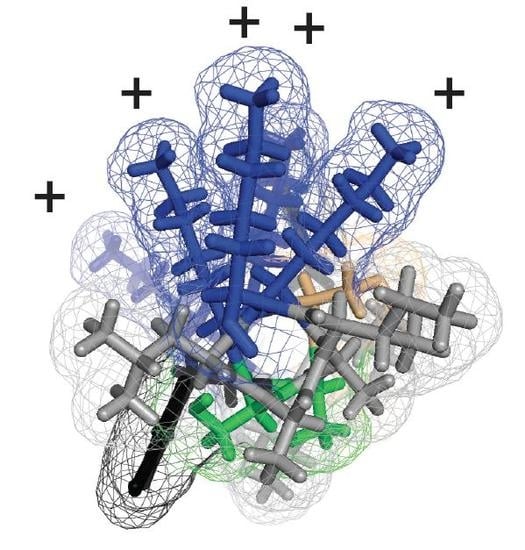
Modeling of the Ebola Virus Delta Peptide Reveals a Potential Lytic Sequence Motif
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Information
2.2. Computational and Graphic Methods
2.3. Modeling
3. Results
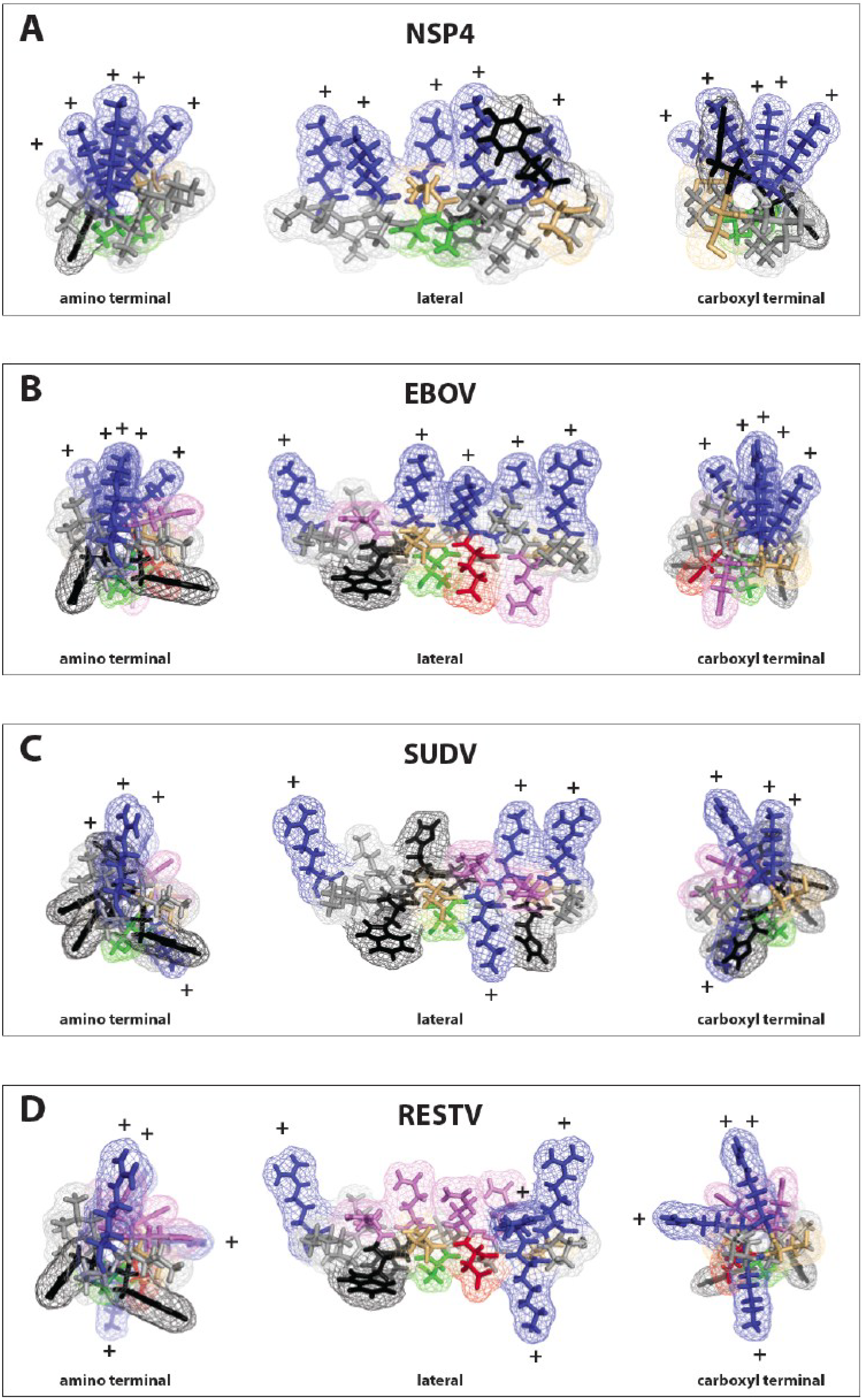
3.1. Delta Peptide is Differentially Conserved amongst Otherwise Disparate Ebolavirus Glycoproteins
 |
 |
3.2. Identification of a Lytic Sequence Motif in Ebolavirus Delta Peptide

3.3. Delta Peptides of Less Pathogenic Ebolaviruses Have Less Focal Display of Positive Charges
3.4. Model for Ebola Virus Delta Peptide
| Peptide | Sequence | WWIHS Score | Hydrophobic Moment | Subtended Angle Arginines/Lysines |
|---|---|---|---|---|
| Viral lytic peptides | ||||
| LLP-1 | VVQGACRAIRHIPRRIRQGLERI | 0 | 7.06 @ 53° | 180° |
| NSP4 | PTMKIALKASKCSYKVIKYCVV | 0 | 3.68 @ 132° | 100° |
| ebolaviruses | ||||
| EBOV/SLE14-EM95 | WLQKIPLQWFKCTVKEGKLQCRI | 3.66 | 4.16 @ 95° | 80° |
| EBOV/Yam76-May | WLQKIPLQWFKCTVKEGKLQCRI | 3.66 | 4.16 @ 95° | 80° |
| SUDV/Gul-808892 | WFQRIPLGWFHCTYQKGKQHCRL | 4.98 | 4.14 @ 100° | 180° |
| BDBV/But-811250 | WFQRIPLQWFKCETSRGKTQCRP | 4.41 | 3.02 @ 79° | 160° |
| TAFV/Pau94-CCI | WFQRIPLQWFQCSLQDGQRKCRP | 4.41 | 3.33 @ 52° | 160° |
| RESTV/Phi89-Pen | WFQRIPLQWFRCKTSRERTQCQP | 4.41 | 3.53 @ 126° | 180° |
| cuevovirus | ||||
| LLOV/Ast03 | YIQAIPLVKFRCHWEGLRHVCRRYPSW | 1.60 | 5.92 @ 147° | 200° |
