An Insight into Cotton Leaf Curl Multan Betasatellite, the Most Important Component of Cotton Leaf Curl Disease Complex
Abstract
:1. Introduction−Begomoviruses and Betasatellites
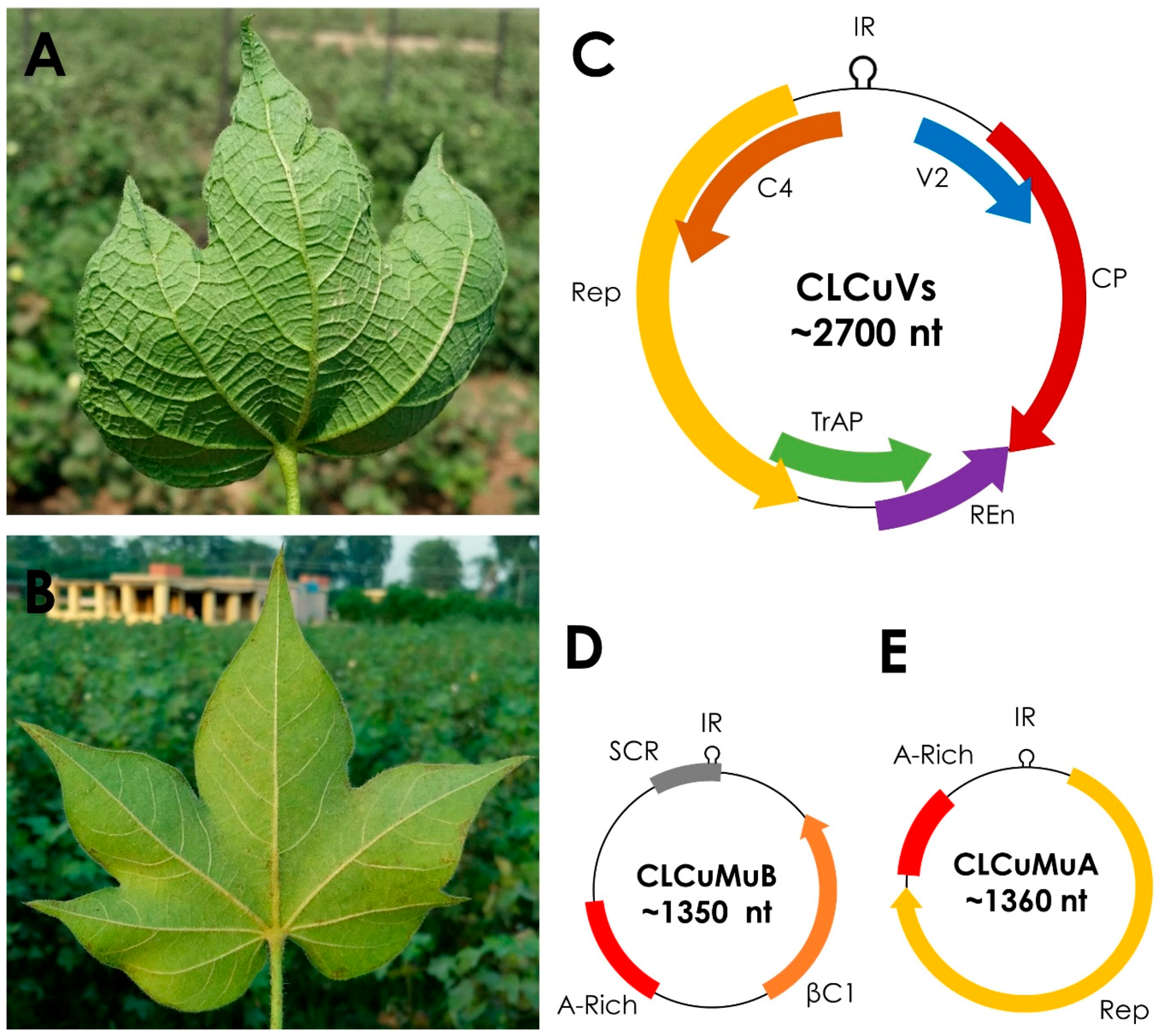
2. Cotton Leaf Curl Virus Disease
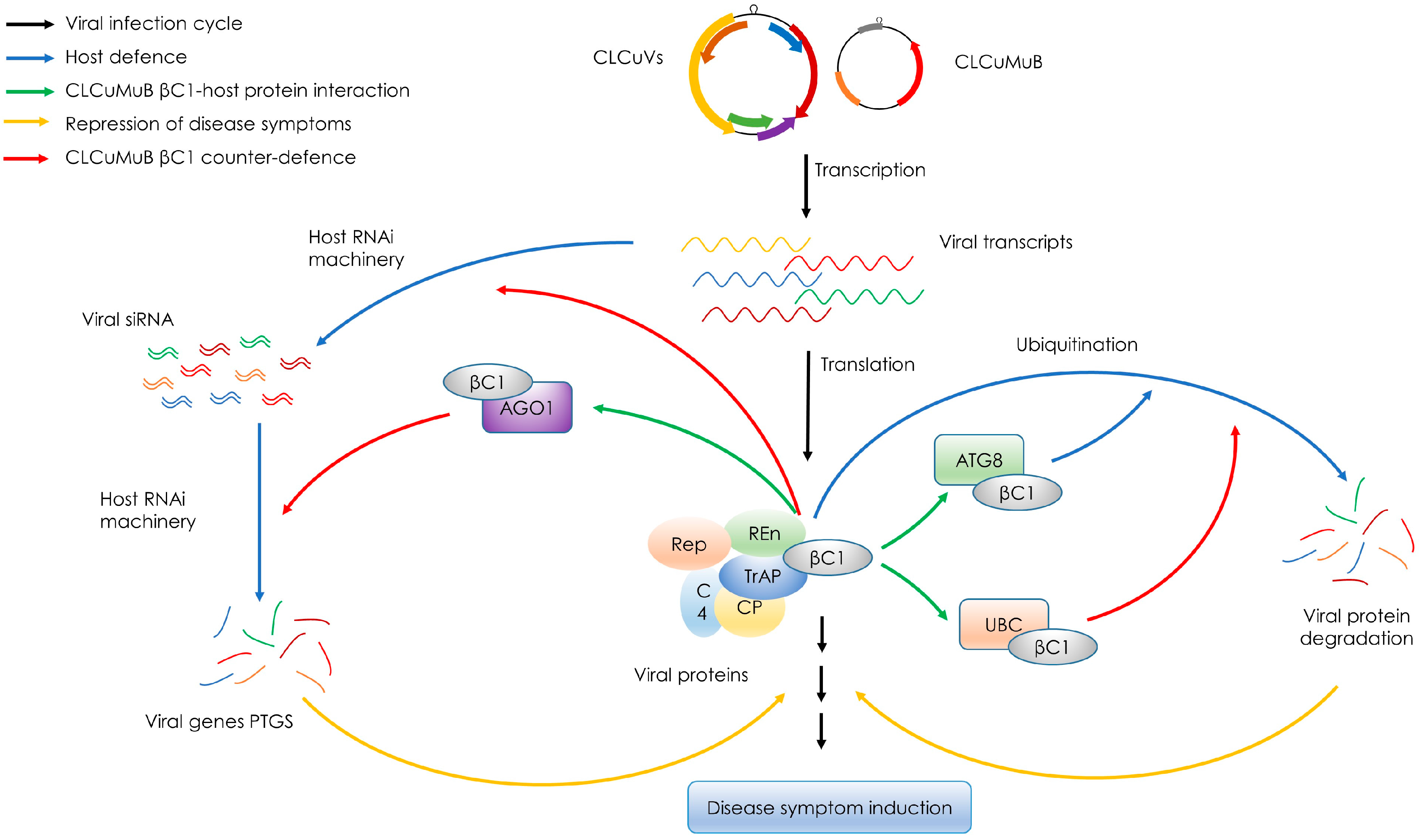
3. Functions of βC1 and Interaction with Host Proteins
4. Trans-Replication of Cotton Leaf Curl Multan Betasatellite and Its Association with Bipartite Tomato Leaf Curl New Delhi Virus
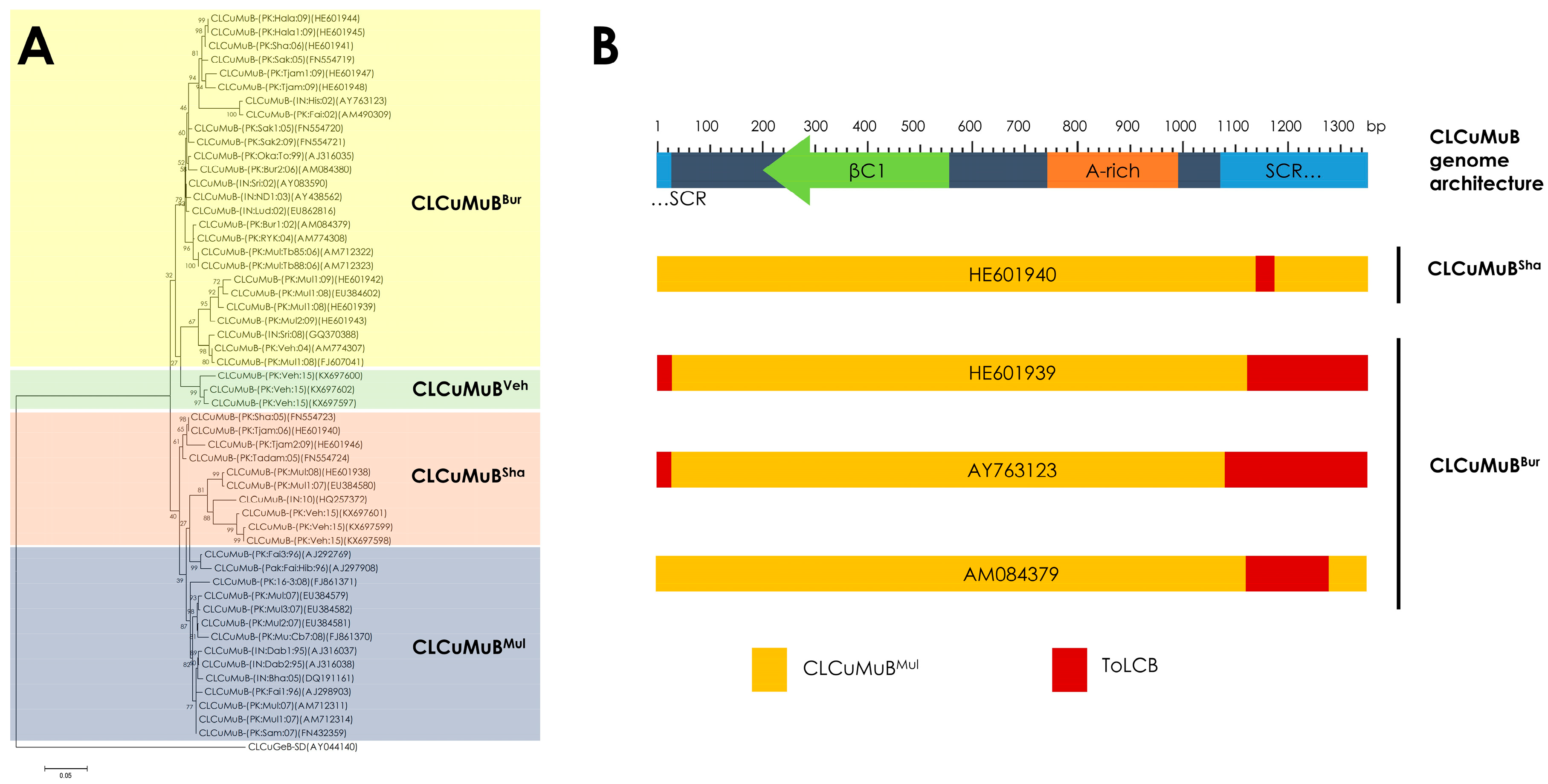
5. Recombination and Phylogeny of Cotton Leaf Curl Multan Betasatellite
6. Conclusions and Future Prospects
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Attar, M.N.; Kvarnheden, A.; Saeed, M.; Briddon, R.W. Cotton leaf curl disease-an emerging threat to cotton production worldwide. J. Gen. Virol. 2013, 94, 695–710. [Google Scholar]
- Sattar, M.N.; Iqbal, Z.; Tahir, M.N.; Ullah, S. The prediction of a new CLCuD epidemic in the Old World. Front. Microbiol. 2017, 8, 631. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Khan, S.H.; Bashir, A.; Saeed, M.; Zafar, Y.; Malik, K.A.; Briddon, R.; Stanley, J.; Markham, P.G. Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology 1999, 259, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Shafiq, M.; Amin, I.; Scheffler, B.E.; Scheffler, J.A.; Briddon, R.W.; Mansoor, S. Frequent occurrence of Tomato leaf curl New Delhi virus in cotton leaf curl disease affected cotton in Pakistan. PLoS ONE 2016, 11, e0155520. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; ICTV Report Consortium. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.W.; Rivera-Bustamante, R.; Murilo Zerbini, F.; Martin, D.P. Capulavirus and Grablovirus: Two New Genera in the Family Geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Bedford, I.D.; Briddon, R.W.; Brown, J.K.; Rosell, R.C.; Markham, P.G. Geminivirus transmission and biological characterisation of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Ann. Appl. Biol. 1994, 125, 311–325. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Padidam, M.; Beachy, R.N.; Fauquet, C.M. Classification and identification of geminiviruses using sequence comparisons. J. Gen. Virol. 1995, 76, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Nawaz-ul-Rehman, M.S.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Maintenance of an Old World betasatellite by a New World helper begomovirus and possible rapid adaptation of the betasatellite. J. Virol. 2009, 83, 9347–9355. [Google Scholar] [CrossRef] [PubMed]
