Pediatric and Adult High-Grade Glioma Stem Cell Culture Models Are Permissive to Lytic Infection with Parvovirus H-1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Culture
2.3. Generation of the Subclones and Cell Authentication
2.4. Virus Production and Infection
2.5. Detection of Infectious H-1PV Particles by Propagation in NB-324k Cells
2.6. Flow Cytometric Analysis of CD133 Expression
2.7. Immunofluorescence Staining, Microscopy and Documentation
2.8. Western Blot Analysis
2.9. Assessment of Cell Viability and Lysis
2.10. Gene Expression Analyses
2.11. Statistical Analysis
2.12. In Vivo Models
3. Results
3.1. The Neurosphere Cultures Analyzed Represent a Broad Variety of HGG Subtypes But Consistently Display Neural Stem Cell Features
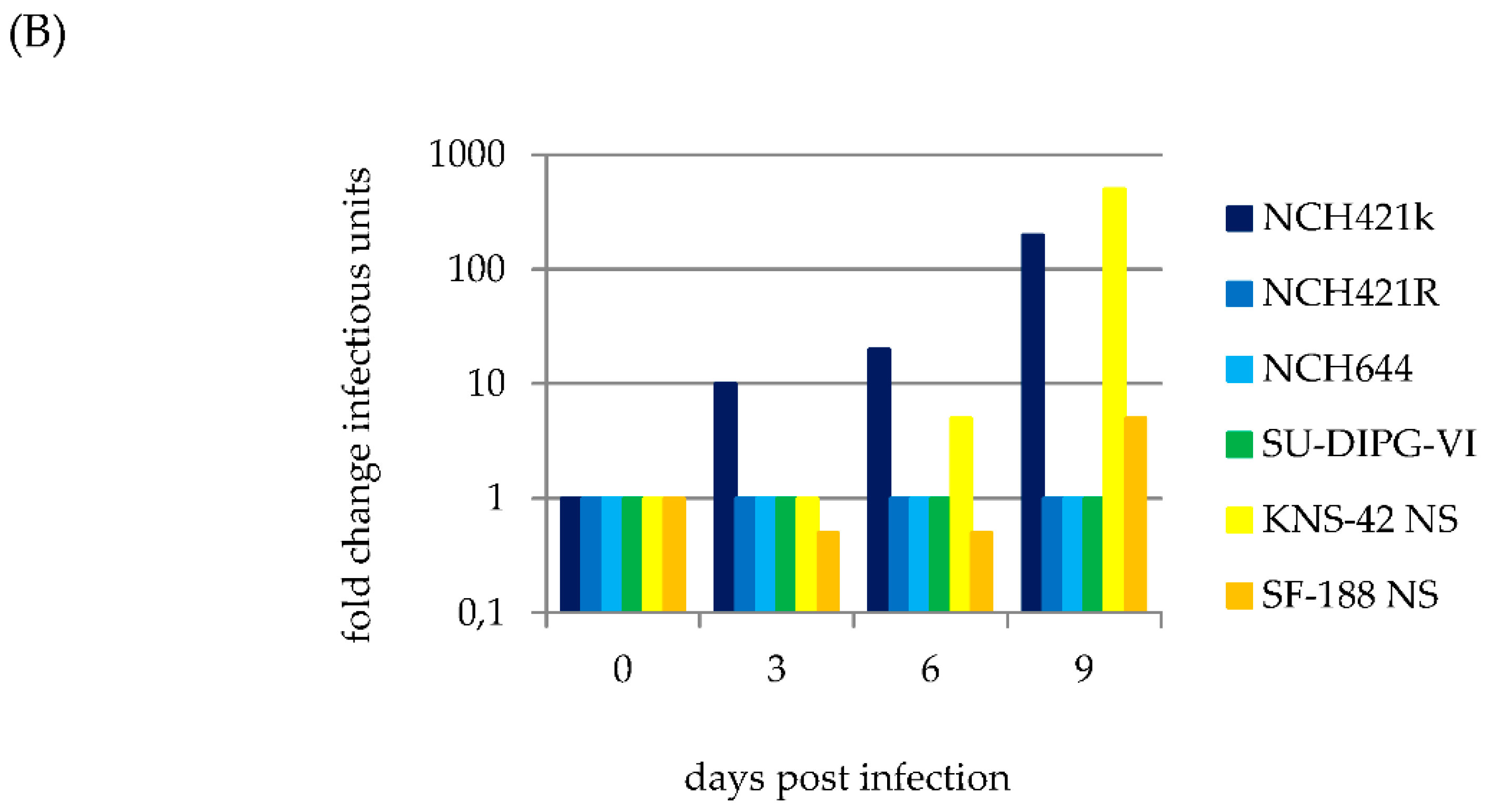
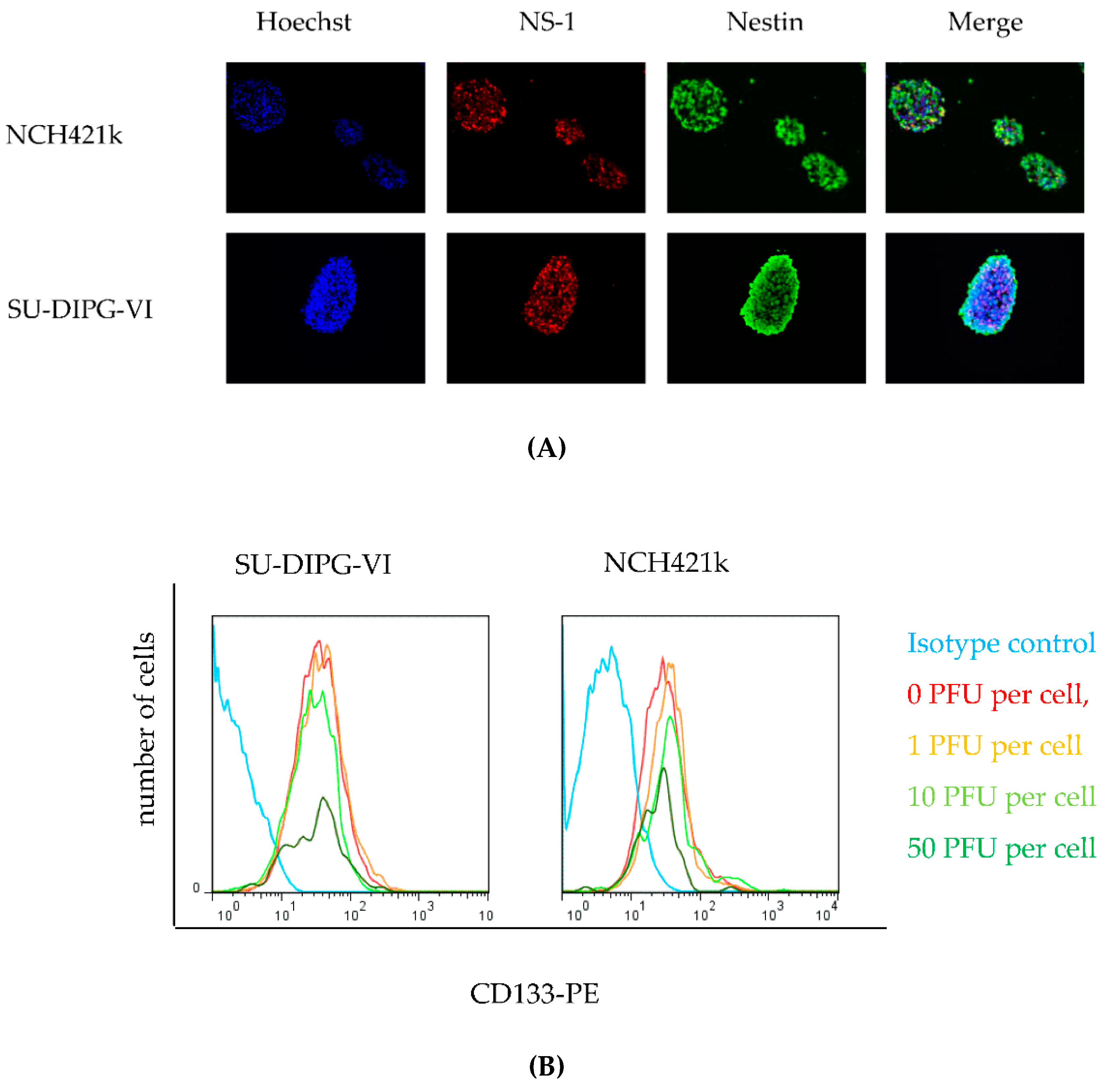
3.2. H-1PV Efficiently Transduces and Replicates in Pediatric and Adult HGG Neurosphere Cell Culture Models
3.3. H-1PV Productive Infection is Restricted to a Subset of Pediatric and Adult HGG Neurosphere Cultures
3.4. Infection of Pediatric and Adult HGG with H-1PV Induces Complete Cell Death in All Neurosphere Cultures
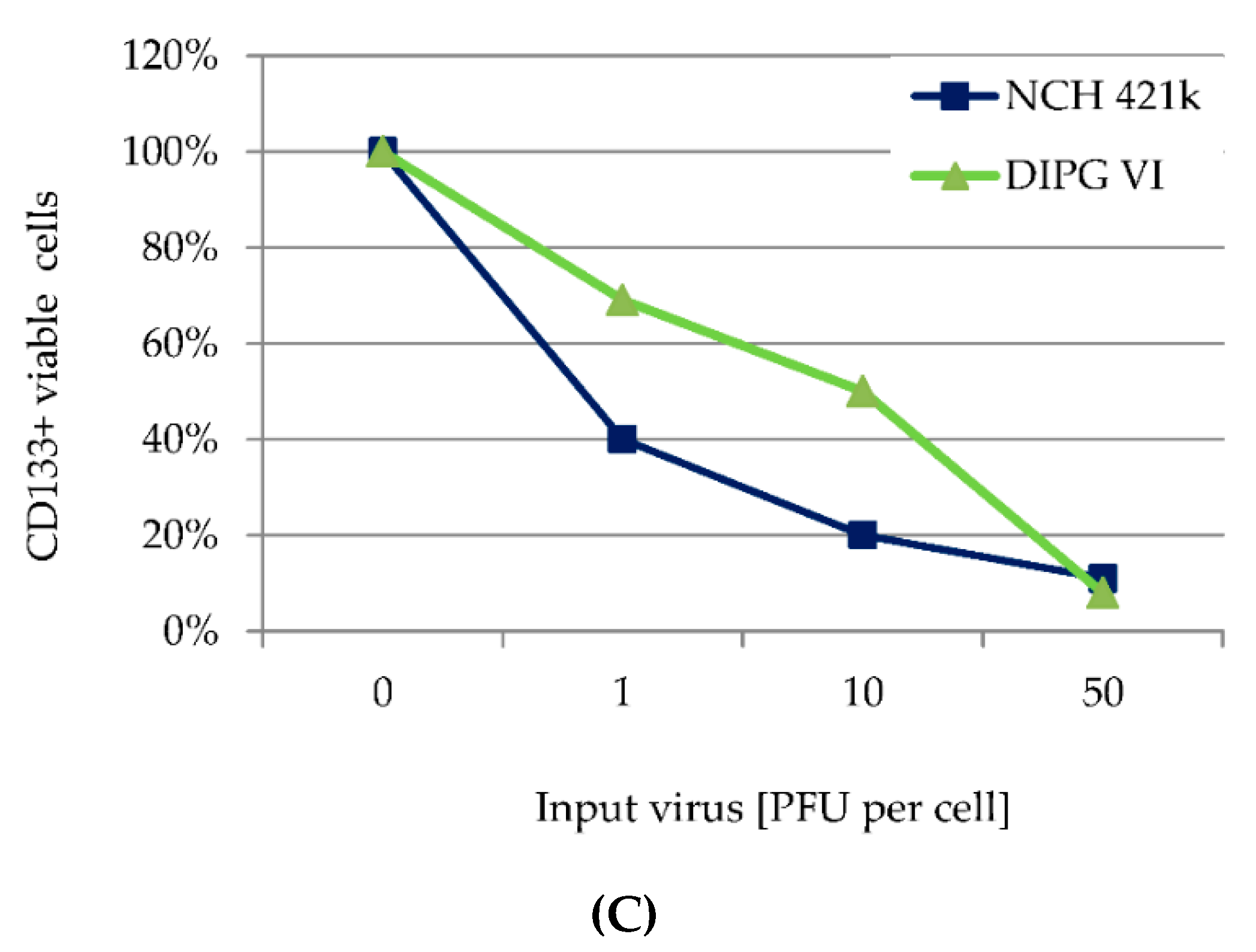
3.5. H-1PV Targets Both CD 133 Positive and CD133 Negative HGG Cells
3.6. H-1PV Resistance Correlates with Dysregulation of Distinct Cellular Genes
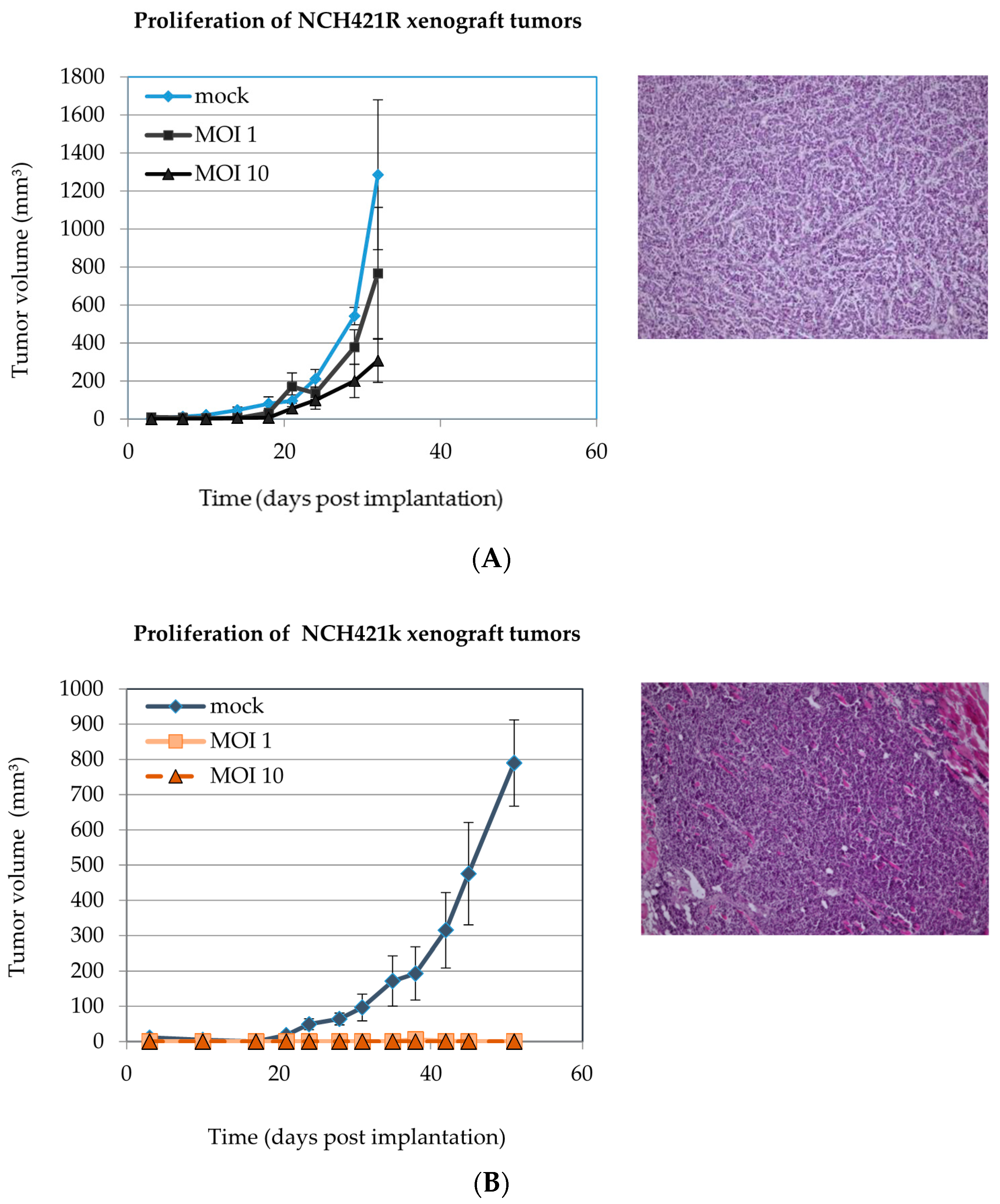
3.7. H-1PV Infection of NCH421k HGG Stem-Like Cells Suppresses Tumor Formation in NOD/SCID Mice
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sturm, D.; Bender, S.; Jones, D.T.; Lichter, P.; Grill, J.; Becher, O.; Hawkins, C.; Majewski, J.; Jones, C.; Costello, J.F.; et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 2014, 14, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Fangusaro, J.; Warren, K.E. Unclear standard of care for pediatric high grade glioma patients. J. Neurooncol. 2013, 113, 341–342. [Google Scholar] [CrossRef] [PubMed]
