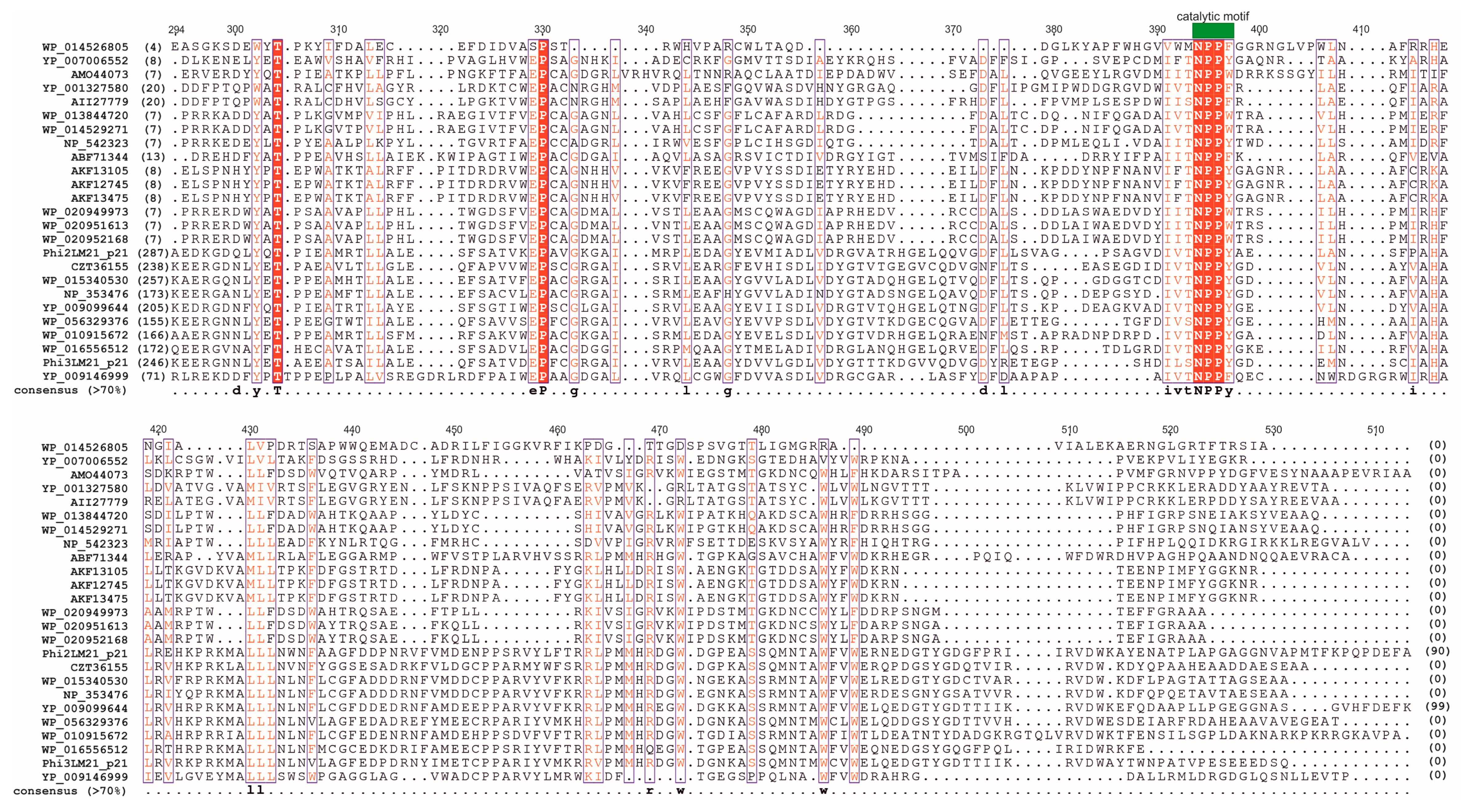
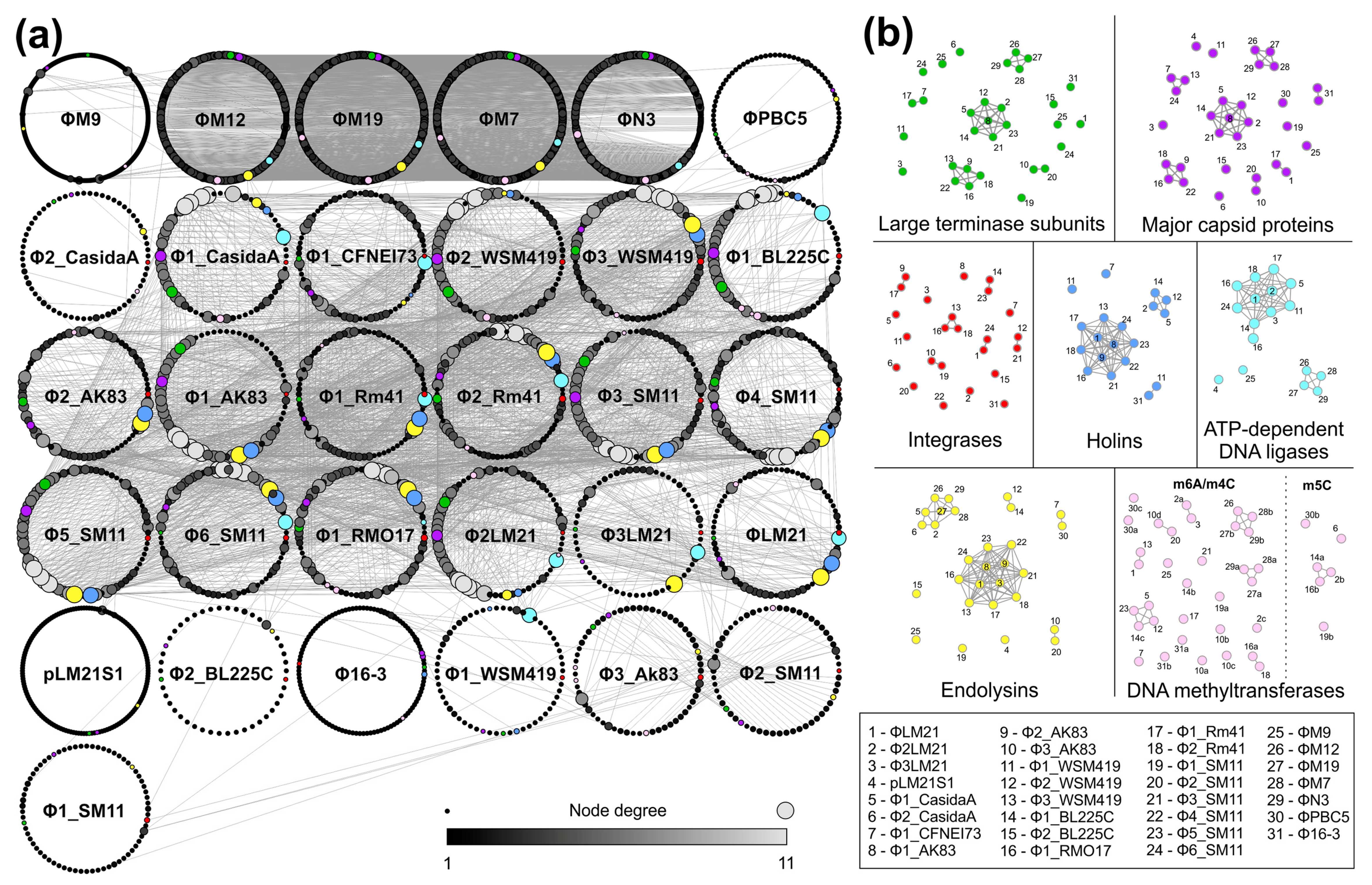
| (Pro)phage Name | (Pro)phage Host | Host/Phage Accession Number | Coordinates in Host’s Genome | (Pro)phage Size (bp) | Number of Genes | Integration Site |
|---|---|---|---|---|---|---|
| Φ1_CasidaA | E. adherens Casida A | NZ_CP015880.1 | 2834796..2878945 | 44,150 | 62 | tRNA-Leu (CAA) |
| (chromosome) | ||||||
| Φ2_CasidaA | E. adherens Casida A | NZ_CP015880.1 | 2972802..3012361 | 39,560 | 63 | Intergenic region |
| (chromosome) | ||||||
| Φ1_CFNEI73 | S. americanum CFNEI73 | NZ_CP013107.1 | 1807693..1861334 | 53,642 | 84 | tRNA-Cys(GCA) |
| (chromosome) | ||||||
| Φ1_WSM419 | S. medicae WSM419 | NC_009636.1 (chromosome) | 1392222..1433999 | 41,778 | 58 | tRNA-Ser(TGA) |
| Φ2_WSM419 | S. medicae WSM419 | NC_009636.1 (chromosome) | 1717421..1768019 | 50,599 | 66 | tRNA-dihydrouridine synthase A (DusA) |
| Φ3_WSM419 | S. medicae WSM419 | NC_009636.1 (chromosome) | 1934112..1984910 | 50,799 | 72 | tRNA-Lys(CTT) |
| Φ1_Rm41 | S. meliloti Rm41 | NC_018700.1 (chromosome) | 742114..794018 | 53,565 | 80 | tRNA-Ser(GCT) |
| Φ2_Rm41 | S. meliloti Rm41 | NC_018700.1 (chromosome) | 1833694..1887258 | 51,921 | 86 | tRNA-Lys(CTT) |
| Φ1_RMO17 | S. meliloti RMO17 | NZ_CP009144.1 (chromosome) | 2233094..2285133 | 52,040 | 76 | tRNA-Lys(CTT) |
| Φ1_BL225C | S. meliloti BL225C | NC_017322.1 (chromosome) | 1366482..1418152 | 51,671 | 67 | tRNA-Asn(GTT) |
| Φ2_BL225C | S. meliloti BL225C | NC_017323.1 | 1651701..1686916 | 35,216 | 46 | tRNA-Arg(CCG) |