3.5. Hypothetical Mechanism by Which EBOV Delta Peptide May Generate Pores within Target Membranes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Malherbe, H.; Strickland-Cholmley, M. Human disease from monkeys (Marburg virus). Lancet 1968, 1, 1434. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Ebola haemorrhagic fever in Zaire, 1976. Bull. World Health Organ. 1978, 56, 271–293. [Google Scholar]
- Muyembe, T.; Kipasa, M. Ebola haemorrhagic fever in Kikwit, Zaire. InteRNAtional scientific and technical committee and who collaborating centre for haemorrhagic fevers. Lancet 1995, 345, 1448. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.G.; Schafer, I.J. Ebola viral disease outbreak—West Africa, 2014. MMWR Morb. Mortal. Wkly. Rep. 2014, 63, 548–551. [Google Scholar] [PubMed]
- Baize, S.; Pannetier, D.; Oestereich, L.; Rieger, T.; Koivogui, L.; Magassouba, N.; Soropogui, B.; Sow, M.S.; Keita, S.; de Clerck, H.; et al. Emergence of Zaire Ebola virus disease in Guinea—Preliminary report. N. Engl. J. Med. 2014, 371, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Fowler, R.A.; Fletcher, T.; Fischer, W.A., 2nd; Lamontagne, F.; Jacob, S.; Brett-Major, D.; Lawler, J.V.; Jacquerioz, F.A.; Houlihan, C.; O’Dempsey, T.; et al. Caring for critically ill patients with Ebola virus disease. Perspectives from West Africa. Am. J. Respir. Crit. Care Med. 2014, 190, 733–737. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Ebola Response Roadmap—Situation Report. Available online: http://www.who.int/csr/disease/Ebola/situation-reports/en/ (accessed on 31 December 2014).