- Mahatma, L.; Mahatma, M.K.; Pandya, J.R.; Solanki, R.K.; Solanki, V.A. Epidemiology of begomoviruses: A Global Perspective. In Plant Viruses: Evolution and Management; Gaur, R.K., Petrov, N.M., Eds.; Springer: Singapore, 2016; pp. 171–188. [Google Scholar]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.; Fiallo-Olive, E.; Briddon, R.W.; Hernandez-Zepeda, C.; Idris, A.; et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef] [PubMed]
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Heyraud, F.; Matzeit, V.; Schaefer, S.; Schell, J.; Gronenborn, B. The conserved nonanucleotide motif of the geminivirus stem-loop sequence promotes replicational release of virus molecules from redundant copies. Biochimie 1993, 75, 605–615. [Google Scholar] [CrossRef]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olive, E.; Tovar, R.; Navas-Castillo, J. Deciphering the biology of deltasatellites from the New World: Maintenance by New World Begomoviruses and Whitefly Transmission. New Phytol. 2016, 212, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Lozano, G.; Trenado, H.P.; Fiallo-Olive, E.; Chirinos, D.; Geraud-Pouey, F.; Briddon, R.W.; Navas-Castillo, J. Characterization of non-coding DNA satellites associated with sweepoviruses (genus Begomovirus, Geminiviridae)-Definition of a distinct class of begomovirus-associated satellites. Front. Microbiol. 2016, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Behjatnia, S.A.; Mansoor, S.; Zafar, Y.; Hasnain, S.; Rezaian, M.A. A single complementary-sense transcript of a geminiviral DNA β satellite is determinant of pathogenicity. Mol. Plant Microbe. Interact. 2005, 18, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch. Virol. 2017, 162, 2505–2538. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Bedford, I.D.; Briddon, R.W.; Markham, P.G.; Wong, S.M.; Stanley, J. A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA 2000, 97, 6890–6895. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Zhou, X. Phloem specific promoter from a satellite associated with a DNA virus. Virus Res. 2006, 115, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Idris, A.M.; Mansoor, S.; Bedford, I.D.; Dhawan, P.; Rishi, N.; Siwatch, S.S.; Abdel-Salam, A.M.; et al. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology 2003, 312, 106–121. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Zafar, Y.; Briddon, R.W. Geminivirus disease complexes: the Threat is Spreading. Trends Plant Sci. 2006, 11, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Kirthi, N.; Priyadarshini, C.G.; Sharma, P.; Maiya, S.P.; Hemalatha, V.; Sivaraman, P.; Dhawan, P.; Rishi, N.; Savithri, H.S. Genetic variability of begomoviruses associated with cotton leaf curl disease originating from India. Arch. Virol. 2004, 149, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, Y.; Robinson, D.J.; Harrison, B.D. Four DNA-A variants among Pakistani isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J. Gen. Virol. 1998, 79, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Briddon, R.W.; Bull, S.E.; Bedford, I.D.; Bashir, A.; Hussain, M.; Saeed, M.; Zafar, Y.; Malik, K.A.; Fauquet, C.; et al. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA β. Arch. Virol. 2003, 148, 1969–1986. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Akbar, F.; Iqbal, Z.; Amrao, L.; Amin, I.; Saeed, M.; Mansoor, S. Effects of genetic changes to the begomovirus/betasatellite complex causing cotton leaf curl disease in South Asia post-resistance breaking. Virus Res. 2014, 186, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Markham, P.G. Cotton leaf curl virus disease. Virus Res. 2000, 71, 151–159. [Google Scholar] [CrossRef]
- Amrao, L.; Amin, I.; Shahid, M.S.; Briddon, R.W.; Mansoor, S. Cotton leaf curl disease in resistant cotton is associated with a single begomovirus that lacks an intact transcriptional activator protein. Virus Res. 2010, 152, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Mansoor, S.; Amrao, L.; Hussain, M.; Irum, S.; Zafar, Y.; Bull, S.E.; Briddon, R.W. Mobilisation into cotton and spread of a recombinant cotton leaf curl disease satellite. Arch. Virol. 2006, 151, 2055–2065. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, P.A.; Naik, A.; Katturi, P.; Kurulekar, M.; Kankanallu, R.S.; Anandalakshmi, R. Dominance of resistance-breaking Cotton leaf curl Burewala virus (CLCuBuV) in northwestern India. Arch. Virol. 2017, 157, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Saleem, H.; Nahid, N.; Shakir, S.; Ijaz, S.; Murtaza, G.; Khan, A.A.; Mubin, M.; Nawaz-Ul-Rehman, M.S. Diversity, mutation and recombination analysis of cotton leaf curl geminiviruses. PLoS ONE 2016, 11, e0151161. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Gunapati, S.; Alok, A.; Lalit, A.; Gadre, R.; Sharma, N.C.; Roy, J.K.; Singh, S.P. Cotton leaf curl Burewala virus with intact or mutant transcriptional activator proteins: Complexity of Cotton Leaf Curl Disease. Arch. Virol. 2015, 160, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Hassan, I.; Amin, I.; Mansoor, S.; Briddon, R.W. Further changes in the cotton leaf curl disease complex: An Indication of Things to Come? Virus Genes 2017. [CrossRef] [PubMed]