- Hardesty, D.A.; Sanai, N. The value of glioma extent of resection in the modern neurosurgical era. Front. Neurol. 2012, 3, 140. [Google Scholar] [CrossRef] [PubMed]
- Young, R.M.; Jamshidi, A.; Davis, G.; Sherman, J.H. Current trends in the surgical management and treatment of adult glioblastoma. Ann. Transl. Med. 2015, 3, 121. [Google Scholar] [PubMed]
- Warren, K.E. Diffuse intrinsic pontine glioma: poised for progress. Front. Oncol. 2012, 2, 205. [Google Scholar] [CrossRef] [PubMed]
- Fangusaro, J. Pediatric high grade glioma: a review and update on tumor clinical characteristics and biology. Front. Oncol. 2012, 2, 105. [Google Scholar] [CrossRef] [PubMed]
- Reardon, D.A.; Wen, P.Y. Glioma in 2014: unravelling tumour heterogeneity-implications for therapy. Nat. Rev. Clin. Oncol. 2015, 12, 69–70. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.E.; Mazor, T.; Hong, C.; Barnes, M.; Aihara, K.; McLean, C.Y.; Fouse, S.D.; Yamamoto, S.; Ueda, H.; Tatsuno, K.; et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014, 343, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Gronych, J.; Pfister, S.M.; Jones, D.T. Connect four with glioblastoma stem cell factors. Cell 2014, 157, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Moehler, M.; Goepfert, K.; Heinrich, B.; Breitbach, C.J.; Delic, M.; Galle, P.R.; Rommelaere, J. Oncolytic virotherapy as emerging immunotherapeutic modality: potential of parvovirus h-1. Front. Oncol. 2014, 4, 92. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, J.K.; Chiocca, E.A. Glioma virus therapies between bench and bedside. Neuro. Oncol. 2014, 16, 334–351. [Google Scholar] [CrossRef] [PubMed]
