Inference of a Geminivirus−Host Protein−Protein Interaction Network through Affinity Purification and Mass Spectrometry Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids and Cloning
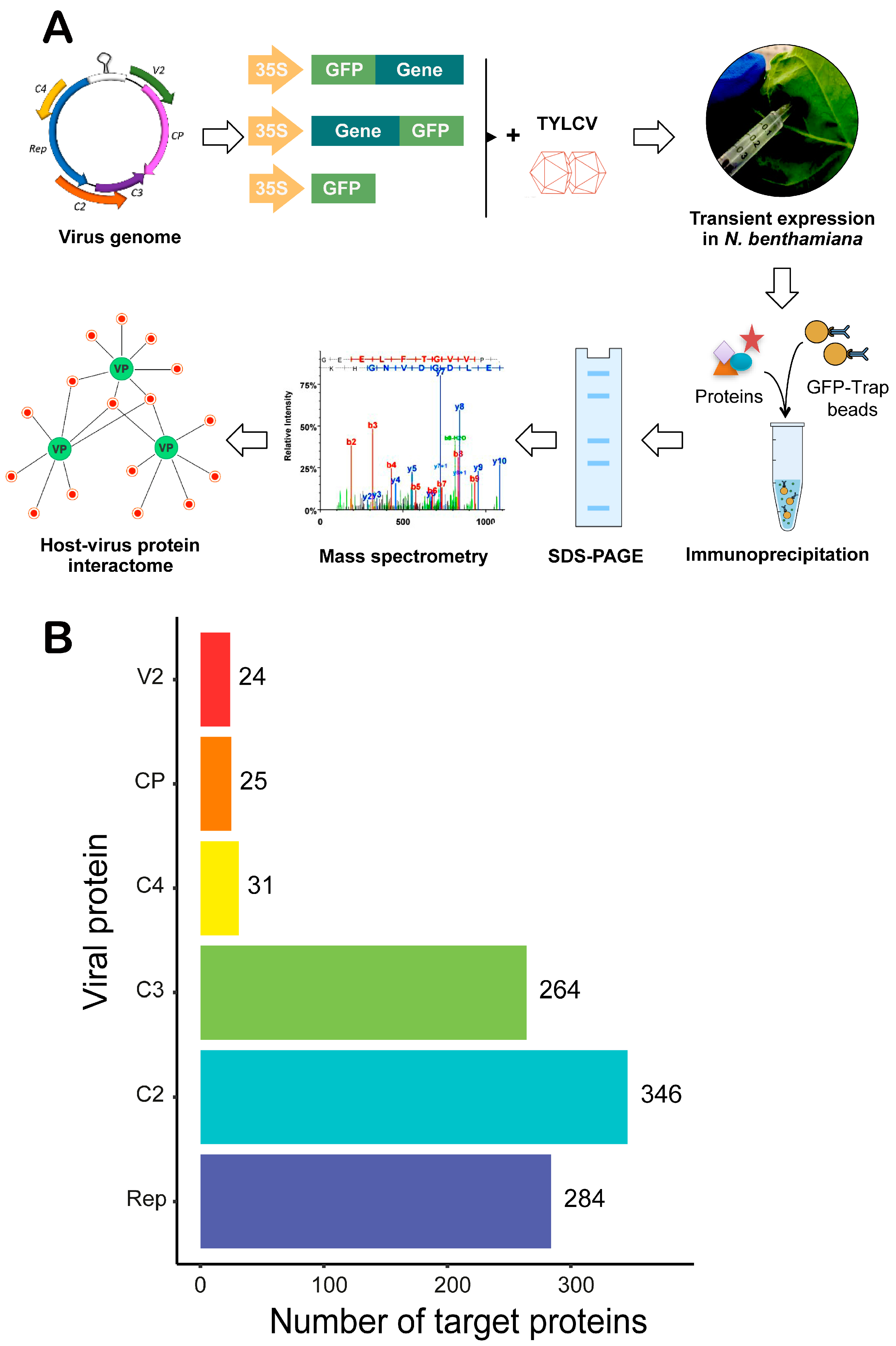
2.2. Affinity Purification-Mass Spectrometry Analysis
2.3. PPI Network Analysis
2.4. Functional Analysis of TYLCV-Targeted Proteins
2.5. Evolutionary Analysis of TYLCV-Targeted Proteins
3. Results
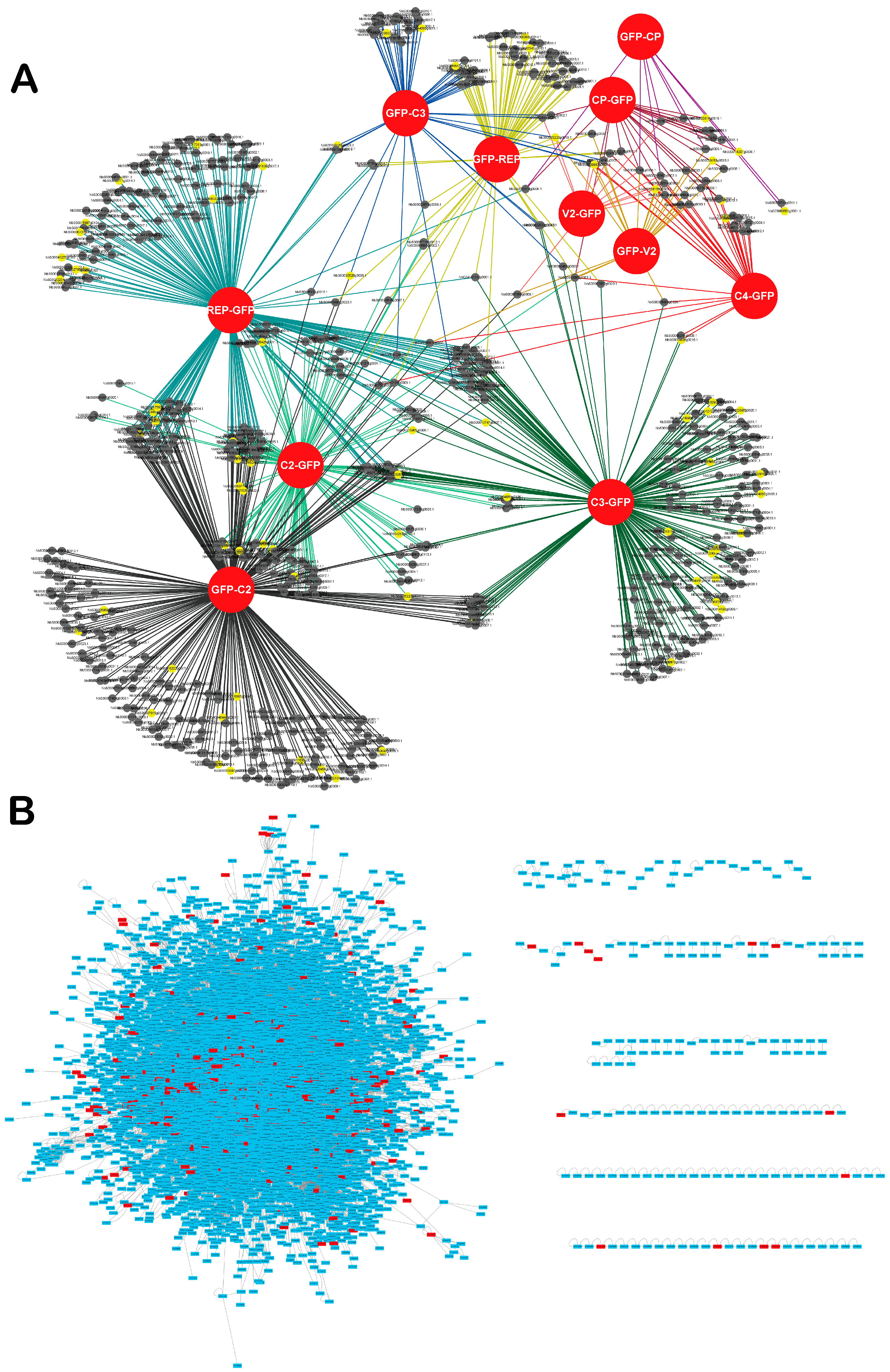
3.1. Identification of Nicotiana benthamiana Proteins Associating with GFP-Tagged TYLCV Proteins
3.2. Inference of the TYLCV-N. benthamiana Protein-Protein Interaction Network and Its Properties
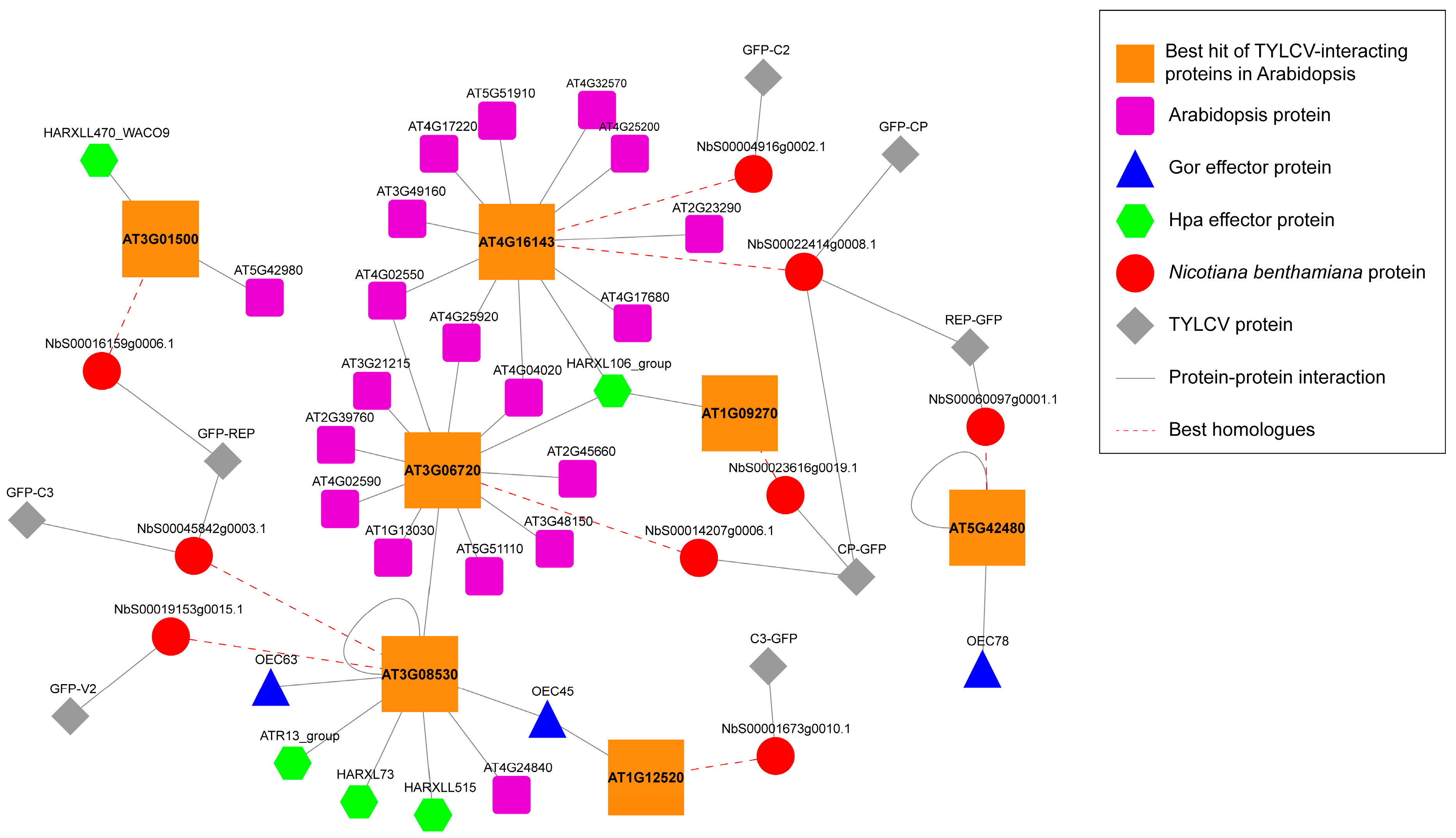
3.3. TYLCV-Targeted Proteins Are Convergently Targeted by Other Pathogens
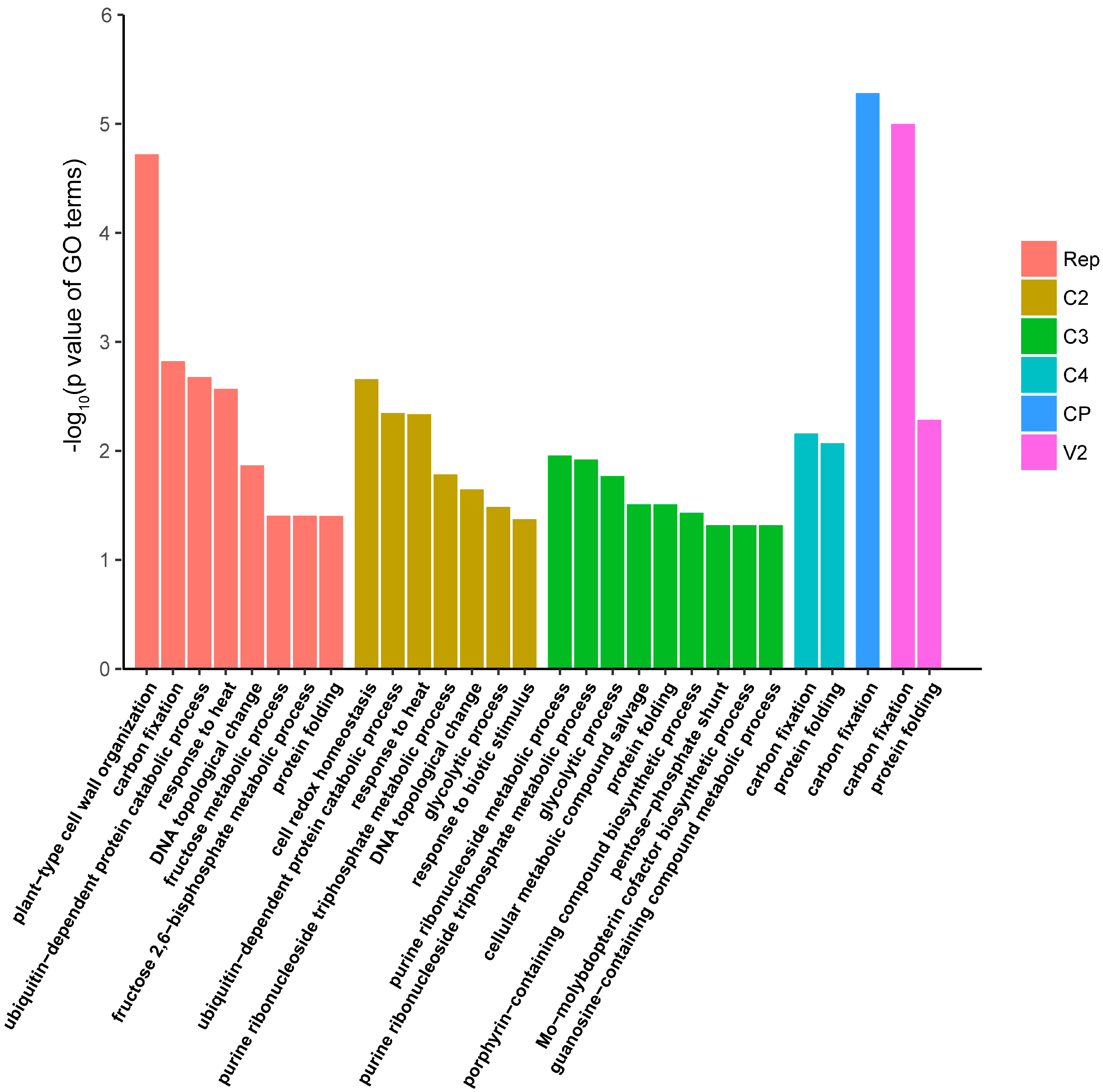
3.4. Functional Enrichment Analysis of the N. benthamiana Proteins Identified as Interactors of TYLCV Proteins
3.5. TYLCV-Targeted Proteins Are Evolutionarily Constrained
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Davis, Z.H.; Verschueren, E.; Jang, G.M.; Kleffman, K.; Johnson, J.R.; Park, J.; Von Dollen, J.; Maher, M.C.; Johnson, T.; Newton, W.; et al. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol. Cell 2015, 57, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Germain, M.A.; Chatel-Chaix, L.; Gagne, B.; Bonneil, E.; Thibault, P.; Pradezynski, F.; de Chassey, B.; Meyniel-Schicklin, L.; Lotteau, V.; Baril, M.; et al. Elucidating novel hepatitis C virus-host interactions using combined mass spectrometry and functional genomics approaches. Mol. Cell Proteom. 2014, 13, 184–203. [Google Scholar] [CrossRef] [PubMed]
