Biophysical Mode-of-Action and Selectivity Analysis of Allosteric Inhibitors of Hepatitis C Virus (HCV) Polymerase
Abstract
:1. Introduction
2. Materials and Methods
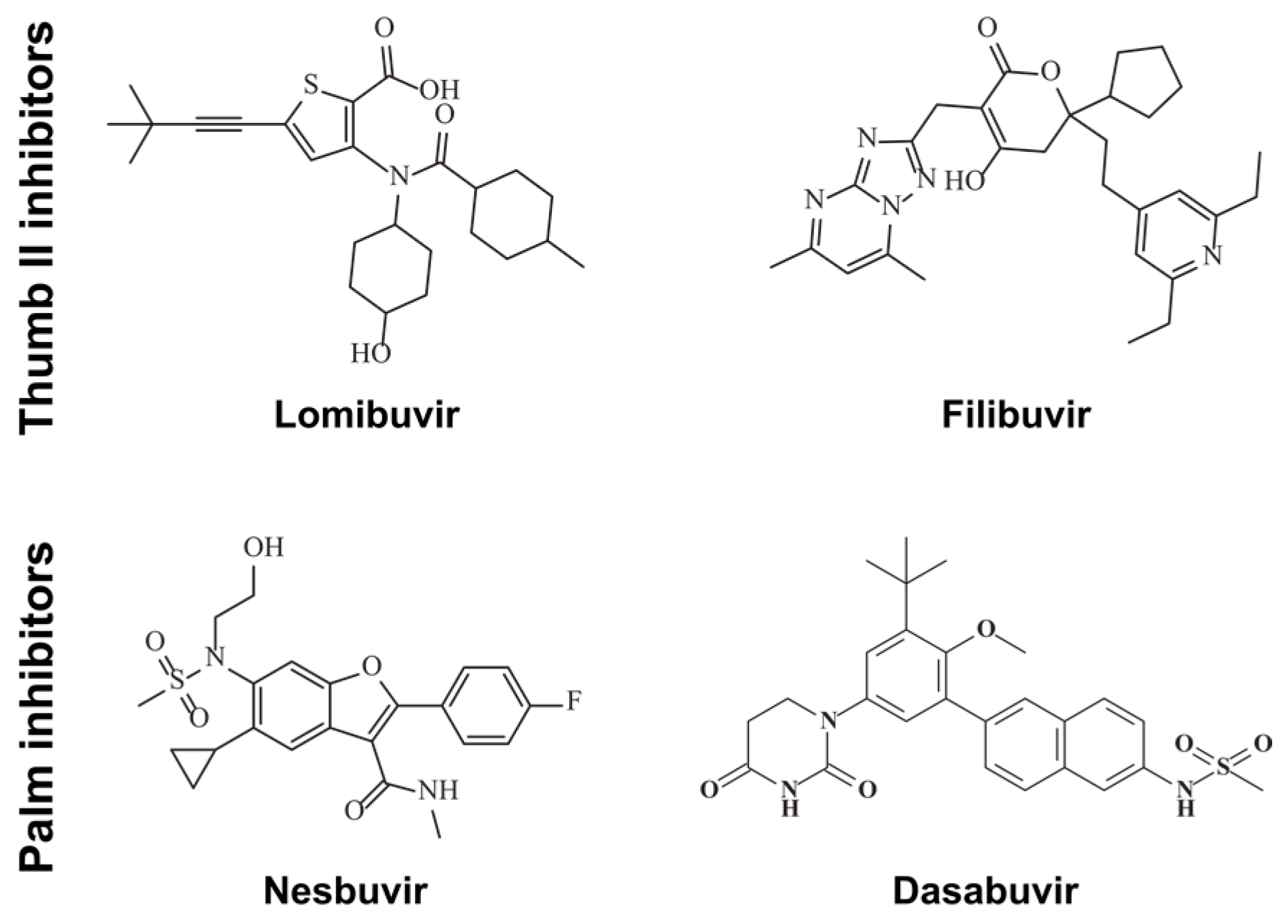
2.1. Inhibitors
2.2. Protein Production
2.3. Differential Scanning Fluorimetry
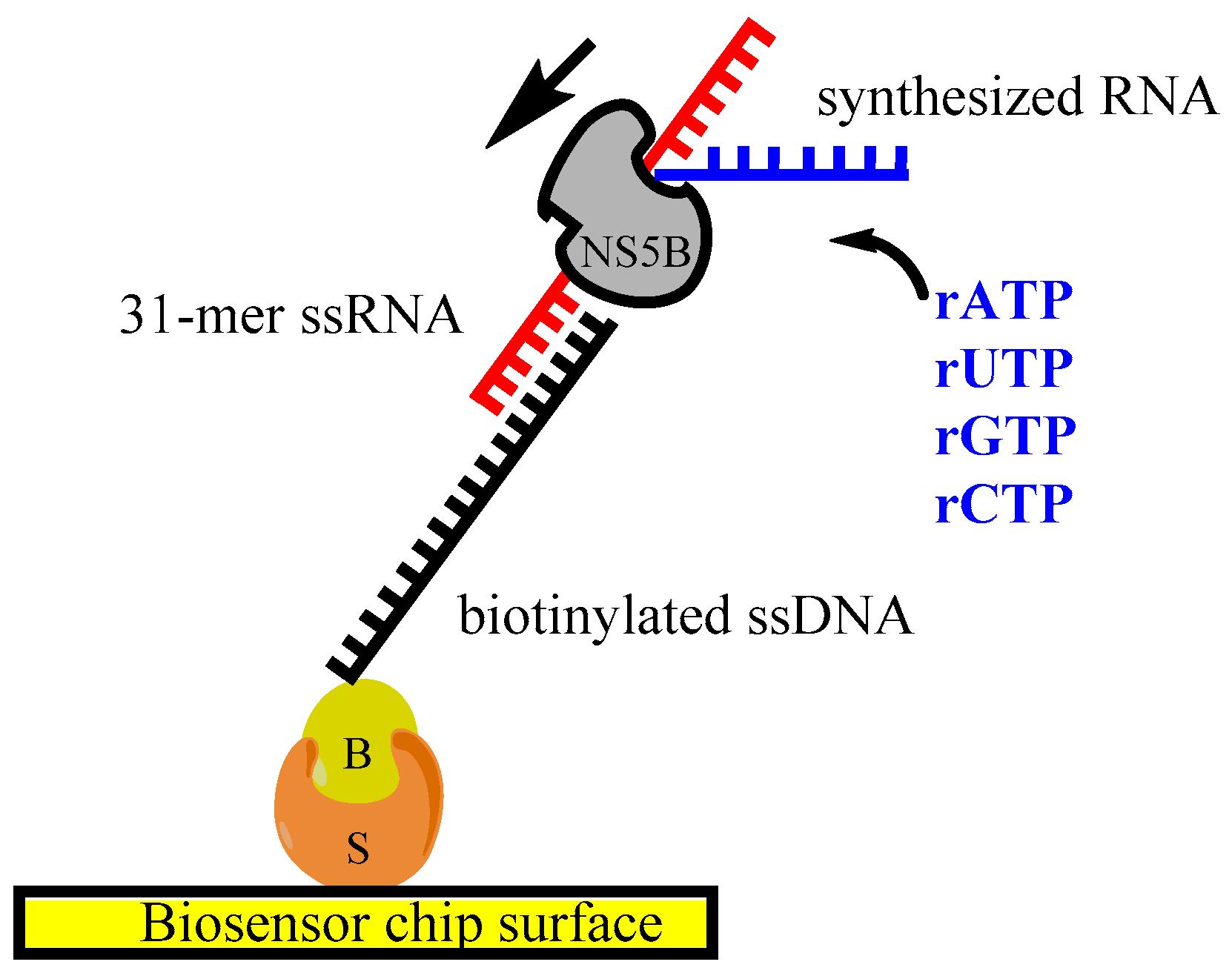
2.4. Biosensor Analysis of Inhibitor–NS5B Interactions
2.5. Analysis of RNA–NS5B Interactions
2.6. In Vitro RNA Synthesis
2.7. Polymerase Activity Assay
2.8. Sequence and Structure Analysis
3. Results
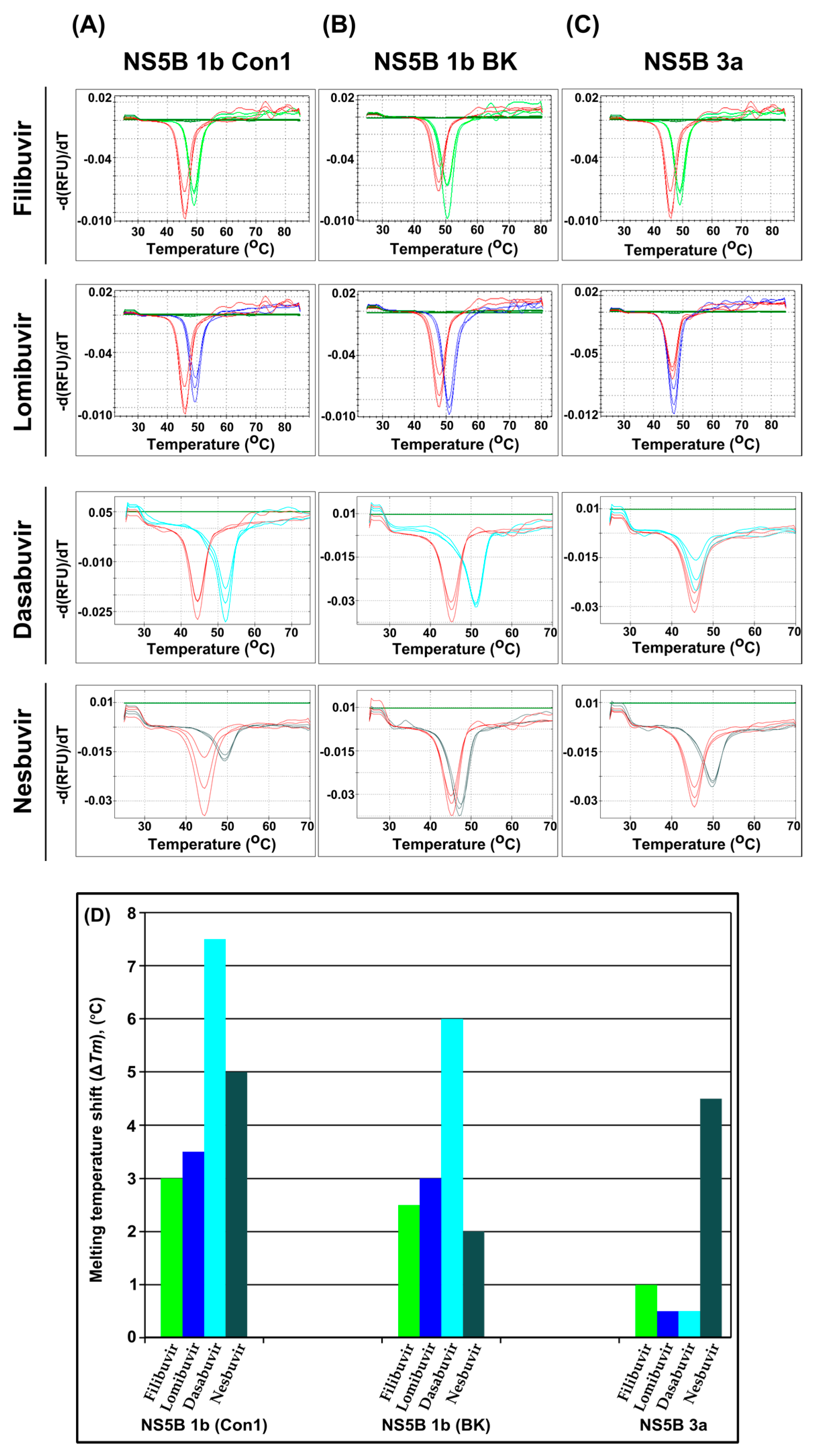
3.1. Stabilization of the Thermal Unfolding of NS5B by Allosteric Inhibitors
3.2. Kinetic Analysis of Interactions between Inhibitors and NS5B
3.3. Effects of Allosteric Inhibitors on Interactions between NS5B and RNA
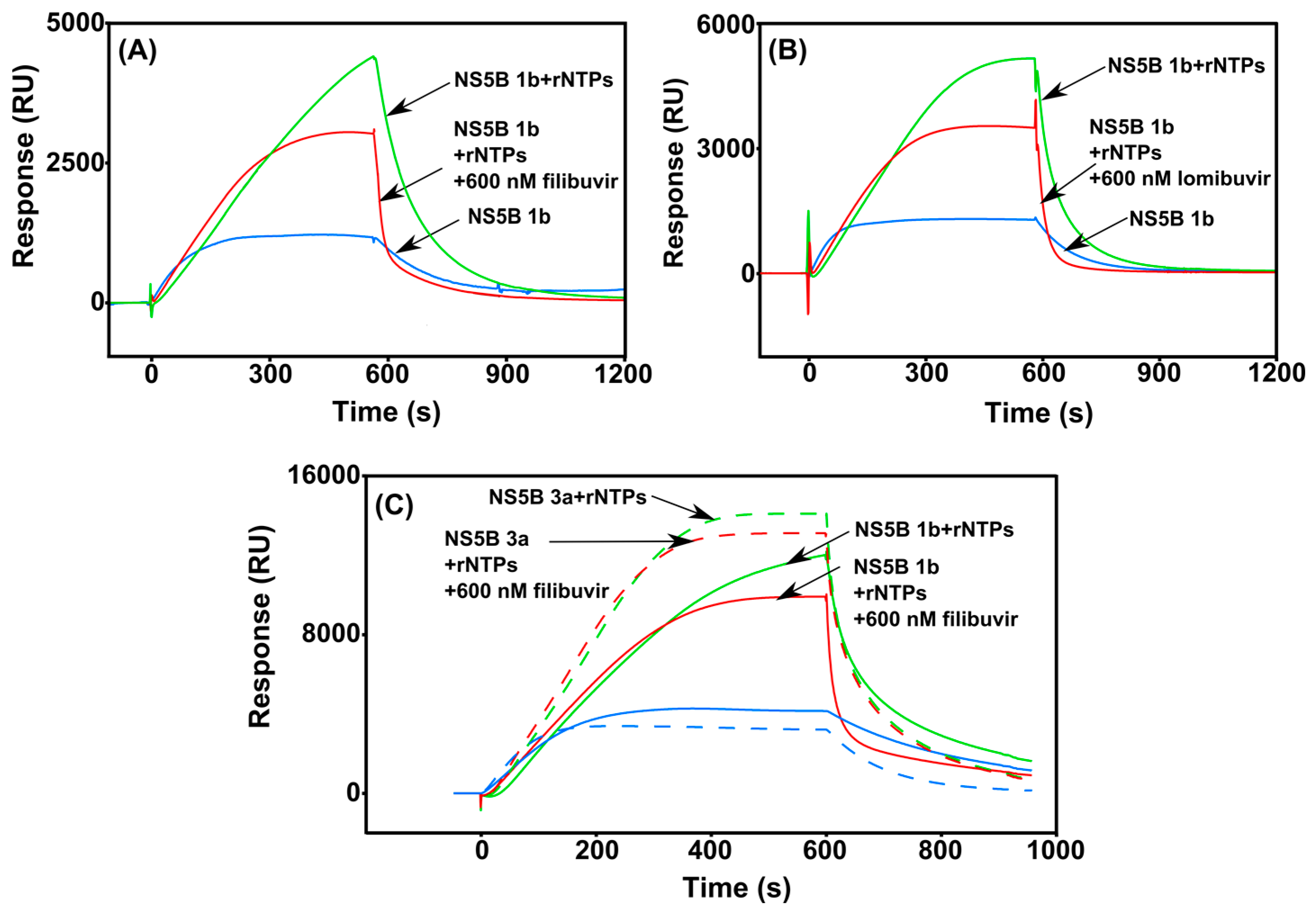
3.4. Real-Time Monitoring of the RNA Polymerization by HCV NS5B
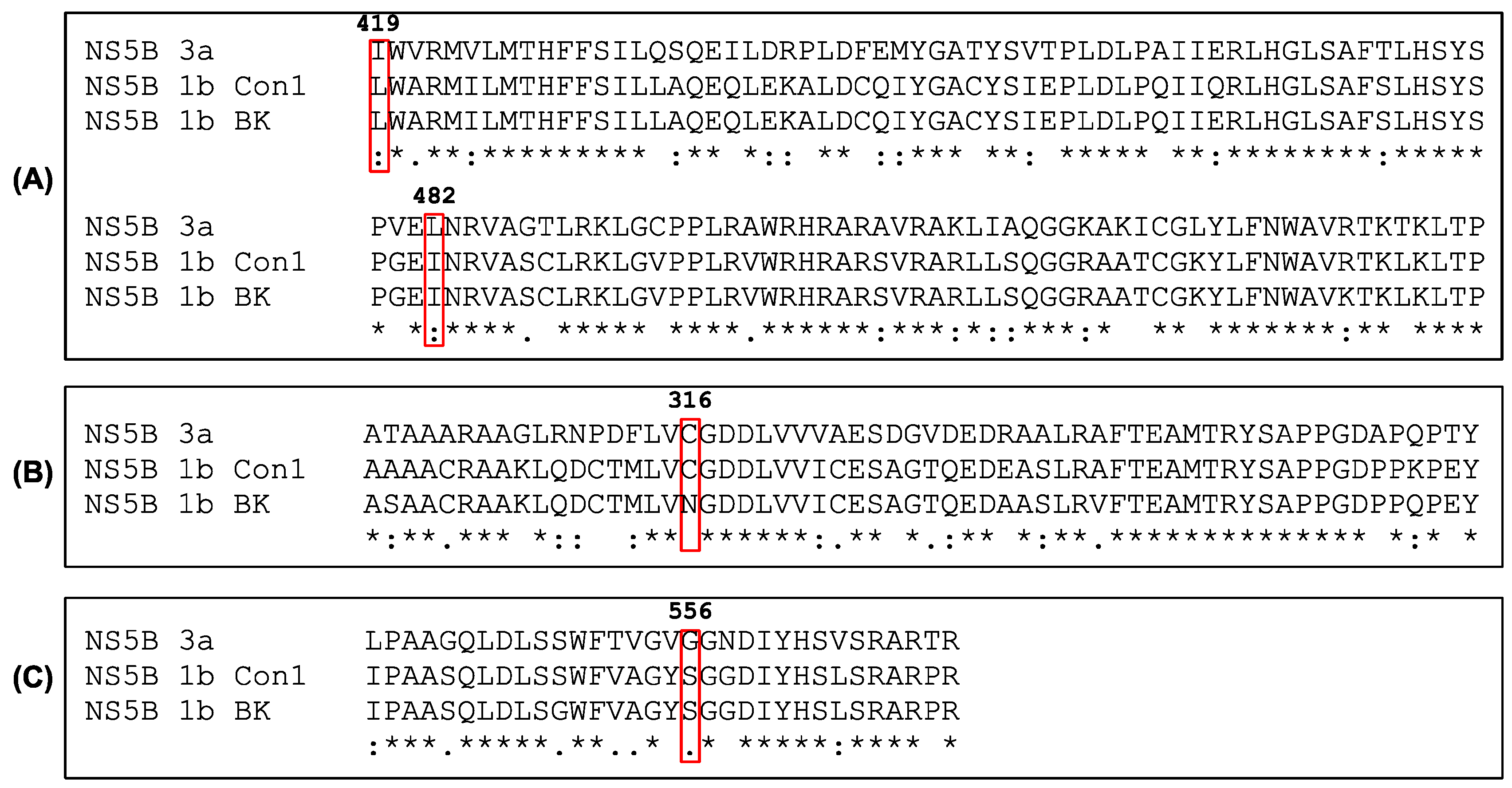
3.5. Sequence Comparison and Structure Analysis
4. Discussion
4.1. Binding
4.2. Effect on RNA-Binding Only or Catalytic Mechanism?
4.2.1. Filibuvir and Lomibuvir
4.2.2. Dasabuvir and Nesbuvir
4.3. Inhibition
4.3.1. Assay Development
4.3.2. Application of the Assay for Inhibition Studies
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization. Hepatitis C, Fact sheet N°164. Available online: http://www.who.int/mediacentre/factsheets/fs164/en/ (accessed on 26 September 2015).
- Messina, J.P.; Humphreys, I.; Flaxman, A.; Brown, A.; Cooke, G.S.; Pybus, O.G.; Barnes, E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015, 61, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Aziz, H.; Raza, A.; Murtaza, S.; Waheed, Y.; Khalid, A.; Irfan, J.; Samra, Z.; Athar, M.A. Molecular epidemiology of hepatitis C virus genotypes in different geographical regions of Punjab Province in Pakistan and a phylogenetic analysis. Int. J. Infect. Dis. 2013, 17, e247–e253. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.L. Hepatitis C virus genotypes and quasispecies. Am. J. Med. 1999, 107, 21–26. [Google Scholar] [CrossRef]
