Transport of the Influenza Virus Genome from Nucleus to Nucleus
Abstract
:1. Introduction
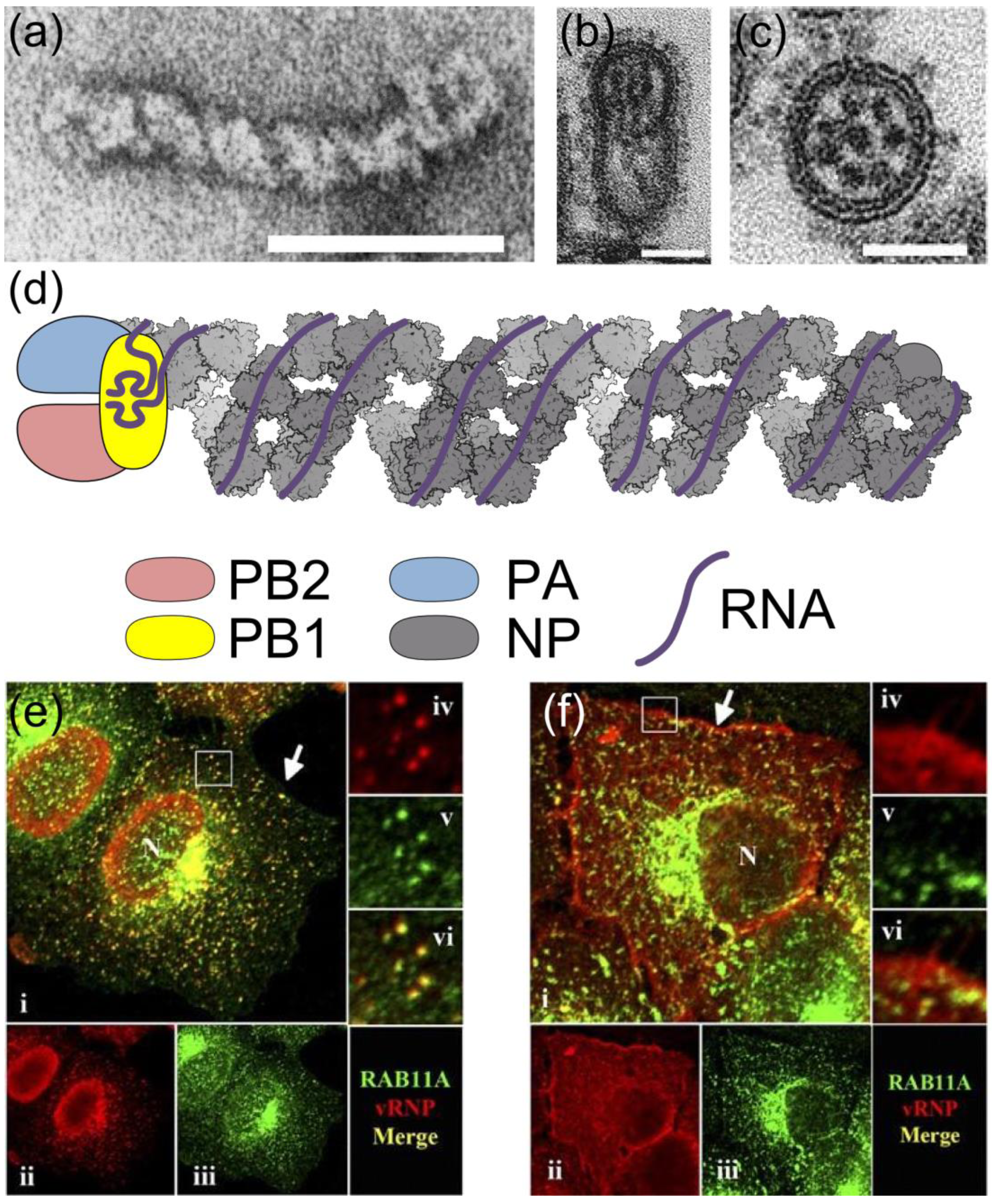
2. RNPs: The Basic Units of Orthomyxoviruses
3. Orthomyxoviruses: The Nuclear Family
4. From Synthesis to Virions
4.1. Synthesis
4.2. Nuclear Export

4.3. Trafficking to the Plasma Membrane
4.4. Virion Assembly
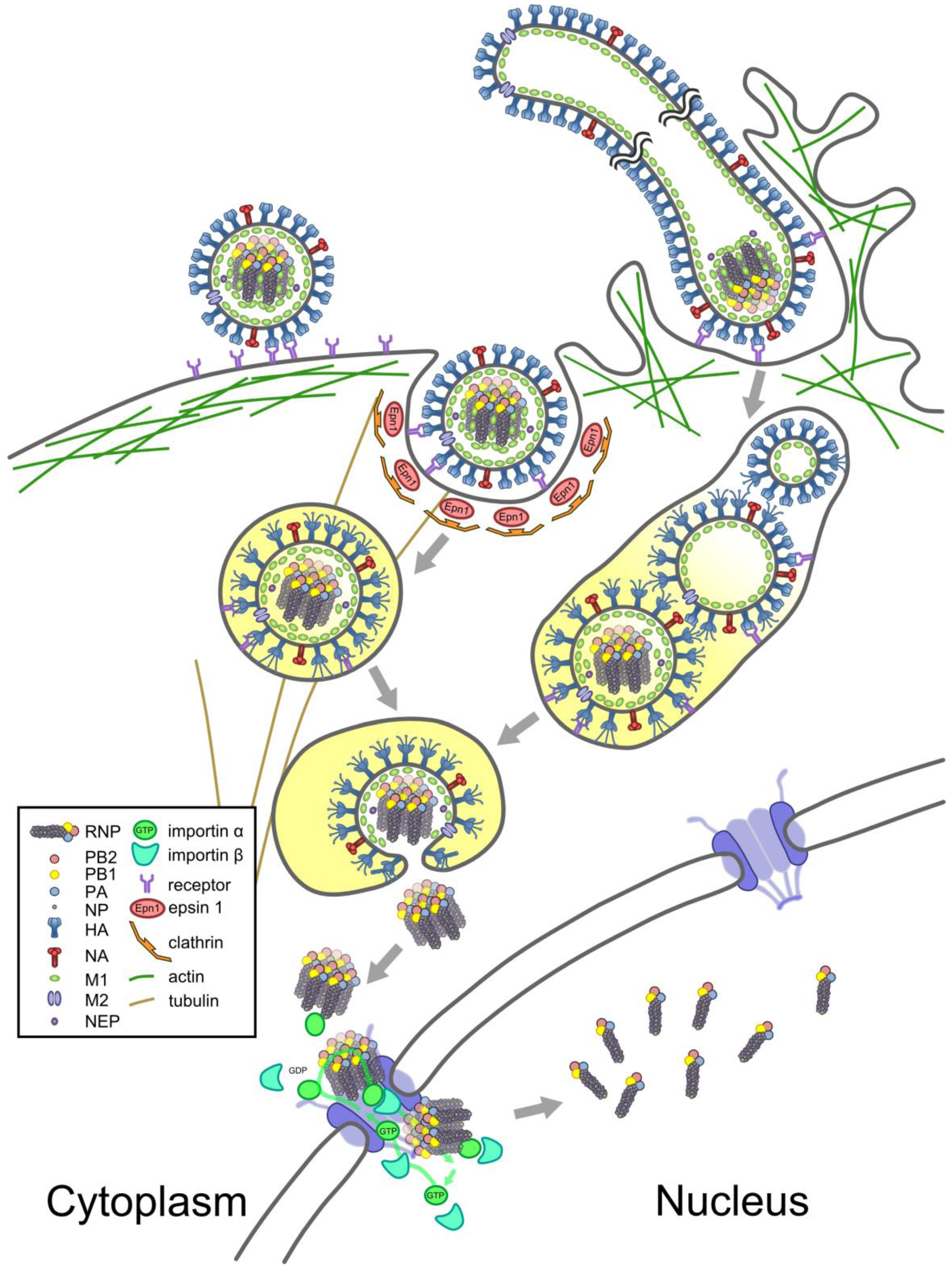
5. From Virions to the Nucleus
5.1. Entry
5.2. Nuclear Import
5.3. A New Cycle of Infection
Acknowledgments
Conflicts of Interest
References and Notes
- Shaw, M.; Palese, P. Orthomyxoviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Chapter 40; Volume 1, pp. 1151–1185. [Google Scholar]
- Pleschka, S.; Jaskunas, R.; Engelhardt, O.G.; Zurcher, T.; Palese, P.; Garcia-Sastre, A. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 1996, 70, 4188–4192. [Google Scholar]
- Fodor, E.; Crow, M.; Mingay, L.J.; Deng, T.; Sharps, J.; Fechter, P.; Brownlee, G.G. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 2002, 76, 8989–9001. [Google Scholar] [CrossRef]
- Mullin, A.E.; Dalton, R.M.; Amorim, M.J.; Elton, D.; Digard, P. Increased amounts of the influenza virus nucleoprotein do not promote higher levels of viral genome replication. J. Gen. Virol. 2004, 85, 3689–3698. [Google Scholar] [CrossRef]
- Kemler, I.; Whittaker, G.; Helenius, A. Nuclear import of microinjected influenza virus ribonucleoproteins. Virology 1994, 202, 1028–1033. [Google Scholar] [CrossRef]
- Fodor, E.; Devenish, L.; Engelhardt, O.G.; Palese, P.; Brownlee, G.G.; Garcia-Sastre, A. Rescue of influenza A virus from recombinant DNA. J. Virol. 1999, 73, 9679–9682. [Google Scholar]
- Neumann, G.; Watanabe, T.; Ito, H.; Watanabe, S.; Goto, H.; Gao, P.; Hughes, M.; Perez, D.R.; Donis, R.; Hoffmann, E.; et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 1999, 96, 9345–9350. [Google Scholar] [CrossRef]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef]
- Arranz, R.; Coloma, R.; Chichon, F.J.; Conesa, J.J.; Carrascosa, J.L.; Valpuesta, J.M.; Ortin, J.; Martin-Benito, J. The structure of native influenza virion ribonucleoproteins. Science 2012, 338, 1634–1637. [Google Scholar] [CrossRef]
- Moeller, A.; Kirchdoerfer, R.N.; Potter, C.S.; Carragher, B.; Wilson, I.A. Organization of the influenza virus replication machinery. Science 2012, 338, 1631–1634. [Google Scholar] [CrossRef]
- Ye, Q.; Krug, R.M.; Tao, Y.J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 2006, 444, 1078–1082. [Google Scholar] [CrossRef]
- Portela, A.; Digard, P. The influenza virus nucleoprotein: A multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002, 83, 723–734. [Google Scholar]
- Gorlich, D.; Kutay, U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol. 1999, 15, 607–660. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar]