- Jager, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV-human protein complexes. Nature 2011, 481, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Kandasamy, K.; Alvisi, G.; Mulhern, O.; Sacco, R.; Habjan, M.; Binder, M.; Stefanovic, A.; Eberle, C.A.; Goncalves, A.; et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 2012, 487, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Ramage, H.R.; Kumar, G.R.; Verschueren, E.; Johnson, J.R.; Von Dollen, J.; Johnson, T.; Newton, B.; Shah, P.; Horner, J.; Krogan, N.J.; et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol. Cell 2015, 57, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Deo, R.C.; Padi, M.; Adelmant, G.; Calderwood, M.A.; Rolland, T.; Grace, M.; Dricot, A.; Askenazi, M.; Tavares, M.; et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 2012, 487, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, F.; Rodrigo, G.; Aragones, V.; Ruiz, M.; Lodewijk, I.; Fernandez, U.; Elena, S.F.; Daros, J.A. Interaction network of tobacco etch potyvirus NIa protein with the host proteome during infection. BMC Genom. 2016, 17, 87. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.J. The use of RNAi-based screens to identify host proteins involved in viral replication. Future Microbiol. 2010, 5, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Navratil, V.; De Chassey, B.; Combe, C.R.; Lotteau, V. When the human viral infectome and diseasome networks collide: Towards a Systems Biology Platform for the Aetiology of Human Diseases. BMC Syst. Biol. 2011, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Albert, R.; Jeong, H.; Barabasi, A.L. Error and attack tolerance of complex networks. Nature 2000, 406, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Han, J.D.; Bertin, N.; Hao, T.; Goldberg, D.S.; Berriz, G.F.; Zhang, L.V.; Dupuy, D.; Walhout, A.J.; Cusick, M.E.; Roth, F.P.; et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 2004, 430, 88–93. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006, 2, e88. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Mason, S.P.; Barabasi, A.L.; Oltvai, Z.N. Lethality and centrality in protein networks. Nature 2001, 411, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Dyer, M.D.; Murali, T.M.; Sobral, B.W. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008, 4, e32. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.S.; Carvunis, A.R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T.; et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Sangsuriya, P.; Huang, J.Y.; Chu, Y.F.; Phiwsaiya, K.; Leekitcharoenphon, P.; Meemetta, W.; Senapin, S.; Huang, W.P.; Withyachumnarnkul, B.; Flegel, T.W.; et al. Construction and application of a protein interaction map for white spot syndrome virus (WSSV). Mol. Cell Proteom. 