- Zubair, M.; Zaidi, S.S.; Shakir, S.; Farooq, M.; Amin, I.; Scheffler, J.A.; Scheffler, B.E.; Mansoor, S. Multiple begomoviruses found associated with cotton leaf curl disease in Pakistan in early 1990 are back in cultivated cotton. Sci. Rep. 2017, 7, 680. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.N.; Amin, I.; Briddon, R.W.; Mansoor, S. The merging of two dynasties-identification of an African cotton leaf curl disease-associated begomovirus with cotton in Pakistan. PLoS ONE 2011, 6, e20366. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.A.; Iqbal, Z.; Amin, I.; Mansoor, S. First report of Tomato leaf curl Gujarat virus, a bipartite begomovirus on cotton showing leaf curl symptoms in Pakistan. Plant Dis. 2015, 99, 1655. [Google Scholar] [CrossRef]
- Akhtar, S.; Tahir, M.N.; Baloch, G.R.; Javaid, S.; Khan, A.Q.; Amin, I.; Briddon, R.W.; Mansoor, S. Regional changes in the sequence of cotton leaf curl multan betasatellite. Viruses 2014, 6, 2186–2203. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S.; Saunders, K.; Stanley, J.; Zafar, Y.; Malik, K.A.; Markham, P.G. Identification of DNA components required for induction of cotton leaf curl disease. Virology 2001, 285, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Qazi, J.; Amin, I.; Mansoor, S.; Iqbal, M.J.; Briddon, R.W. Contribution of the satellite encoded gene βC1 to cotton leaf curl disease symptoms. Virus Res. 2007, 128, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Zafar, Y.; Randles, J.W.; Rezaian, M.A. A monopartite begomovirus-associated DNA β satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 2007, 88, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Tabein, S.; Behjatnia, S.A.; Anabestani, A.; Izadpanah, K. Whitefly-mediated transmission of cotton leaf curl Multan betasatellite: evidence for betasatellite encapsidation in coat protein of helper begomoviruses. Arch. Virol. 2013, 158, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Akbergenov, R.; Si-Ammour, A.; Blevins, T.; Amin, I.; Kutter, C.; Vanderschuren, H.; Zhang, P.; Gruissem, W.; Meins, F., Jr.; Hohn, T.; et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006, 34, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, P.; Vanitharani, R.; Pita, J.; Fauquet, C.M. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J. Virol. 2004, 78, 7465–7477. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Hussain, K.; Akbergenov, R.; Yadav, J.S.; Qazi, J.; Mansoor, S.; Hohn, T.; Fauquet, C.M.; Briddon, R.W. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant Microbe Interact. 2011, 24, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Dogra, S.C.; Dry, I.B.; Randles, J.W. Silencing suppressor activity of a begomovirus DNA β encoded protein and its effect on heterologous helper virus replication. Virus Res 2012, 167, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Eini, O. A betasatellite-encoded protein regulates key components of gene silencing system in plants. Mol. Biol. 2017, 51, 579–585. [Google Scholar] [CrossRef]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 2017, 6, e23897. [Google Scholar] [CrossRef] [PubMed]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Dogra, S.; Selth, L.A.; Dry, I.B.; Randles, J.W.; Rezaian, M.A. Interaction with a host ubiquitin-conjugating enzyme is required for the pathogenicity of a geminiviral DNA β satellite. Mol. Plant Microbe Interact. 2009, 22, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, N.; Sharma, P.K.; Malathi, V.G. Functional characterization of βC1 gene of Cotton leaf curl Multan betasatellite. Virus Genes 2013, 46, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 1999, 18, 71–106. [Google Scholar] [CrossRef]
- Alberter, B.; Ali Rezaian, M.; Jeske, H. Replicative intermediates of Tomato leaf curl virus and its satellite DNAs. Virology 2005, 331, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.L.; Fauquet, C.M. Differential interaction between cassava mosaic geminiviruses and geminivirus satellites. J. Gen. Virol. 2010, 91, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Hameed, U.; Zia-Ur-Rehman, M.; Herrmann, H.W.; Haider, M.S.; Brown, J.K. First report of Okra enation leaf curl virus and associated Cotton leaf curl Multan betasatellite and cotton leaf curl Multan alphasatellite Infecting cotton in Pakistan: A new member of the cotton leaf curl disease complex. Plant Dis. 2014, 98, 1447. [Google Scholar] [CrossRef]
- Singh, R.M.; Sharma, S.; Zaidi, A.A.; Hamid, A.; Hallan, V. Association of Bhendi yellow vein mosaic virus and Cotton leaf curl Multan betasatellite with Capsicum annuum from Kashmir valley, India. New Dis. Rep. 2015, 32, 9. [Google Scholar] [CrossRef]