- Jennings, P.A.; Finch, J.T.; Winter, G.; Robertson, J.S. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell 1983, 34, 619–627. [Google Scholar] [CrossRef]
- Noda, T.; Sagara, H.; Yen, A.; Takada, A.; Kida, H.; Cheng, R.H.; Kawaoka, Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 2006, 439, 490–492. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Kawakami, E.; Watanabe, T.; Neumann, G.; Kawaoka, Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 2011, 85, 6117–6126. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Tan, C.P.; Goubau, D.; Schulz, O.; Pichlmair, A.; Bier, K.; Robb, N.; Vreede, F.; Barclay, W.; Fodor, E.; et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 2010, 140, 397–408. [Google Scholar] [CrossRef]
- Engelhardt, O.G.; Smith, M.; Fodor, E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 2005, 79, 5812–5818. [Google Scholar] [CrossRef]
- Mayer, D.; Molawi, K.; Martinez-Sobrido, L.; Ghanem, A.; Thomas, S.; Baginsky, S.; Grossmann, J.; Garcia-Sastre, A.; Schwemmle, M. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 2007, 6, 672–682. [Google Scholar] [CrossRef]
- Rameix-Welti, M.A.; Tomoiu, A.; dos Santos Afonso, E.; van der Werf, S.; Naffakh, N. Avian influenza A virus polymerase association with nucleoprotein, but not polymerase assembly, is impaired in human cells during the course of infection. J. Virol. 2009, 83, 1320–1331. [Google Scholar]
- Rodriguez, A.; Perez-Gonzalez, A.; Nieto, A. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J. Virol. 2007, 81, 5315–5324. [Google Scholar] [CrossRef]
- Chan, A.Y.; Vreede, F.T.; Smith, M.; Engelhardt, O.G.; Fodor, E. Influenza virus inhibits RNA polymerase II elongation. Virology 2006, 351, 210–217. [Google Scholar] [CrossRef]
- Rodriguez, A.; Perez-Gonzalez, A.; Hossain, M.J.; Chen, L.M.; Rolling, T.; Perez-Brena, P.; Donis, R.; Kochs, G.; Nieto, A. Attenuated strains of influenza A viruses do not induce degradation of RNA polymerase II. J. Virol. 2009, 83, 11166–11174. [Google Scholar] [CrossRef]
- Vreede, F.T.; Chan, A.Y.; Sharps, J.; Fodor, E. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 2010, 396, 125–134. [Google Scholar] [CrossRef]
- Vreede, F.T.; Fodor, E. The role of the influenza virus RNA polymerase in host shut-off. Virulence 2010, 1, 436–439. [Google Scholar] [CrossRef]
- York, A.; Fodor, E. Biogenesis, assembly and export of viral messenger ribonucleoproteins in the influenza A virus infected cell. RNA Biol. 2013, 10, 1274–1282. [Google Scholar] [CrossRef]
- Lamb, R.A.; Choppin, P.W. Identification of a second protein (M2) encoded by RNA segment 7 of influenza virus. Virology 1981, 112, 729–737. [Google Scholar] [CrossRef]