- Piccioni, D.E.; Kesari, S. Clinical trials of viral therapy for malignant gliomas. Expert. Rev. Anticancer. Ther. 2013, 13, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Geletneky, K.; Kiprianova, I.; Ayache, A.; Koch, R.; Herrero, Y.C.; Deleu, L.; Sommer, C.; Thomas, N.; Rommelaere, J.; Schlehofer, J.R. Regression of advanced rat and human gliomas by local or systemic treatment with oncolytic parvovirus H-1 in rat models. Neuro. Oncol. 2010, 12, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Kiprianova, I.; Thomas, N.; Ayache, A.; Fischer, M.; Leuchs, B.; Klein, M.; Rommelaere, J.; Schlehofer, J.R. Regression of glioma in rat models by intranasal application of parvovirus H-1. Clin. Cancer Res. 2011, 17, 5333–5342. [Google Scholar] [CrossRef] [PubMed]
- Geletneky, K.; Huesing, J.; Rommelaere, J.; Schlehofer, J.R.; Leuchs, B.; Dahm, M.; Krebs, O.; von Knebel, D.M.; Huber, B.; Hajda, J. Phase I/IIa study of intratumoral/intracerebral or intravenous/intracerebral administration of Parvovirus H-1 (ParvOryx) in patients with progressive primary or recurrent glioblastoma multiforme: ParvOryx01 protocol. BMC Cancer 2012, 12, 99. [Google Scholar] [CrossRef] [PubMed]
- Angelova, A.L.; Geletneky, K.; Nuesch, J.P.; Rommelaere, J. Tumor Selectivity of Oncolytic Parvoviruses: From in vitro and Animal Models to Cancer Patients. Front. Bioeng. Biotechnol. 2015, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Wakimoto, H.; Mohapatra, G.; Kanai, R.; Curry, W.T., Jr.; Yip, S.; Nitta, M.; Patel, A.P.; Barnard, Z.R.; Stemmer-Rachamimov, A.O.; Louis, D.N.; et al. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro. Oncol. 2012, 14, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Edlund, K.; Bergenheim, A.T.; Wadell, G. Adenoviruses 16 and CV23 efficiently transduce human low-passage brain tumor and cancer stem cells. Mol. Ther. 2007, 15, 2140–2145. [Google Scholar] [CrossRef] [PubMed]