- Rubin, E.J.; Baden, L.R. Out of Africa—Caring for patients with Ebola. N. Engl. J. Med. 2014, 371, 2430–2432. [Google Scholar] [CrossRef] [PubMed]
- Frieden, T.R.; Damon, I.; Bell, B.P.; Kenyon, T.; Nichol, S. Ebola 2014—New challenges, new global response and responsibility. N. Engl. J. Med. 2014, 371, 1177–1180. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Ebola virus disease in West Africa—The first 9 months of the epidemic and forward projections. N. Engl. J. Med. 2014, 371, 1481–1495. [Google Scholar]
- Sanchez, A.; Kiley, M.P.; Holloway, B.P.; Auperin, D.D. Sequence analysis of the Ebola virus genome: Organization, genetic elements, and comparison with the genome of marburg virus. Virus Res. 1993, 29, 215–240. [Google Scholar] [CrossRef] [PubMed]
- Volchkova, V.A.; Dolnik, O.; Martinez, M.J.; Reynard, O.; Volchkov, V.E. Genomic RNA editing and its impact on Ebola virus adaptation during serial passages in cell culture and infection of guinea pigs. J. Infect. Dis. 2011, 204 (Suppl. 3), S941–S946. [Google Scholar] [CrossRef] [PubMed]
- Mehedi, M.; Hoenen, T.; Robertson, S.; Ricklefs, S.; Dolan, M.A.; Taylor, T.; Falzarano, D.; Ebihara, H.; Porcella, S.F.; Feldmann, H.; et al. Ebola virus RNA editing depends on the primary editing site sequence and an upstream secondary structure. PLoS Pathog. 2013, 9, e1003677. [Google Scholar] [CrossRef] [PubMed]
- Shabman, R.S.; Jabado, O.J.; Mire, C.E.; Stockwell, T.B.; Edwards, M.; Mahajan, M.; Geisbert, T.W.; Basler, C.F. Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells. mBio 2014, 5, e02011–e02014. [Google Scholar] [CrossRef] [PubMed]
- Radoshitzky, S.R.; Warfield, K.L.; Chi, X.; Dong, L.; Kota, K.; Bradfute, S.B.; Gearhart, J.D.; Retterer, C.; Kranzusch, P.J.; Misasi, J.N.; et al. Ebolavirus delta-peptide immunoadhesins inhibit marburgvirus and ebolavirus cell entry. J. Virol. 2011, 85, 8502–8513. [Google Scholar] [CrossRef] [PubMed]
- Mehedi, M.; Falzarano, D.; Seebach, J.; Hu, X.; Carpenter, M.S.; Schnittler, H.J.; Feldmann, H. A new Ebola virus nonstructural glycoprotein expressed through RNA editing. J. Virol. 2011, 85, 5406–5414. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Trappier, S.G.; Mahy, B.W.; Peters, C.J.; Nichol, S.T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. USA 1996, 93, 3602–3607. [Google Scholar] [CrossRef] [PubMed]
- Volchkov, V.E.; Becker, S.; Volchkova, V.A.; Ternovoj, V.A.; Kotov, A.N.; Netesov, S.V.; Klenk, H.D. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology 1995, 214, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Volchkov, V.E.; Volchkova, V.A.; Muhlberger, E.; Kolesnikova, L.V.; Weik, M.; Dolnik, O.; Klenk, H.D. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 2001, 291, 1965–1969. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, H.; Volchkov, V.E.; Volchkova, V.A.; Klenk, H.D. The glycoproteins of Marburg and Ebola virus and their potential roles in pathogenesis. Arch. Virol. Suppl. 1999, 15, 159–169. [Google Scholar] [PubMed]