- Wise, H.M.; Hutchinson, E.C.; Jagger, B.W.; Stuart, A.D.; Kang, Z.H.; Robb, N.; Schwartzman, L.M.; Kash, J.C.; Fodor, E.; Firth, A.E.; et al. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012, 8, e1002998. [Google Scholar] [CrossRef]
- Lamb, R.A.; Choppin, P.W. Segment 8 of the influenza virus genome is unique in coding for two polypeptides. Proc. Natl. Acad. Sci. USA 1979, 76, 4908–4912. [Google Scholar] [CrossRef]
- Selman, M.; Dankar, S.; Forbes, N.; JIa, J.-J.; Brown, E. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg. Microbes Infect. 2012, 1, e42. [Google Scholar] [CrossRef]
- David, A.; Dolan, B.P.; Hickman, H.D.; Knowlton, J.J.; Clavarino, G.; Pierre, P.; Bennink, J.R.; Yewdell, J.W. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J. Cell Biol. 2012, 197, 45–57. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; Fodor, E. Nuclear import of the influenza A virus transcriptional machinery. Vaccine 2012, 30, 7353–7358. [Google Scholar] [CrossRef]
- Resa-Infante, P.; Gabriel, G. The nuclear import machinery is a determinant of influenza virus host adaptation. Bioessays 2013, 35, 23–27. [Google Scholar] [CrossRef]
- Deng, T.; Sharps, J.; Fodor, E.; Brownlee, G.G. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J. Virol. 2005, 79, 8669–8674. [Google Scholar] [CrossRef]
- Fodor, E.; Smith, M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 2004, 78, 9144–9153. [Google Scholar] [CrossRef]
- Huet, S.; Avilov, S.V.; Ferbitz, L.; Daigle, N.; Cusack, S.; Ellenberg, J. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J. Virol. 2010, 84, 1254–1264. [Google Scholar] [CrossRef]
- Jones, I.M.; Reay, P.A.; Philpott, K.L. Nuclear location of all three influenza polymerase proteins and a nuclear signal in polymerase PB2. EMBO J. 1986, 5, 2371–2376. [Google Scholar]
- Loucaides, E.M.; von Kirchbach, J.C.; Foeglein, A.; Sharps, J.; Fodor, E.; Digard, P. Nuclear dynamics of influenza A virus ribonucleoproteins revealed by live-cell imaging studies. Virology 2009, 394, 154–163. [Google Scholar] [CrossRef]
- Resa-Infante, P.; Jorba, N.; Zamarreno, N.; Fernandez, Y.; Juarez, S.; Ortin, J. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One 2008, 3, e3904. [Google Scholar]
- Fislova, T.; Thomas, B.; Graef, K.M.; Fodor, E. Association of the influenza virus RNA polymerase subunit PB2 with the host chaperonin CCT. J. Virol. 2010, 84, 8691–8699. [Google Scholar] [CrossRef]
- Naito, T.; Momose, F.; Kawaguchi, A.; Nagata, K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 2007, 81, 1339–1349. [Google Scholar] [CrossRef]
- Momose, F.; Naito, T.; Yano, K.; Sugimoto, S.; Morikawa, Y.; Nagata, K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 2002, 277, 45306–45314. [Google Scholar]
- Chase, G.; Deng, T.; Fodor, E.; Leung, B.W.; Mayer, D.; Schwemmle, M.; Brownlee, G. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology 2008, 377, 431–439. [Google Scholar] [CrossRef]