- Bach, P.; Abel, T.; Hoffmann, C.; Gal, Z.; Braun, G.; Voelker, I.; Ball, C.R.; Johnston, I.C.; Lauer, U.M.; Herold-Mende, C.; et al. Specific elimination of CD133+ tumor cells with targeted oncolytic measles virus. Cancer Res. 2013, 73, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Friedman, G.K.; Langford, C.P.; Coleman, J.M.; Cassady, K.A.; Parker, J.N.; Markert, J.M.; Yancey, G.G. Engineered herpes simplex viruses efficiently infect and kill CD133+ human glioma xenograft cells that express CD111. J. Neurooncol. 2009, 95, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Wakimoto, H.; Kesari, S.; Farrell, C.J.; Curry, W.T., Jr.; Zaupa, C.; Aghi, M.; Kuroda, T.; Stemmer-Rachamimov, A.; Shah, K.; Liu, T.C.; et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009, 69, 3472–3481. [Google Scholar] [CrossRef] [PubMed]
- Cheema, T.A.; Kanai, R.; Kim, G.W.; Wakimoto, H.; Passer, B.; Rabkin, S.D.; Martuza, R.L. Enhanced antitumor efficacy of low-dose Etoposide with oncolytic herpes simplex virus in human glioblastoma stem cell xenografts. Clin. Cancer. Res. 2011, 17, 7383–7393. [Google Scholar] [CrossRef] [PubMed]
- Friedman, G.K.; Nan, L.; Haas, M.C.; Kelly, V.M.; Moore, B.P.; Langford, C.P.; Xu, H.; Han, X.; Beierle, E.A.; Markert, J.M.; et al. gamma(1)34.5-deleted HSV-1-expressing human cytomegalovirus IRS1 gene kills human glioblastoma cells as efficiently as wild-type HSV-1 in normoxia or hypoxia. Gene Ther. 2015, 22, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.R.; Mackay, A.; Truffaux, N.; Butterfield, Y.S.; Morozova, O.; Philippe, C.; Castel, D.; Grasso, C.S.; Vinci, M.; Carvalho, D.; et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat. Genet. 2014, 46, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Caretti, V.; Sewing, A.C.; Lagerweij, T.; Schellen, P.; Bugiani, M.; Jansen, M.H.; van Vuurden, D.G.; Navis, A.C.; Horsman, I.; Vandertop, W.P.; et al. Human pontine glioma cells can induce murine tumors. Acta. Neuropathol. 2014, 127, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Campos, B.; Wan, F.; Farhadi, M.; Ernst, A.; Zeppernick, F.; Tagscherer, K.E.; Ahmadi, R.; Lohr, J.; Dictus, C.; Gdynia, G.; et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin. Cancer Res. 2010, 16, 2715–2728. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Mitra, S.S.; Monje, M.; Henrich, K.N.; Bangs, C.D.; Nitta, R.T.; Wong, A.J. Expression of epidermal growth factor variant III (EGFRvIII) in pediatric diffuse intrinsic pontine gliomas. J. Neurooncol. 2012, 108, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Pawlita, M. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Res. 2009, 37, e119. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.; Dirks, W.G.; Fahnrich, S.; Hotz-Wagenblatt, A.; Pawlita, M.; Schmitt, M. High-throughput SNP-based authentication of human cell lines. Int. J. Cancer 2013, 132, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Hovestadt, V.; Remke, M.; Kool, M.; Pietsch, T.; Northcott, P.A.; Fischer, R.; Cavalli, F.M.; Ramaswamy, V.; Zapatka, M.; Reifenberger, G.; et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol. 2013, 125, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Bjerke, L.; Mackay, A.; Nandhabalan, M.; Burford, A.; Jury, A.; Popov, S.; Bax, D.A.; Carvalho, D.; Taylor, K.R.; Vinci, M. Histone H3.3 Mutations Drive Pediatric Glioblastoma through Upregulation of MYCN. Cancer Discov. 2013, 3, 512–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bageritz, J.; Puccio, L.; Piro, R.M.; Hovestadt, V.; Phillips, E.; Pankert, T.; Lohr, J.; Herold-Mende, C.; Lichter, P.; Goidts, V. Stem cell characteristics in glioblastoma are maintained by the ecto-nucleotidase E-NPP1. Cell Death Differ. 2014, 21, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Trent, J.; Meltzer, P.; Rosenblum, M.; Harsh, G.; Kinzler, K.; Mashal, R.; Feinberg, A.; Vogelstein, B. Evidence for rearrangement, amplification, and expression of c-myc in a human glioblastoma. Pro. Natl. Acad. Sci. USA 1986, 83, 470–473. [Google Scholar] [CrossRef]