2014, 13, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Wessling, R.; Epple, P.; Altmann, S.; He, Y.; Yang, L.; Henz, S.R.; McDonald, N.; Wiley, K.; Bader, K.C.; Glasser, C.; et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 2014, 16, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Wuchty, S.; Almaas, E. Evolutionary cores of domain co-occurrence networks. BMC Evol. Biol. 2005, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at Redirecting and Reprogramming Plant Processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Jeske, H. Geminiviruses. Curr. Top. Microbiol. Immunol. 2009, 331, 185–226. [Google Scholar] [PubMed]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Wang, Y.; Ni, H.; Cazale, A.C.; She, Y.M.; Peeters, N.; Macho, A.P. The Ralstonia solanacearum type III effector RipAY targets plant redox regulators to suppress immune responses. Mol. Plant Pathol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Verschueren, E.; Von Dollen, J.; Cimermancic, P.; Gulbahce, N.; Sali, A.; Krogan, N.J. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr. Protoc. Bioinform. 2015, 49, 1–16. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Geisler-Lee, J.; O’Toole, N.; Ammar, R.; Provart, N.J.; Millar, A.H.; Geisler, M. A predicted interactome for Arabidopsis. Plant Physiol. 2007, 145, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML: A Program Package for Phylogenetic Analysis by Maximum Likelihood. Comput. Appl. Biosci. 1997, 13, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Fondong, V.N.; Reddy, R.V.; Lu, C.; Hankoua, B.; Felton, C.; Czymmek, K.; Achenjang, F. The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol. Plant Microbe Interact. 2007, 20, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Diaz, T.; Zhang, D.; Fan, P.; Wang, L.; Ding, X.; Jiang, Y.; Jimenez-Gongora, T.; Medina-Puche, L.; Zhao, X.; Feng, Z.; et al. A plant receptor-like kinase promotes cell-to-cell spread of RNAi and is targeted by a virus. BioRxiv 2017. [Google Scholar] [CrossRef]
- Morilla, G.; Janssen, D.; Garcia-Andres, S.; Moriones, E.; Cuadrado, I.M.; Bejarano, E.R. Pepper (Capsicum annuum) Is a Dead-End Host for Tomato yellow leaf curl virus. Phytopathology 2005, 95, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science 2011, 333, 601–607. [Google Scholar]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: Understanding the Cell’s Functional Organization. Nat. Rev. Genet. 2004, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Jiang, H.; Salati, R.; Xoconostle-Cazares, B.; Sudarshana, M.R.; Lucas, W.J.; Gilbertson, R.L. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 2001, 291, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Duran, R.; Rosas-Diaz, T.; Gusmaroli, G.; Luna, A.P.; Taconnat, L.; Deng, X.W.; Bejarano, E.R. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 2011, 23, 1014–1032. [Google Scholar] [CrossRef] [PubMed]