- Boulo, S.; Akarsu, H.; Lotteau, V.; Muller, C.W.; Ruigrok, R.W.; Baudin, F. Human importin alpha and RNA do not compete for binding to influenza A virus nucleoprotein. Virology 2011, 409, 84–90. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; Orr, O.E.; Man Liu, S.; Engelhardt, O.G.; Fodor, E. Characterization of the interaction between the influenza A virus polymerase subunit PB1 and the host nuclear import factor Ran-binding protein 5. J. Gen. Virol. 2011, 92 (Pt 8), 1859–1869. [Google Scholar]
- Fodor, E. The RNA polymerase of influenza a virus: Mechanisms of viral transcription and replication. Acta Virol. 2013, 57, 113–122. [Google Scholar] [CrossRef]
- Ye, Q.; Guu, T.S.; Mata, D.A.; Kuo, R.L.; Smith, B.; Krug, R.M.; Tao, Y.J. Biochemical and structural evidence in support of a coherent model for the formation of the double-helical influenza A virus ribonucleoprotein. mBio 2012, 4, e00467-12. [Google Scholar]
- Shapiro, G.I.; Gurney, T., Jr.; Krug, R.M. Influenza virus gene expression: Control mechanisms at early and late times of infection and nuclear-cytoplasmic transport of virus-specific RNAs. J. Virol. 1987, 61, 764–773. [Google Scholar]
- Jorba, N.; Coloma, R.; Ortin, J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 2009, 5, e1000462. [Google Scholar] [CrossRef]
- Vreede, F.T.; Jung, T.E.; Brownlee, G.G. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 2004, 78, 9568–9572. [Google Scholar] [CrossRef]
- Hatada, E.; Hasegawa, M.; Mukaigawa, J.; Shimizu, K.; Fukuda, R. Control of influenza virus gene expression: Quantitative analysis of each viral RNA species in infected cells. J. Biochem. 1989, 105, 537–546. [Google Scholar]
- Robb, N.C.; Smith, M.; Vreede, F.T.; Fodor, E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 2009, 90, 1398–1407. [Google Scholar] [CrossRef]
- Tchatalbachev, S.; Flick, R.; Hobom, G. The packaging signal of influenza viral RNA molecules. Rna 2001, 7, 979–989. [Google Scholar] [CrossRef]
- Ye, Z.; Robinson, D.; Wagner, R.R. Nucleus-Targeting domain of the matrix protein (M1) of influenza virus. J. Virol. 1995, 69, 1964–1970. [Google Scholar]
- Baudin, F.; Petit, I.; Weissenhorn, W.; Ruigrok, R.W. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 2001, 281, 102–108. [Google Scholar] [CrossRef]
- Noton, S.L.; Medcalf, E.; Fisher, D.; Mullin, A.E.; Elton, D.; Digard, P. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 2007, 88, 2280–2290. [Google Scholar] [CrossRef]
- Ye, Z.; Liu, T.; Offringa, D.P.; McInnis, J.; Levandowski, R.A. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 1999, 73, 7467–7473. [Google Scholar]
- Wu, C.Y.; Jeng, K.S.; Lai, M.M. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J. Virol. 2011, 85, 6618–6628. [Google Scholar] [CrossRef]