- Volchkova, V.A.; Klenk, H.D.; Volchkov, V.E. Delta-peptide is the carboxy-terminal cleavage fragment of the nonstructural small glycoprotein sGP of Ebola virus. Virology 1999, 265, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Hyser, J.M.; Collinson-Pautz, M.R.; Utama, B.; Estes, M.K. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio 2010, 1. [Google Scholar] [CrossRef]
- Giorda, K.M.; Hebert, D.N. Viroporins customize host cells for efficient viral propagation. DNA Cell Biol. 2013, 32, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Microbiol. 2012, 10, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Ball, J.M.; Tian, P.; Zeng, C.Q.; Morris, A.P.; Estes, M.K. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 1996, 272, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Ball, J.M.; Schroeder, M.E.; Williams, C.V.; Schroeder, F.; Parr, R.D. Mutational analysis of the rotavirus NSP4 enterotoxic domain that binds to caveolin-1. Virol. J. 2013, 10, 336. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.E.; Estes, M.K. Viroporin-mediated calcium-activated autophagy. Autophagy 2013, 9, 797–798. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hyser, J.M.; Utama, B.; Crawford, S.E.; Broughman, J.R.; Estes, M.K. Activation of the endoplasmic reticulum calcium sensor stim1 and store-operated calcium entry by rotavirus requires NSP4 viroporin activity. J. Virol. 2013, 87, 13579–13588. [Google Scholar] [CrossRef] [PubMed]
- Hyser, J.M.; Utama, B.; Crawford, S.E.; Estes, M.K. Genetic divergence of rotavirus nonstructural protein 4 results in distinct serogroup-specific viroporin activity and intracellular punctate structure morphologies. J. Virol. 2012, 86, 4921–4934. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Meyer, J.C.; Bellamy, A.R.; Taylor, J.A. Rotavirus nonstructural glycoprotein NSP4 alters plasma membrane permeability in mammalian cells. J. Virol. 1997, 71, 9458–9465. [Google Scholar] [PubMed]
- Chacko, A.R.; Arifullah, M.; Sastri, N.P.; Jeyakanthan, J.; Ueno, G.; Sekar, K.; Read, R.J.; Dodson, E.J.; Rao, D.C.; Suguna, K.; et al. Novel pentameric structure of the diarrhea-inducing region of the rotavirus enterotoxigenic protein NSO4. J. Virol. 2011, 85, 12721–12732. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The clustal_x windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. Expasy: Sib bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Helical Wheel Projections. Available online: http://rzlab.ucr.edu/scripts/wheel/wheel.cgi (accessed on 11 November 2014).
- Snider, C.; Jayasinghe, S.; Hristova, K.; White, S.H. Mpex: A tool for exploring membrane proteins. Protein Sci. 2009, 18, 2624–2628. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L. The PyMol Molecular Graphics System. Available online: http://www.pymol.org/ (accessed on 11 November 2014).