- Bochud, P.-Y.; Cai, T.; Overbeck, K.; Bochud, M.; Dufour, J.-F.; Müllhaupt, B.; Borovicka, J.; Heim, M.; Moradpour, D.; Cerny, A.; et al. Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J. Hepatol. 2009, 51, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, F.; Kramer, J.R.; Ilyas, J.; Duan, Z.; El-Serag, H.B. HCV genotype 3 is associated with an increased risk of cirrhosis and hepatocellular cancer in a national sample of U.S. Veterans with HCV. Hepatology 2014, 60, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Kottilil, S.; Wilson, E. Sofosbuvir/velpatasvir: A pangenotypic drug to simplify HCV therapy. Hepatol. Int. 2017, 11, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Bertino, G.; Ardiri, A.; Proiti, M.; Rigano, G.; Frazzetto, E.; Demma, S.; Ruggeri, M.I.; Scuderi, L.; Malaguarnera, G.; Bertino, N.; et al. Chronic hepatitis C: This and the new era of treatment. World J. Hepatol. 2016, 8, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Sanford, M. Simeprevir: A Review of its use in patients with chronic Hepatitis C virus infection. Drugs 2015, 75, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Kanda, T.; Haga, Y.; Sasaki, R.; Wu, S.; Nakamoto, S.; Yasui, S.; Arai, M.; Imazeki, F.; Yokosuka, O. Sofosbuvir treatment and hepatitis C virus infection. World J. Hepatol. 2016, 8, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Sofia, M.J.; Bao, D.; Chang, W.; Du, J.; Nagarathnam, D.; Rachakonda, S.; Reddy, P.G.; Ross, B.S.; Wang, P.; Zhang, H.-R.; et al. Discovery of a β-d-2′-Deoxy-2′-α-fluoro-2′-β-C-methyluridine Nucleotide Prodrug (PSI-7977) for the Treatment of Hepatitis C Virus. J. Med. Chem. 2010, 53, 7202–7218. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lim, B.H.; Jiang, W.W.; Flentge, C.A.; Hutchinson, D.K.; Madigan, D.L.; Randolph, J.T.; Wagner, R.; Maring, C.J.; Kati, W.M.; et al. Identification of aryl dihydrouracil derivatives as palm initiation site inhibitors of HCV NS5B polymerase. Bioorgan. Med. Chem. Lett. 2012, 22, 3747–3750. [Google Scholar] [CrossRef] [PubMed]
- Kati, W.; Koev, G.; Irvin, M.; Beyer, J.; Liu, Y.; Krishnan, P.; Reisch, T.; Mondal, R.; Wagner, R.; Molla, A.; et al. In Vitro Activity and Resistance Profile of Dasabuvir, a Nonnucleoside Hepatitis C Virus Polymerase Inhibitor. Antimicrob. Agents Chemother. 2015, 59, 1505–1511. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.M.; Espiritu, C.; Bansal, S.; Micolochick Steuer, H.M.; Niu, C.; Zennou, V.; Keilman, M.; Zhu, Y.; Lan, S.; Otto, M.J.; et al. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of Hepatitis C virus. Antimicrob. Agents Chemother. 2012, 56, 3359–3368. [Google Scholar] [CrossRef] [PubMed]