- Wakefield, L.; Brownlee, G.G. RNA-Binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 1989, 17, 8569–8580. [Google Scholar] [CrossRef]
- Akarsu, H.; Burmeister, W.P.; Petosa, C.; Petit, I.; Muller, C.W.; Ruigrok, R.W.; Baudin, F. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J. 2003, 22, 4646–4655. [Google Scholar] [CrossRef]
- Shimizu, T.; Takizawa, N.; Watanabe, K.; Nagata, K.; Kobayashi, N. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP. FEBS Lett. 2011, 585, 41–46. [Google Scholar] [CrossRef]
- Ward, A.C.; Castelli, L.A.; Lucantoni, A.C.; White, J.F.; Azad, A.A.; Macreadie, I.G. Expression and analysis of the NS2 protein of influenza A virus. Arch. Virol. 1995, 140, 2067–2073. [Google Scholar] [CrossRef]
- Yasuda, J.; Nakada, S.; Kato, A.; Toyoda, T.; Ishihama, A. Molecular assembly of influenza virus: Association of the NS2 protein with virion matrix. Virology 1993, 196, 249–255. [Google Scholar] [CrossRef]
- Neumann, G.; Hughes, M.T.; Kawaoka, Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 2000, 19, 6751–6758. [Google Scholar] [CrossRef]
- O'Neill, R.E.; Talon, J.; Palese, P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998, 17, 288–296. [Google Scholar] [CrossRef]
- Huang, S.; Chen, J.; Chen, Q.; Wang, H.; Yao, Y.; Chen, J.; Chen, Z. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 2013, 87, 767–778. [Google Scholar] [CrossRef]
- Paterson, D.; Fodor, E. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 2012, 8, e1003019. [Google Scholar] [CrossRef]
- Bui, M.; Wills, E.G.; Helenius, A.; Whittaker, G.R. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 2000, 74, 1781–1786. [Google Scholar] [CrossRef]
- Elton, D.; Simpson-Holley, M.; Archer, K.; Medcalf, L.; Hallam, R.; McCauley, J.; Digard, P. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 2001, 75, 408–419. [Google Scholar] [CrossRef]
- Cao, S.; Liu, X.; Yu, M.; Li, J.; Jia, X.; Bi, Y.; Sun, L.; Gao, G.F.; Liu, W. A nuclear export signal in the matrix protein of Influenza A virus is required for efficient virus replication. J. Virol. 2012, 86, 4883–4891. [Google Scholar] [CrossRef]
- Yu, M.; Liu, X.; Cao, S.; Zhao, Z.; Zhang, K.; Xie, Q.; Chen, C.; Gao, S.; Bi, Y.; Sun, L.; et al. Identification and characterization of three novel nuclear export signals in the influenza A virus nucleoprotein. J. Virol. 2012, 86, 4970–4980. [Google Scholar]
- Chase, G.P.; Rameix-Welti, M.A.; Zvirbliene, A.; Zvirblis, G.; Gotz, V.; Wolff, T.; Naffakh, N.; Schwemmle, M. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS Pathog. 2011, 7, e1002187. [Google Scholar] [CrossRef]
- Hinshaw, V.S.; Olsen, C.W.; Dybdahl-Sissoko, N.; Evans, D. Apoptosis: A mechanism of cell killing by influenza A and B viruses. J. Virol. 1994, 68, 3667–3673. [Google Scholar]