- Takeshita, I.; Takaki, T.; Kuramitsu, M.; Nagasaka, S.; Machi, T.; Ogawa, H.; Egami, H.; Mannoji, H.; Fukui, M.; Kitamura, K. Characteristics of an established human glioma cell line, KNS-42. Neurol. Med. Chir. (Tokyo) 1987, 27, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Lawn, S.; Krishna, N.; Pisklakova, A.; Qu, X.; Fenstermacher, D.A.; Fournier, M.; Vrionis, F.D.; Tran, N.; Chan, J.A.; Kenchappa, R.S.; et al. Neurotrophin signaling via TrkB and TrkC receptors promotes the growth of brain tumor-initiating cells. J. Biol. Chem. 2015, 290, 3814–3824. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, H.; Zong, G.; Li, P. The role of IFITM3 in the growth and migration of human glioma cells. BMC Neurol. 2013, 13, 210. [Google Scholar] [CrossRef] [PubMed]
- Reiss, K.; Stencel, J.E.; Liu, Y.; Blaum, B.S.; Reiter, D.M.; Feizi, T.; Dermody, T.S.; Stehle, T. The GM2 glycan serves as a functional coreceptor for serotype 1 reovirus. PLoS Pathog. 2012, 8, e1003078. [Google Scholar] [CrossRef] [PubMed]
- Caja, L.; Bellomo, C.; Moustakas, A. Transforming growth factor beta and bone morphogenetic protein actions in brain tumors. FEBS Lett. 2015, 589, 1588–1597. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Wang, Y.Y. Innate immune responses to DNA viruses. Protein Cell 2013, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Narvaiza, I.; Linfesty, D.C.; Greener, B.N.; Hakata, Y.; Pintel, D.J.; Logue, E.; Landau, N.R.; Weitzman, M.D. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009, 5, e1000439. [Google Scholar] [CrossRef] [PubMed]
- Pertel, T.; Hausmann, S.; Morger, D.; Zuger, S.; Guerra, J.; Lascano, J.; Reinhard, C.; Santoni, F.A.; Uchil, P.D.; Chatel, L.; et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 2011, 472, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhang, W.; Tan, L.; Yang, H.; Ge, M.; Zhu, C.; Zhang, R.; Cao, Y.; Chen, J.; Luo, Z.; et al. Hepatitis B virus hijacks CTHRC1 to evade host immunity and maintain replication. J. Mol. Cell Biol. 