- Mendizabal, M.; Reddy, K.R. Chronic Hepatitis C and chronic kidney disease: Advances, limitations and unchartered territories. J. Viral Hepat. 2017. [Google Scholar] [CrossRef] [PubMed]
- Moradpour, D.; Penin, F.; Rice, C.M. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007, 5, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Ago, H.; Adachi, T.; Yoshida, A.; Yamamoto, M.; Habuka, N.; Yatsunami, K.; Miyano, M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 1999, 7, 1417–1426. [Google Scholar] [CrossRef]
- Wang, Q.M.; Hockman, M.A.; Staschke, K.; Johnson, R.B.; Case, K.A.; Lu, J.; Parsons, S.; Zhang, F.; Rathnachalam, R.; Kirkegaard, K.; et al. Oligomerization and cooperative RNA synthesis activity of Hepatitis C virus RNA-dependent RNA Polymerase. J. Virol. 2002, 76, 3865–3872. [Google Scholar] [CrossRef] [PubMed]
- Le Pogam, S.; Seshaadri, A.; Kosaka, A.; Chiu, S.; Kang, H.; Hu, S.; Rajyaguru, S.; Symons, J.; Cammack, N.; Nájera, I. Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. J. Antimicrob. Chemother. 2008, 61, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Khungar, V.; Han, S.-H. A Systematic Review of Side Effects of Nucleoside and Nucleotide Drugs Used for Treatment of Chronic Hepatitis B. Curr. Hepat. Rep. 2010, 9, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Gentile, I.; Buonomo, A.R.; Zappulo, E.; Borgia, G. Discontinued drugs in 2012–2013: Hepatitis C virus infection. Expert Opin. Investig. Drugs 2014, 24, 239–251. [Google Scholar] [CrossRef] [PubMed]
- TREK Therapeutics. TREK Therapeutics News. Available online: http://trektx.com/news/ (accessed on 18 September 2016).
- U.S. Food and Drug Administration. FDA Approves Viekira Pak to Treat Hepatitis C. 2014. Available online: http:// www.idsociety.org/FDA_20141219/ (accessed on 3 November 2015).
- Deredge, D.; Li, J.; Johnson, K.A.; Wintrode, P.L. Hydrogen/Deuterium Exchange Kinetics Demonstrate Long Range Allosteric Effects of Thumb Site 2 Inhibitors of Hepatitis C Viral RNA-dependent RNA Polymerase. J. Biol. Chem. 2016. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tatlock, J.; Linton, A.; Gonzalez, J.; Jewell, T.; Patel, L.; Ludlum, S.; Drowns, M.; Rahavendran, S.V.; Skor, H.; et al. Discovery of (R)-6-Cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl)-4-hydroxy-5,6-dihydropyran-2-one (PF-00868554) as a potent and orally available Hepatitis C virus polymerase inhibitor. J. Med. Chem. 2009, 52, 1255–1258. [Google Scholar] [CrossRef] [PubMed]