| (pSINMEB02) | ||||||
| Φ1_SM11 | S. meliloti SM11 | NC_017325.1 (chromosome) | 912263..969433 | 57,171 | 69 | tRNA-Met(CAT) |
| Φ2_SM11 | S. meliloti SM11 | NC_017325.1 (chromosome) | 1084292..1130096 | 45,805 | 57 | N/A |
| Φ3_SM11 | S. meliloti SM11 | NC_017325.1 (chromosome) | 1453058..1501481 | 48,424 | 63 | tRNA-dihydrouridine synthase A (DusA) |
| Φ4_SM11 | S. meliloti SM11 | NC_017325.1 (chromosome) | 1795391..1849554 | 54,164 | 81 | tRNA-Leu(TAA) |
| Φ5_SM11 | S. meliloti SM11 | NC_017325.1 (chromosome) | 1864579..1915967 | 51,389 | 72 | tRNA-Asn(GTT) |
| Φ6_SM11 | S. meliloti SM11 | NC_017325.1 (chromosome) | 2351730..2402613 | 50,865 | 76 | tRNA-Pro(GGG) |
| Φ1_AK83 | S. meliloti AK83 | NC_015590.1 (chromosome 1) | 264329..313309 | 48,981 | 62 | tRNA-Thr(GGT) |
| Φ2_AK83 | S. meliloti AK83 | NC_015590.1 (chromosome 1) | 795050..847939 | 52,890 | 78 | tRNA-Ser(GCT) |
| Φ3_AK83 | S. meliloti AK83 | NC_015590.1 (chromosome 1) | 2309762..2355282 | 45,521 | 61 | tRNA-Met(CAT) |
| pS1LM21 | Sinorhizobium sp. LM21 | KM659098 | N/A | 117,539 | 150 | N/A |
| ΦLM21 | Sinorhizobium sp. LM21 | KJ743987 | N/A | 50,827 | 72 | tRNA-Pro(GGG) |
| Φ2LM21 | Sinorhizobium sp. LM21 | SAMN06765771 | 550879..597478 (Contig_2) | 46,599 | 69 | tRNA-Phe(GAA) |
| Φ3LM21 | Sinorhizobium sp. LM21 | SAMN06765771 | 78163..119610 (Contig_16) | 41,447 | 59 | tRNA-Pro(CGG) |
| Φ16-3 | S. meliloti Rm41 | DQ500118 | N/A | 60,195 | 110 | tRNA-Pro(CGG) |
| ΦPBC5 | S. meliloti 2011 | NC_003324 | N/A | 57,416 | 83 | N/A |