2015, 7, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.M.; Yang, Q.; Zhang, J.; Liu, S.M.; Zhang, Y.; Lin, H.; Huang, Z.F.; Wang, Y.Y.; Zhang, X.D.; Zhong, B.; et al. TRIM38 inhibits TNFalpha- and IL-1beta-triggered NF-kappaB activation by mediating lysosome-dependent degradation of TAB2/3. Proc. Natl. Acad. Sci. USA 2014, 111, 1509–1514. [Google Scholar] [CrossRef] [PubMed]
- Bulliard, Y.; Narvaiza, I.; Bertero, A.; Peddi, S.; Rohrig, U.F.; Ortiz, M.; Zoete, V.; Castro-Diaz, N.; Turelli, P.; Telenti, A.; et al. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J. Virol. 2011, 85, 1765–1776. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Clarke, I.D.; Hide, T.; Dirks, P.B. Cancer stem cells in nervous system tumors. Oncogene 2004, 23, 7267–7273. [Google Scholar] [CrossRef] [PubMed]
- Nuesch, J.P.; Lacroix, J.; Marchini, A.; Rommelaere, J. Molecular pathways: rodent parvoviruses-mechanisms of oncolysis and prospects for clinical cancer treatment. Clin. Cancer Res. 2012, 18, 3516–3523. [Google Scholar] [CrossRef] [PubMed]
- Herrero, Y.C.; Cornelis, J.J.; Herold-Mende, C.; Rommelaere, J.; Schlehofer, J.R.; Geletneky, K. Parvovirus H-1 infection of human glioma cells leads to complete viral replication and efficient cell killing. Int. J. Cancer 2004, 109, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Di Piazza, M.; Mader, C.; Geletneky, K.; Herrero y Calle, M.; Weber, E.; Schlehofer, J.; Deleu, L.; Rommelaere, J. Cytosolic activation of cathepsins mediates parvovirus H-1-induced killing of cisplatin and TRAIL-resistant glioma cells. J. Virol. 2007, 81, 4186–4198. [Google Scholar] [CrossRef] [PubMed]
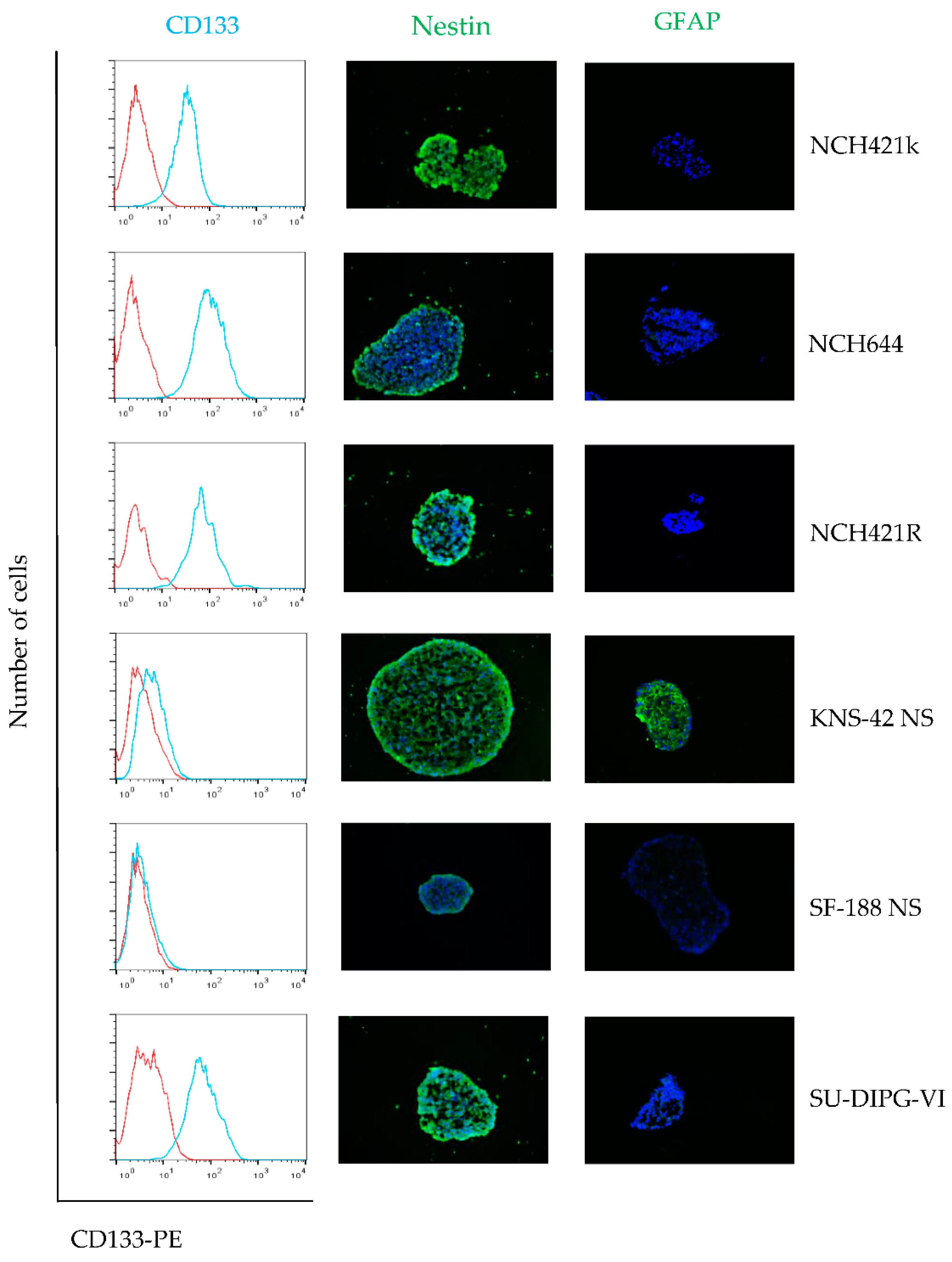
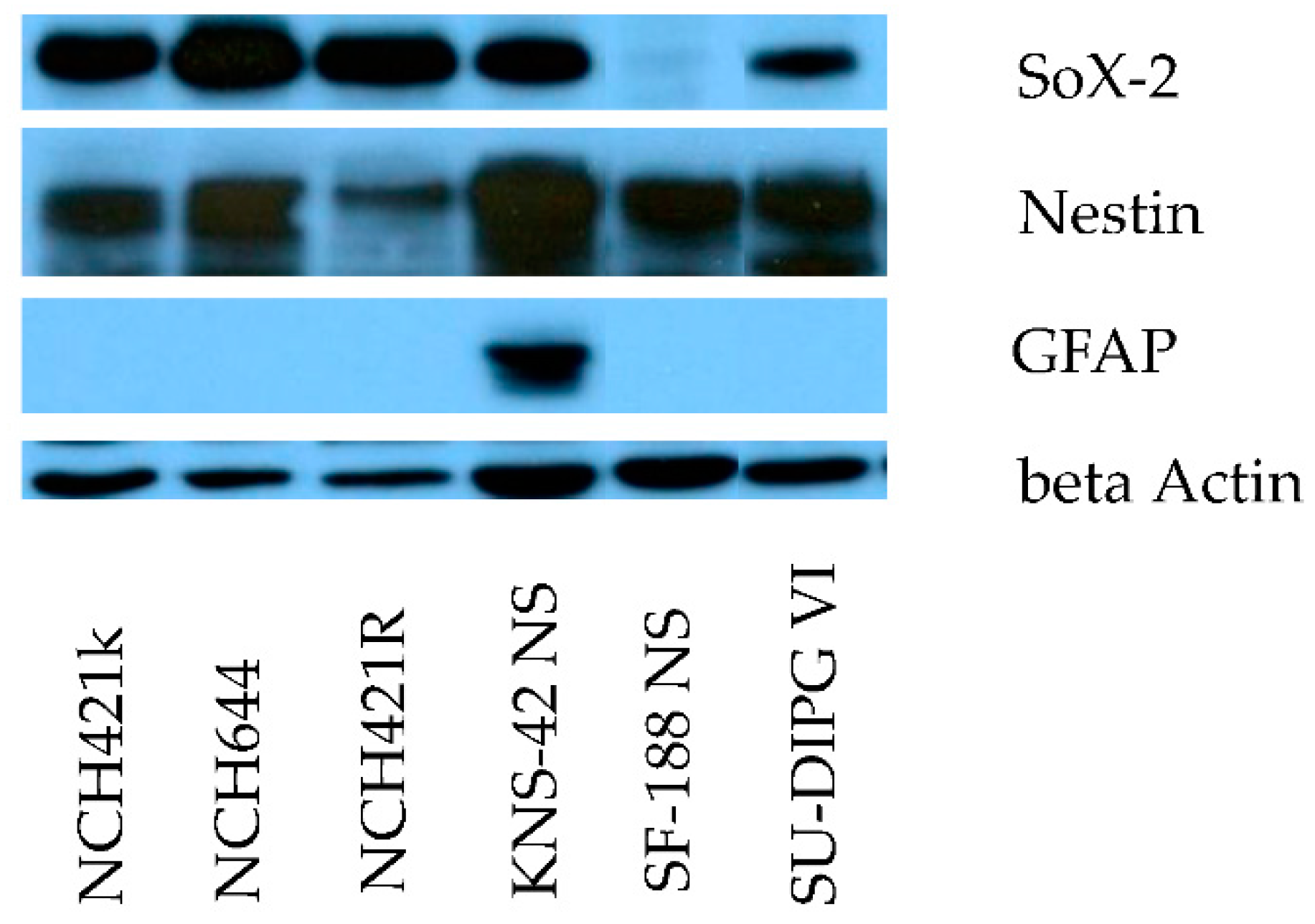




| Cell culture | Diagnosis | WHO Grade | Age | Sex | Survival [months] | Ref. |
|---|---|---|---|---|---|---|
| KNS-42 NS | pGBM | IV | 16 | male | 12 | [35] |
| SU-DIPG-IV | DIPG | IV | 3 | female | 6 | [25] |