- Hang, J.Q.; Yang, Y.; Harris, S.F.; Leveque, V.; Whittington, H.J.; Rajyaguru, S.; Ao-Ieong, G.; McCown, M.F.; Wong, A.; Giannetti, A.M.; et al. Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of Hepatitis C virus. J. Biol. Chem. 2009, 284, 15517–15529. [Google Scholar] [CrossRef] [PubMed]
- Wagner, F.; Thompson, R.; Kantaridis, C.; Simpson, P.; Troke, P.J.F.; Jagannatha, S.; Neelakantan, S.; Purohit, V.S.; Hammond, J.L. Antiviral activity of the hepatitis C virus polymerase inhibitor filibuvir in genotype 1–infected patients. Hepatology 2011, 54, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.; Deval, J.; Fan, B.; Cai, H.; Soulard, C.; Ranjith-Kumar, C.T.; Smith, D.B.; Blatt, L.; Beigelman, L.; Kao, C.C. Biochemical study of the comparative inhibition of Hepatitis C virus RNA polymerase by VX-222 and Filibuvir. Antimicrob. Agents Chemother. 2012, 56, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Eltahla, A.A.; Lackovic, K.; Marquis, C.; Eden, J.-S.; White, P.A. A fluorescence-based high-throughput screen to identify small compound inhibitors of the genotype 3a Hepatitis C virus RNA polymerase. J. Biomol. Screening 2013. [Google Scholar] [CrossRef] [PubMed]
- Winquist, J.; Abdurakhmanov, E.; Baraznenok, V.; Henderson, I.; Vrang, L.; Danielson, U.H. Resolution of the interaction mechanisms and characteristics of non-nucleoside inhibitors of hepatitis C virus polymerase. Antivir. Res. 2013, 97, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Ehrenberg, A.E.; Schmuck, B.; Anwar, M.I.; Gustafsson, S.S.; Stenberg, G.; Danielson, U.H. Accounting for strain variations and resistance mutations in the characterization of hepatitis C NS3 protease inhibitors. J. Enzym. Inhib. Med. Chem. 2014, 29, 868–876. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Källberg, M.; Wang, H.; Wang, S.; Peng, J.; Wang, Z.; Lu, H.; Xu, J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012, 7, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Caillet-Saguy, C.; Simister, P.C.; Bressanelli, S. An objective assessment of conformational variability in complexes of Hepatitis C virus polymerase with non-nucleoside inhibitors. J. Mol. Biol. 2011, 414, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Buckle, M.; Williams, R.M.; Negroni, M.; Buc, H. Real time measurements of elongation by a reverse transcriptase using surface plasmon resonance. Proc. Natl. Acad. Sci. USA 1996, 93, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Howe, A.Y.M.; Cheng, H.; Johann, S.; Mullen, S.; Chunduru, S.K.; Young, D.C.; Bard, J.; Chopra, R.; Krishnamurthy, G.; Mansour, T.; et al. Molecular mechanism of Hepatitis C virus replicon variants with reduced susceptibility to a Benzofuran inhibitor, HCV-796. Antimicrob. Agents Chemother. 2008, 52, 3327–3338. [Google Scholar] [CrossRef] [PubMed]
- Le Pogam, S.; Kang, H.; Harris, S.F.; Leveque, V.; Giannetti, A.M.; Ali, S.; Jiang, W.-R.; Rajyaguru, S.; Tavares, G.; Oshiro, C.; et al. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the Hepatitis C virus. J. Virol. 2006, 80, 6146–6154. [Google Scholar] [CrossRef] [PubMed]
- Barreca, M.L.; Iraci, N.; Manfroni, G.; Cecchetti, V. Allosteric inhibition of the hepatitis C virus NS5B polymerase: In silico strategies for drug discovery and development. Future Med. Chem. 2011, 3, 1027–1055. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, M.; Eng, S.; Leavitt, S.A.; Lee, T.-J.; Mabery, E.M.; Tian, Y.; Byun, D.; Canales, E.; Clarke, M.O.; Doerffler, E.; et al. Preclinical characterization of GS-9669, a Thumb Site II Inhibitor of the Hepatitis C Virus NS5B Polymerase. Antimicrob. Agents Chemother. 2012, 57, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Boyce, S.E.; Tirunagari, N.; Niedziela-Majka, A.; Perry, J.; Wong, M.; Kan, E.; Lagpacan, L.; Barauskas, O.; Hung, M.; Fenaux, M.; et al. Structural and regulatory elements of HCV NS5B polymerase—β-Loop and C-Terminal tail—Are required for activity of allosteric thumb site II inhibitors. PLoS ONE 2014, 9, e84808. [Google Scholar] [CrossRef] [PubMed]