- Mubin, M.; Shahid, M.S.; Tahir, M.N.; Briddon, R.W.; Mansoor, S. Characterization of begomovirus components from a weed suggests that begomoviruses may associate with multiple distinct DNA satellites. Virus Genes 2010, 40, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, J.; Khan, Z.A.; Yadav, N.; Sinha, V.; Bhatnagar, D.; Khan, J.A. Study of betasatellite molecule from leaf curl disease of sunn hemp (Crotalaria juncea) in India. Virus Genes 2010, 41, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, K.P.; Ullah, R.; Saeed, M.; Sarwar, N.; Mansoor, S. China rose (Hibiscus rosa-Sinensis): A New Natural Host of Cotton Leaf Curl Burewala Virus in Pakistan. J. Plant Pathol. 2014, 96, 385–389. [Google Scholar]
- Tahir, M.N.; Mansoor, S.; Briddon, R.W.; Amin, I. Begomovirus and associated satellite components infecting cluster bean (Cyamopsis tetragonoloba) in Pakistan. J. Phytopathol. 2017, 165, 115–122. [Google Scholar] [CrossRef]
- Saeed, M. Tomato leaf curl virus and Cotton leaf curl Multan betasatellite can cause mild transient symptoms in cotton. Australas Plant Dis. Notes 2010, 5, 58. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Martin, D.P.; Amin, I.; Farooq, M.; Mansoor, S. Tomato leaf curl New Delhi virus: A Widespread Bipartite Begomovirus in the Territory of Monopartite Begomoviruses. Mol. Plant Pathol. 2017, 18, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Jyothsna, P.; Haq, Q.M.; Singh, P.; Sumiya, K.V.; Praveen, S.; Rawat, R.; Briddon, R.W.; Malathi, V.G. Infection of Tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus with betasatellites, results in enhanced level of helper virus components and antagonistic interaction between DNA B and betasatellites. App. Microbiol. Biotechnol. 2013, 97, 5457–5471. [Google Scholar] [CrossRef] [PubMed]
- Sivalingam, P.N.; Varma, A. Role of betasatellite in the pathogenesis of a bipartite begomovirus affecting tomato in India. Arch. Virol. 2012, 157, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M. Tomato leaf curl New Delhi virus DNA A component and cotton leaf curl Multan betasatellite can cause mild transient symptoms in cotton. Acta Virol. 2010, 54, 317–318. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.L.; Hidayat, S.H.; Paplomatas, E.J.; Rojas, M.R.; Hou, Y.M.; Maxwell, D.P. Pseudorecombination between infectious cloned DNA components of tomato mottle and bean dwarf mosaic geminiviruses. J. Gen. Virol. 1993, 74, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Gilbertson, R.L. Increased pathogenicity in a pseudorecombinant bipartite geminivirus correlates with intermolecular recombination. J. Virol. 1996, 70, 5430–5436. [Google Scholar] [PubMed]