- Wurzer, W.J.; Planz, O.; Ehrhardt, C.; Giner, M.; Silberzahn, T.; Pleschka, S.; Ludwig, S. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 2003, 22, 2717–2728. [Google Scholar] [CrossRef]
- Faleiro, L.; Lazebnik, Y. Caspases disrupt the nuclear-cytoplasmic barrier. J. Cell Biol. 2000, 151, 951–959. [Google Scholar] [CrossRef]
- Marjuki, H.; Alam, M.I.; Ehrhardt, C.; Wagner, R.; Planz, O.; Klenk, H.D.; Ludwig, S.; Pleschka, S. Membrane accumulation of influenza A virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Calpha-mediated activation of ERK signaling. J. Biol. Chem. 2006, 281, 16707–16715. [Google Scholar]
- Pleschka, S.; Wolff, T.; Ehrhardt, C.; Hobom, G.; Planz, O.; Rapp, U.R.; Ludwig, S. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 2001, 3, 301–305. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; Denham, E.M.; Thomas, B.; Trudgian, D.C.; Hester, S.S.; Ridlova, G.; York, A.; Turrell, L.; Fodor, E. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 2012, 8, e1002993. [Google Scholar] [CrossRef]
- Robb, N.C.; Jackson, D.; Vreede, F.T.; Fodor, E. Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J. Gen. Virol. 2010, 91, 2331–2340. [Google Scholar] [CrossRef]
- Chua, M.A.; Schmid, S.; Perez, J.T.; Langlois, R.A.; Tenoever, B.R. Influenza A virus utilizes suboptimal splicing to coordinate the timing of infection. Cell Rep. 2013, 3, 23–29. [Google Scholar] [CrossRef]
- Rodriguez Boulan, E.; Sabatini, D.D. Asymmetric budding of viruses in epithelial monlayers: A model system for study of epithelial polarity. Proc. Natl. Acad. Sci. USA 1978, 75, 5071–5075. [Google Scholar] [CrossRef]
- Elton, D.; Amorim, M.J.; Medcalf, L.; Digard, P. ‘Genome gating’; polarized intranuclear trafficking of influenza virus RNPs. Biol. Lett. 2005, 1, 113–117. [Google Scholar] [CrossRef]
- Bruce, E.A.; Digard, P.; Stuart, A.D. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 2010, 84, 5848–5859. [Google Scholar] [CrossRef]
- Jo, S.; Kawaguchi, A.; Takizawa, N.; Morikawa, Y.; Momose, F.; Nagata, K. Involvement of vesicular trafficking system in membrane targeting of the progeny influenza virus genome. Microbes Infect./Inst. Pasteur 2010, 12, 1079–1084. [Google Scholar] [CrossRef]
- Momose, F.; Kikuchi, Y.; Komase, K.; Morikawa, Y. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes Infect./Inst. Pasteur 2007, 9, 1422–1433. [Google Scholar] [CrossRef]
- Amorim, M.J.; Bruce, E.A.; Read, E.K.; Foeglein, A.; Mahen, R.; Stuart, A.D.; Digard, P. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 2011, 85, 4143–4156. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Matsumoto, K.; Nagata, K. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. J. Virol. 2012, 86, 11086–11095. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. Human immunodeficiency virus rev-binding protein is essential for influenza a virus replication and promotes genome trafficking in late-stage infection. J. Virol. 2011, 85, 9588–9598. [Google Scholar] [CrossRef]