- McCown, M.F.; Rajyaguru, S.; Kular, S.; Cammack, N.; Nájera, I. GT-1a or GT-1b Subtype-specific resistance profiles for Hepatitis C virus inhibitors telaprevir and HCV-796. Antimicrob. Agents Chemother. 2009, 53, 2129–2132. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Gerick, F.; Thornton, J.M. The structural basis of allosteric regulation in proteins. FEBS Lett. 2009, 583, 1692–1698. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.C.; Thorpe, I.F. Thumb inhibitor binding eliminates functionally important dynamics in the hepatitis C virus RNA polymerase. Proteins Struct. Funct. Bioinf. 2013, 81, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ng, K.K.-S.; Cherney, M.M.; Chan, L.; Yannopoulos, C.G.; Bedard, J.; Morin, N.; Nguyen-Ba, N.; Alaoui-Ismaili, M.H.; Bethell, R.C.; et al. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase: Crystal structures and mechanism of inhibition. J. Biol. Chem. 2003, 278, 9489–9495. [Google Scholar] [CrossRef] [PubMed]
- Reich, S.; Golbik, R.P.; Geissler, R.; Lilie, H.; Behrens, S.-E. Mechanisms of activity and inhibition of the Hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 2010, 285, 13685–13693. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, V.; Roos, A.; Körner, F.; Koch, J.O.; Bartenschlager, R. Biochemical and kinetic analyses of NS5B RNA-Dependent RNA polymerase of the Hepatitis C virus. Virology 1998, 249, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, E.; He, Z.; Palermo, R.E.; Huang, H.-C. Hepatitis C Virus NS5B Polymerase exhibits distinct nucleotide requirements for initiation and elongation. J. Biol. Chem. 2008, 283, 33893–33901. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Hamatake, R.K.; Mathis, D.M.; Racela, J.; Rigat, K.L.; Lemm, J.; Colonno, R.J. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of Hepatitis C virus. J. Virol. 2000, 74, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Sheets, M.P.; Wells, P.A.; Shelly, J.A.; Poorman, R.A.; Epps, D.E.; Morgan, A.G. Continuous-read assay for the detection of de Novo HCV RNA Polymerase activity. US Patent 20,040,209,283, 21 October 2004. [Google Scholar]
- Hwang, S.B.; Park, K.-J.; Kim, Y.-S.; Sung, Y.C.; Lai, M.M.C. Hepatitis C Virus NS5B Protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology 1997, 227, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Kaneko, S.; Shirota, Y.; Qin, W.; Nomura, T.; Kobayashi, K.; Murakami, S. RNA-dependent RNA Polymerase activity of the soluble recombinant Hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 1998, 273, 15479–15486. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, V.; Körner, F.; Herian, U.; Bartenschlager, R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 1997, 71, 8416–8428. [Google Scholar] [PubMed]