- Kumar, P.P.; Usha, R.; Zrachya, A.; Levy, Y.; Spanov, H.; Gafni, Y. Protein-protein interactions and nuclear trafficking of coat protein and βC1 protein associated with bhendi yellow vein mosaic disease. Virus Res. 2006, 122, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Rasheed, M.S.; Randles, J.W. In situ hybridization and promoter analysis reveal that cotton leaf curl Multan betasatellite localizes in the phloem. Acta Virol. 2017, 61, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Bedford, I.D.; Stanley, J. Pathogenicity of a natural recombinant associated with ageratum yellow vein disease: Implications for Geminivirus Evolution and Disease Aetiology. Virology 2001, 282, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zhou, X. Pathogenicity of a naturally occurring recombinant DNA satellite associated with Tomato yellow leaf curl China virus. J. Gen. Virol. 2008, 89, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J.; Saunders, K.; Pinner, M.S.; Wong, S.M. Novel defective interfering DNAs associated with Ageratum yellow vein geminivirus infection of Ageratum conyzoides. Virology 1997, 239, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J. Subviral DNAs associated with geminivirus disease complexes. Vet. Microbiol. 2004, 98, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Briddon, R.W.; Stanley, J. Replication promiscuity of DNA-β satellites associated with monopartite begomoviruses; deletion mutagenesis of the Ageratum yellow vein virus DNA-β satellite localizes sequences involved in replication. J. Gen. Virol. 2008, 89, 3165–3172. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Stanley, J. Subviral agents associated with plant single-stranded DNA viruses. Virology 2006, 344, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Mansoor, S. Viral vectors for plant genome engineering. Front. Plant Sci. 2017, 8, 539. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Baltes, N.J.; Cegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Kharazmi, S.; Behjatnia, S.A.; Hamzehzarghani, H.; Niazi, A. Cotton leaf curl Multan betasatellite as a plant gene delivery vector trans-activated by taxonomically diverse geminiviruses. Arch. Virol. 2012, 157, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Kharazmi, S.; Ataie Kachoie, E.; Behjatnia, S.A. Cotton leaf curl Multan betasatellite DNA as a tool to deliver and express the human B-cell lymphoma 2 (Bcl-2) gene in plants. Mol. Biotechnol. 