- Chandran, S.A.; Levy, Y.; Mett, A.; Belausov, E.; Ramakrishnan, U.; Gafni, Y. Mapping of functional region conferring nuclear localization and karyopherin alpha-binding activity of the C2 protein of bhendi yellow vein mosaic virus. J. Gen. Virol. 2012, 93, 1367–1374. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Duran, R.; Rosas-Diaz, T.; Luna, A.P.; Bejarano, E.R. Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLoS ONE 2011, 6, e22383. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Ding, X.; Xiao, J.; Jiménez-Gόngora, T.; Liu, R.; Lozano-Durán, R. Inference of a Geminivirus−Host Protein−Protein Interaction Network through Affinity Purification and Mass Spectrometry Analysis. Viruses 2017, 9, 275. https://doi.org/10.3390/v9100275
Wang L, Ding X, Xiao J, Jiménez-Gόngora T, Liu R, Lozano-Durán R. Inference of a Geminivirus−Host Protein−Protein Interaction Network through Affinity Purification and Mass Spectrometry Analysis. Viruses. 2017; 9(10):275. https://doi.org/10.3390/v9100275
Chicago/Turabian StyleWang, Liping, Xue Ding, Jiajing Xiao, Tamara Jiménez-Gόngora, Renyi Liu, and Rosa Lozano-Durán. 2017. "Inference of a Geminivirus−Host Protein−Protein Interaction Network through Affinity Purification and Mass Spectrometry Analysis" Viruses 9, no. 10: 275. https://doi.org/10.3390/v9100275