- Ferrari, E.; Wright-Minogue, J.; Fang, J.W.S.; Baroudy, B.M.; Lau, J.Y.N.; Hong, Z. Characterization of soluble Hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 1999, 73, 1649–1654. [Google Scholar] [PubMed]
- Shi, S.T.; Herlihy, K.J.; Graham, J.P.; Nonomiya, J.; Rahavendran, S.V.; Skor, H.; Irvine, R.; Binford, S.; Tatlock, J.; Li, H.; et al. Preclinical characterization of PF-00868554, a potent nonnucleoside inhibitor of the Hepatitis C virus RNA-Dependent RNA polymerase. Antimicrob. Agents Chemother. 2009, 53, 2544–2552. [Google Scholar] [CrossRef] [PubMed]
- Bartenschlager, R.; Lohmann, V.; Penin, F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 2013, 11, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Biswal, B.K.; Cherney, M.M.; Wang, M.; Chan, L.; Yannopoulos, C.G.; Bilimoria, D.; Nicolas, O.; Bedard, J.; James, M.N.G. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of Hepatitis C Virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 2005, 280, 18202–18210. [Google Scholar] [CrossRef] [PubMed]
- Appleby, T.C.; Perry, J.K.; Murakami, E.; Barauskas, O.; Feng, J.; Cho, A.; Fox, D.; Wetmore, D.R.; McGrath, M.E.; Ray, A.S.; et al. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 2015, 347, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Karam, P.; Powdrill, M.H.; Liu, H.-W.; Vasquez, C.; Mah, W.; Bernatchez, J.; Götte, M.; Cosa, G. Dynamics of HCV RNA-dependent RNA Polymerase NS5B in Complex with RNA. J. Biol. Chem. 2014. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Johnson, K.A. Thumb site 2 inhibitors of Hepatitis C viral RNA-dependent RNA polymerase allosterically block the transition from initiation to elongation. J. Biol. Chem. 2016. [Google Scholar] [CrossRef]
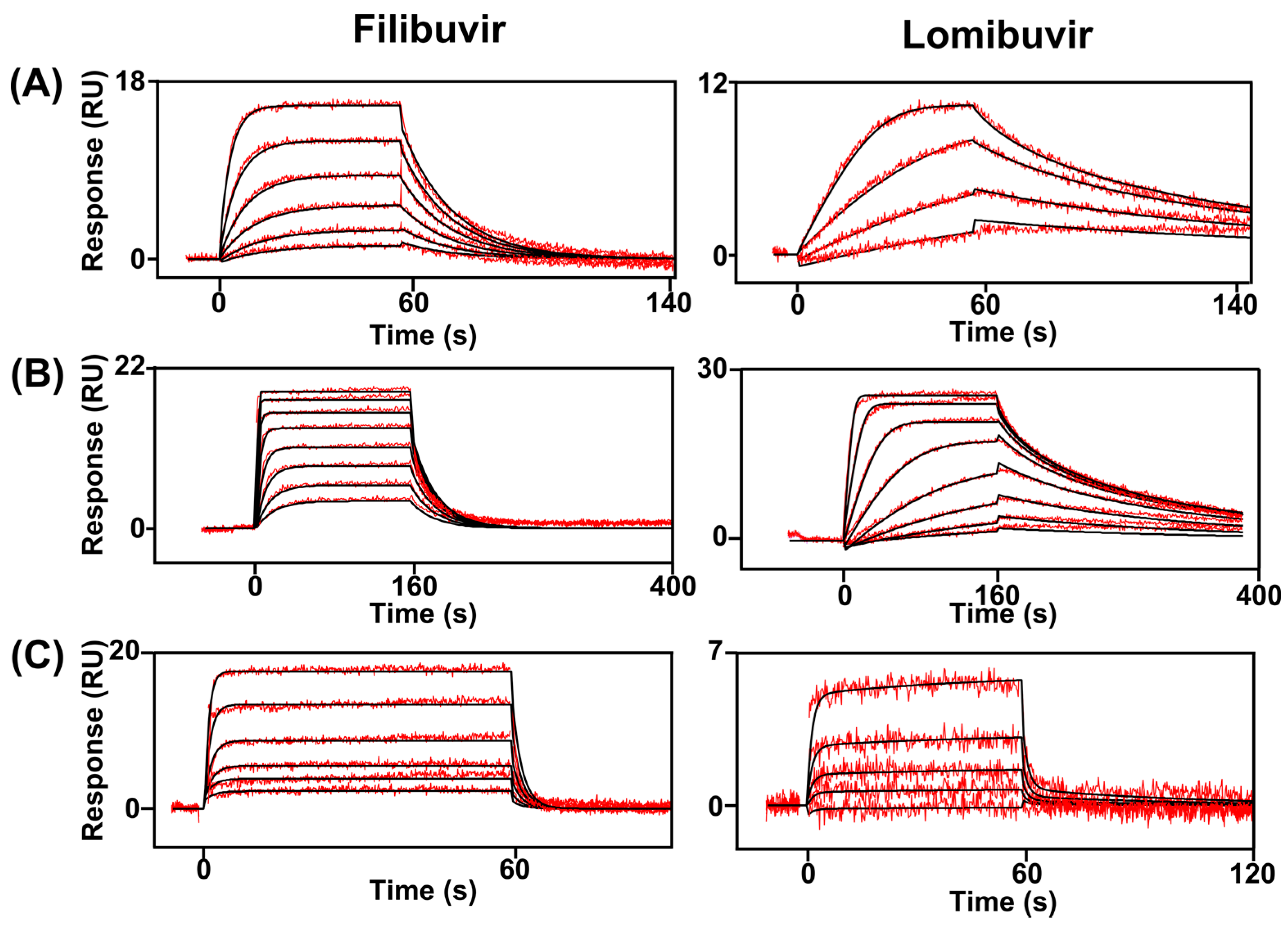

 ) and in complex with filibuvir (
) and in complex with filibuvir (  ), lomibuvir (
), lomibuvir (  ), dasabuvir (
), dasabuvir (  ), and nesbuvir (
), and nesbuvir (  ). The corresponding equilibrium dissociation constants (KD) determined by a 1:1 steady-state affinity model are presented in Table 4.