| ΦM7 | S. meliloti (lh) 1 | KR052480 | virulent | 188,427 | 359 | N/A |
| ΦM9 | S. meliloti (lh) | KP881232 | virulent | 149,218 | 271 | N/A |
| ΦM12 | S. meliloti (lh) | KF381361 | virulent | 194,701 | 377 | N/A |
| ΦM19 | S. meliloti (lh) | KR052481 | virulent | 188,047 | 361 | N/A |
| ΦN3 | S. meliloti (lh) | KR052482 | virulent | 206,713 | 398 | N/A |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Decewicz, P.; Radlinska, M.; Dziewit, L. Characterization of Sinorhizobium sp. LM21 Prophages and Virus-Encoded DNA Methyltransferases in the Light of Comparative Genomic Analyses of the Sinorhizobial Virome. Viruses 2017, 9, 161. https://doi.org/10.3390/v9070161
Decewicz P, Radlinska M, Dziewit L. Characterization of Sinorhizobium sp. LM21 Prophages and Virus-Encoded DNA Methyltransferases in the Light of Comparative Genomic Analyses of the Sinorhizobial Virome. Viruses. 2017; 9(7):161. https://doi.org/10.3390/v9070161
Chicago/Turabian StyleDecewicz, Przemyslaw, Monika Radlinska, and Lukasz Dziewit. 2017. "Characterization of Sinorhizobium sp. LM21 Prophages and Virus-Encoded DNA Methyltransferases in the Light of Comparative Genomic Analyses of the Sinorhizobial Virome" Viruses 9, no. 7: 161. https://doi.org/10.3390/v9070161
APA StyleDecewicz, P., Radlinska, M., & Dziewit, L. (2017). Characterization of Sinorhizobium sp. LM21 Prophages and Virus-Encoded DNA Methyltransferases in the Light of Comparative Genomic Analyses of the Sinorhizobial Virome. Viruses, 9(7), 161. https://doi.org/10.3390/v9070161