| SF-188 NS | pGBM | IV | 8 | male | 4,5 | [34] |
| SU-DIPG-VI | DIPG | IV | 7 | female | 8 | [25] |
| NCH644 | GBM | IV | 67 | female | 7,5 | [33] |
| NCH421k | GBM | IV | 66 | male | 8,5 | [27] |
| NCH421R | GBM | IV | 66 | male | 8,5 | [27] |
| Cell Culture | CD133 | Nestin | SOX-2 | GFAP | LD50 (PFU per cell) |
|---|---|---|---|---|---|
| KNS-42 NS | - | +++ | + | +++ | 1 |
| SU-DIPG-IV | n. d. | n. d. | n. d. | - | 1 |
| SF-188 NS | - | ++ | - | - | 10 |
| SU-DIPG-VI | +++ | ++ | ++ | - | 10 |
| NCH644 | +++ | ++ | ++ | - | 10 |
| NCH421k | +++ | ++ | ++ | - | 10 |
| NCH421R | +++ | + | ++ | - | >100 |
| Cell Culture | Virus Dose [PfU per Cell] | Engraftment Rate | |
|---|---|---|---|
| NCH421k | 0 | 10 | /10 |
| NCH421k | 1 | 0 | /10 * |
| NCH421k | 10 | 0 | /10 ° |
| NCH421R | 0 | 10 | /10 |
| NCH421R | 1 | 10 | /10 |
| NCH421R | 10 | 9 | /10 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Josupeit, R.; Bender, S.; Kern, S.; Leuchs, B.; Hielscher, T.; Herold-Mende, C.; Schlehofer, J.R.; Dinsart, C.; Witt, O.; Rommelaere, J.; et al. Pediatric and Adult High-Grade Glioma Stem Cell Culture Models Are Permissive to Lytic Infection with Parvovirus H-1. Viruses 2016, 8, 138. https://doi.org/10.3390/v8050138
Josupeit R, Bender S, Kern S, Leuchs B, Hielscher T, Herold-Mende C, Schlehofer JR, Dinsart C, Witt O, Rommelaere J, et al. Pediatric and Adult High-Grade Glioma Stem Cell Culture Models Are Permissive to Lytic Infection with Parvovirus H-1. Viruses. 2016; 8(5):138. https://doi.org/10.3390/v8050138
Chicago/Turabian StyleJosupeit, Rafael, Sebastian Bender, Sonja Kern, Barbara Leuchs, Thomas Hielscher, Christel Herold-Mende, Jörg R. Schlehofer, Christiane Dinsart, Olaf Witt, Jean Rommelaere, and et al. 2016. "Pediatric and Adult High-Grade Glioma Stem Cell Culture Models Are Permissive to Lytic Infection with Parvovirus H-1" Viruses 8, no. 5: 138. https://doi.org/10.3390/v8050138