- Gallaher, W.R. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 1996, 85, 477–478. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, W.R.; Ball, J.M.; Garry, R.F.; Griffin, M.C.; Montelaro, R.C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res. Hum. Retrovir. 1989, 5, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, W.R.; DiSimone, C.; Buchmeier, M.J. The viral transmembrane superfamily: Possible divergence of arenavirus and filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 2001, 1, 1. [Google Scholar] [CrossRef] [PubMed]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Honigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. Predictprotein—An open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, W337–W343. [Google Scholar] [CrossRef] [PubMed]
- Costin, J.M.; Rausch, J.M.; Garry, R.F.; Wimley, W.C. Viroporin potential of the lentivirus lytic peptide (llp) domains of the HIV-1 gp41 protein. Virol. J. 2007, 4, 123. [Google Scholar] [CrossRef] [PubMed]
- Baize, S.; Pannetier, D.; Oestereich, L.; Rieger, T.; Koivogui, L.; Magassouba, N.; Soropogui, B.; Sow, M.S.; Keita, S.; de Clerck, H. Emergence of Zaire Ebola virus disease in guinea. N. Engl. J. Med. 2014, 371, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Gire, S.K.; Goba, A.; Andersen, K.G.; Sealfon, R.S.; Park, D.J.; Kanneh, L.; Jalloh, S.; Momoh, M.; Fullah, M.; Dudas, G.; et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 2014, 345, 1369–1372. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.A.; Towner, J.S.; Sealy, T.K.; McMullan, L.K.; Khristova, M.L.; Burt, F.J.; Swanepoel, R.; Rollin, P.E.; Nichol, S.T. Molecular evolution of viruses of the family Filoviridae based on 97 whole-genome sequences. J. Virol. 2013, 87, 2608–2616. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, W.R. Detection of a fusion peptide sequence in the transmembrane protein of human immunodeficiency virus. Cell 1987, 50, 327–328. [Google Scholar] [CrossRef] [PubMed]
- Bucki, R.; Pastore, J.J.; Randhawa, P.; Vegners, R.; Weiner, D.J.; Janmey, P.A. Antibacterial activities of rhodamine B-conjugated gelsolin-derived peptides compared to those of the antimicrobial peptides cathelicidin LL37, magainin II, and melittin. Antimicrob. Agents Chemother. 2004, 48, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.; Bian, X.; Sun, S.; Hu, X.; Klemm, R.W.; Prinz, W.A.; Rapoport, T.A.; Hu, J. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. USA 2012, 109, E2146–E2154. [Google Scholar] [CrossRef] [PubMed]
- Yau, W.M.; Wimley, W.C.; Gawrisch, K.; White, S.H. The preference of tryptophan for membrane interfaces. Biochemistry 1998, 37, 14713–14718. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tolbert, W.D.; Ericksen, B.; Zhan, C.; Wu, X.; Yuan, W.; Li, X.; Pazgier, M.; Lu, W. Single, double and quadruple alanine substitutions at oligomeric interfaces identify hydrophobicity as the key determinant of human neutrophil alpha defensin hnp1 function. PLoS One 2013, 8, e78937. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, M.A.; Wong, G.; Kobinger, G.P.; Qiu, X. The multiple roles of sGP in Ebola pathogenesis. Viral Immunol. 2015, 1, 3–9. [Google Scholar] [CrossRef]
- Patel, H.; Huynh, Q.; Barlehner, D.; Heerklotz, H. Additive and synergistic membrane permeabilization by antimicrobial (lipo)peptides and detergents. Biophys. J. 2014, 106, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Bortolus, M.; Dalzini, A.; Toniolo, C.; Hahm, K.S.; Maniero, A.L. Interaction of hydrophobic and amphipathic antimicrobial peptides with lipid bicelles. J. Pept. Sci. 2014, 20, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.S.; Booth, L.N.; Knudsen, S.M.; Ellington, A.D. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA 2005, 11, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Tews, B.A.; Meyers, G. The pestivirus glycoprotein Erns is anchored in plane in the membrane via an amphipathic helix. J. Biol. Chem. 2007, 282, 32730–32741. [Google Scholar] [CrossRef] [PubMed]