). The corresponding equilibrium dissociation constants (KD) determined by a 1:1 steady-state affinity model are presented in Table 4.
 ) and in complex with filibuvir (
) and in complex with filibuvir (  ), lomibuvir (
), lomibuvir (  ), dasabuvir (
), dasabuvir (  ), and nesbuvir (
), and nesbuvir (  ). The corresponding equilibrium dissociation constants (KD) determined by a 1:1 steady-state affinity model are presented in Table 4.
). The corresponding equilibrium dissociation constants (KD) determined by a 1:1 steady-state affinity model are presented in Table 4.
| Inhibitor | NS5B | 1b Con1 | 1b BK | 3a | |||
|---|---|---|---|---|---|---|---|
| (°C) | Tm | ΔTm | Tm | ΔTm | Tm | ΔTm | |
| - a | 46.0 (44.5) b | 48.0 (45.0) b | 46.5 (45.5) b | ||||
| Filibuvir | 49.0 | 3 | 50.5 | 2.5 | 47.5 | 1 | |
| Lomibuvir | 49.5 | 3.5 | 51.0 | 3 | 47.0 | 0.5 | |
| Dasabuvir | 52.0 | 7.5 | 51.0 | 6 | 46.0 | 0.5 | |
| Nesbuvir | 49.5 | 5 | 47.0 | 2 | 50.0 | 4.5 | |
| Filibuvir | Lomibuvir | |||||
|---|---|---|---|---|---|---|
| 1b Con1 (±SD) d | 1b BK (±SD) | 3a (±SD) | 1b Con1 (±SD) | 1b BK (±SD) | 3a (±SD) | |
| KD (nM) b | 55 ± 11 (30 ± 6) | 54 ± 1.4 (30 ± 11) | 545 ± 7 (800 ± 80) | Nd a (19 ± 7) | Nd a (20 ± 0.4) | >2000 (2600) |
| ka (M−1s−1) | 2.6 × 106 ± 1.1 × 106 | 1.84 × 106 ± 0.22 × 106 | 7.0 × 105 ± 0.2 × 105 | 2.1 × 106 ± 1.1 × 106 | 1.8 × 106 ± 0.22 × 106 | 1.75 × 105 ± 1.05 × 105 |
| kd (s−1) | 0.07 ± 0.02 | 0.03 ± 0.008 | 0.57 ± 0.06 | 0.04 ± 0.03 | 0.03 ± 0.007 | 0.45 ± 0.21 |
| k2 (s−1) | N/A c | N/A | N/A | 7.7 × 10−6 ± 3.1 × 10−6 | 2.6 × 10−6 ± 0.9 × 10−6 | 0.048 ± 0.06 |
| k-2 (s−1) | N/A | N/A | N/A | 0.001 ± 7 × 10−5 | 0.002 ± 0.0007 | 0.025 ± 0.007 |
| χ2 | 0.49 ± 0.19 | 0.5 ± 0.2 | 0.16 ± 0.13 | 0.22 ± 0.2 | 0.66 ± 0.33 | 0.09 ± 0.02 |
| Dasabuvir | |
|---|---|
| NS5B 1b Con1 (±SD) a | |
| ka1 (M−1 s−1) | 1.5 × 104 ± 0.3 × 104 |
| kd1 (s−1) | 3.3 × 10−7 ± 0.2 × 10−7 |
| KD1 (M) | 2.3 × 10−11 ± 0.6 × 10−11 |
| ka2 (M−1 s−1) | 8.6 × 105 ± 1.1 × 106 |
| kd2 (s−1) | 5.1 × 10−3 ± 2.6 × 10−3 |
| KD2 (M) | 3.6 × 10−8 ± 0.9 × 10−8 |
| χ2 | 0.4 ± 0.4 |
| NS5 1b KD (nM) ± SE | Bmax (RU) | NS5B 3a KD (nM) ± SE | Bmax (RU) | |
|---|---|---|---|---|
| NS5B | 390 ± 149 | 8473 | 266 ± 49 | 7003 |
| NS5B + Filibuvir | 950 ± 446 | 5693 | 947 ± 319 | 8158 |
| NS5B + Lomibuvir | 3350 ± 4346 | 17,715 | 553 ± 185 | 8418 |
| NS5B + Dasabuvir | 430 ± 200 | 8340 | 407 ± 127 | 7364 |
| NS5B + Nesbuvir | 530 ± 243 | 8957 | 411 ± 134 | 7107 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdurakhmanov, E.; Øie Solbak, S.; Danielson, U.H. Biophysical Mode-of-Action and Selectivity Analysis of Allosteric Inhibitors of Hepatitis C Virus (HCV) Polymerase. Viruses 2017, 9, 151. https://doi.org/10.3390/v9060151
Abdurakhmanov E, Øie Solbak S, Danielson UH. Biophysical Mode-of-Action and Selectivity Analysis of Allosteric Inhibitors of Hepatitis C Virus (HCV) Polymerase. Viruses. 2017; 9(6):151. https://doi.org/10.3390/v9060151
Chicago/Turabian StyleAbdurakhmanov, Eldar, Sara Øie Solbak, and U. Helena Danielson. 2017. "Biophysical Mode-of-Action and Selectivity Analysis of Allosteric Inhibitors of Hepatitis C Virus (HCV) Polymerase" Viruses 9, no. 6: 151. https://doi.org/10.3390/v9060151