2016, 58, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Mansoor, S.; Ali, Z.; Tashkandi, M.; Mahfouz, M.M. Engineering plants for geminivirus resistance with CRISPR/Cas9 system. Trends Plant Sci. 2016, 21, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Tashkandi, M.; Mansoor, S.; Mahfouz, M.M. Engineering plant immunity: Using CRISPR/Cas9 to generate virus resistance. Front. Plant Sci. 2016, 7, 1673. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Tashkandi, M.; Mahfouz, M.M. Engineering molecular immunity against plant viruses. Prog. Mol. Biol. Transl. Sci. 2017, 149, 167–186. [Google Scholar] [PubMed]
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.; Mahfouz, M.M. CRISPR/Cas9-mediated immunity to geminiviruses: Differential Interference and Evasion. Sci. Rep. 2016, 6, 26912. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 2015, 1, 15145. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Briddon, R.W.; Mansoor, S. Engineering dual Begomovirus-Bemisia tabaci resistance in plants. Trends Plant Sci. 2017, 22, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Akmal, M.; Khan, J.A. Resistance to cotton leaf curl disease in transgenic tobacco expressing βC1 gene derived intron-hairpin RNA. Indian J. Biotechnol. 2017, 16, 56–62. [Google Scholar]
- Ahmad, A.; Zia-Ur-Rehman, M.; Hameed, U.; Qayyum Rao, A.; Ahad, A.; Yasmeen, A.; Akram, F.; Bajwa, K.S.; Scheffler, J.; Nasir, I.A.; et al. Engineered disease resistance in cotton using RNA-interference to knock down Cotton leaf curl Kokhran virus-Burewala and Cotton leaf curl Multan betasatellite expression. Viruses 2017, 9, 257. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Mahfouz, M.M.; Mansoor, S. CRISPR-Cpf1: A new tool for plant genome editing. Trends Plant Sci. 2017, 22, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, M.M. Genome editing: The efficient tool CRISPR-Cpf1. Nat. Plants 2017, 3, 17028. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zubair, M.; Zaidi, S.S.-e.-A.; Shakir, S.; Amin, I.; Mansoor, S. An Insight into Cotton Leaf Curl Multan Betasatellite, the Most Important Component of Cotton Leaf Curl Disease Complex. Viruses 2017, 9, 280. https://doi.org/10.3390/v9100280
Zubair M, Zaidi SS-e-A, Shakir S, Amin I, Mansoor S. An Insight into Cotton Leaf Curl Multan Betasatellite, the Most Important Component of Cotton Leaf Curl Disease Complex. Viruses. 2017; 9(10):280. https://doi.org/10.3390/v9100280
Chicago/Turabian StyleZubair, Muhammad, Syed Shan-e-Ali Zaidi, Sara Shakir, Imran Amin, and Shahid Mansoor. 2017. "An Insight into Cotton Leaf Curl Multan Betasatellite, the Most Important Component of Cotton Leaf Curl Disease Complex" Viruses 9, no. 10: 280. https://doi.org/10.3390/v9100280