- Garry, R.F. Unexpected similarity between the carboxyl termini of lentivirus and pestivirus envelope glycoproteins. AIDS 2003, 17, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.J.; Ballinger, M.J.; Zhan, J.J.; Hanzly, L.E.; Bruenn, J.A. Evidence that ebolaviruses and cuevaviruses have been diverging from marburgviruses since the miocene. PeerJ 2014, 2, e556. [Google Scholar] [CrossRef] [PubMed]
- Groseth, A.; Marzi, A.; Hoenen, T.; Herwig, A.; Gardner, D.; Becker, S.; Ebihara, H.; Feldmann, H. The Ebola virus glycoprotein contributes to but is not sufficient for virulence in vivo. PLoS Pathog. 2012, 8, e1002847. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.; Kobinger, G.P.; Qiu, X. Characterization of host immune responses in Ebola virus infections. Expert Rev. Clin. Immunol. 2014, 10, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Mohan, G.S.; Li, W.; Ye, L.; Compans, R.W.; Yang, C. Antigenic subversion: A novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 2012, 8, e1003065. [Google Scholar] [CrossRef] [PubMed]
- Kimberlin, C.R.; Bornholdt, Z.A.; Li, S.; Woods, V.L., Jr.; MacRae, I.J.; Saphire, E.O. Ebolavirus vp35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. USA 2010, 107, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Volchkov, V.E.; Blinov, V.M.; Netesov, S.V. The envelope glycoprotein of Ebola virus contains an immunosuppressive-like domain similar to oncogenic retroviruses. FEBS Lett. 1992, 305, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Perez, B.; Volchkova, V.A.; Dolnik, O.; Lawrence, P.; Volchkov, V.E. Shed GP of Ebola virus triggers immune activation and increased vascular permeability. PLoS Pathog. 2014, 10, e1004509. [Google Scholar] [CrossRef] [PubMed]
- Bratt, M.A.; Clavell, L.A. Hemolytic interaction of Newcastle disease virus and chicken erythrocytes. I. Quantitative comparison procedure. Appl. Microbol. 1972, 23, 454–460. [Google Scholar]
- Polos, P.G.; Gallaher, W.R. A quantitative assay for cytolysis induced by newcastle disease virus. J. Gen. Virol. 1981, 52, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Schafer, W.; Stroh, A.; Berghofer, S.; Seiler, J.; Vey, M.; Kruse, M.L.; Kern, H.F.; Klenk, H.D.; Garten, W. Two independent targeting signals in the cytoplasmic domain determine trans-golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 1995, 14, 2424–2435. [Google Scholar] [PubMed]
- Hu, F.; Luo, W.; Hong, M. Mechanisms of proton conduction and gating in influenza m2 proton channels from solid-state NMR. Science 2010, 330, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Dessen, A.; Volchkov, V.; Dolnik, O.; Klenk, H.D.; Weissenhorn, W. Crystal structure of the matrix protein vp40 from Ebola virus. EMBO J. 2000, 19, 4228–4236. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallaher, W.R.; Garry, R.F. Modeling of the Ebola Virus Delta Peptide Reveals a Potential Lytic Sequence Motif. Viruses 2015, 7, 285-305. https://doi.org/10.3390/v7010285
Gallaher WR, Garry RF. Modeling of the Ebola Virus Delta Peptide Reveals a Potential Lytic Sequence Motif. Viruses. 2015; 7(1):285-305. https://doi.org/10.3390/v7010285
Chicago/Turabian StyleGallaher, William R., and Robert F. Garry. 2015. "Modeling of the Ebola Virus Delta Peptide Reveals a Potential Lytic Sequence Motif" Viruses 7, no. 1: 285-305. https://doi.org/10.3390/v7010285
APA StyleGallaher, W. R., & Garry, R. F. (2015). Modeling of the Ebola Virus Delta Peptide Reveals a Potential Lytic Sequence Motif. Viruses, 7(1), 285-305. https://doi.org/10.3390/v7010285