- Momose, F.; Sekimoto, T.; Ohkura, T.; Jo, S.; Kawaguchi, A.; Nagata, K.; Morikawa, Y. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS One 2011, 6, e21123. [Google Scholar]
- Avilov, S.V.; Moisy, D.; Naffakh, N.; Cusack, S. Influenza A virus progeny vRNP trafficking in live infected cells studied with the virus-encoded fluorescently tagged PB2 protein. Vaccine 2012, 30, 7411–7417. [Google Scholar] [CrossRef]
- Bruce, E.A.; Stuart, A.; McCaffrey, M.W.; Digard, P. Role of the Rab11 pathway in negative-strand virus assembly. Biochem. Soc. Trans. 2012, 40, 1409–1415. [Google Scholar] [CrossRef]
- Konig, R.; Stertz, S.; Zhou, Y.; Inoue, A.; Hoffmann, H.H.; Bhattacharyya, S.; Alamares, J.G.; Tscherne, D.M.; Ortigoza, M.B.; Liang, Y.; et al. Human host factors required for influenza virus replication. Nature 2010, 463, 813–817. [Google Scholar] [CrossRef]
- Babcock, H.P.; Chen, C.; Zhuang, X. Using single-particle tracking to study nuclear trafficking of viral genes. Biophys. J. 2004, 87, 2749–2758. [Google Scholar] [CrossRef]
- Shaw, M.L.; Stone, K.L.; Colangelo, C.M.; Gulcicek, E.E.; Palese, P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008, 4, e1000085. [Google Scholar] [CrossRef]
- Chou, Y.Y.; Heaton, N.S.; Gao, Q.; Palese, P.; Singer, R.; Lionnet, T. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog. 2013, 9, e1003358. [Google Scholar] [CrossRef]
- Fournier, E.; Moules, V.; Essere, B.; Paillart, J.C.; Sirbat, J.D.; Cavalier, A.; Rolland, J.P.; Thomas, D.; Lina, B.; Isel, C.; et al. Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine 2012, 30, 7359–7367. [Google Scholar] [CrossRef]
- Fournier, E.; Moules, V.; Essere, B.; Paillart, J.C.; Sirbat, J.D.; Isel, C.; Cavalier, A.; Rolland, J.P.; Thomas, D.; Lina, B.; et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 2012, 40, 2197–2209. [Google Scholar] [CrossRef]
- Gavazzi, C.; Isel, C.; Fournier, E.; Moules, V.; Cavalier, A.; Thomas, D.; Lina, B.; Marquet, R. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: Comparison with a human H3N2 virus. Nucleic Acids Res. 2013, 41, 1241–1254. [Google Scholar]
- Noda, T.; Sugita, Y.; Aoyama, K.; Hirase, A.; Kawakami, E.; Miyazawa, A.; Sagara, H.; Kawaoka, Y. Three-Dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 2012, 3, 639. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; von Kirchbach, J.C.; Gog, J.R.; Digard, P. Genome packaging in influenza A virus. J. Gen. Virol. 2010, 91, 313–328. [Google Scholar] [CrossRef]
- Goto, H.; Muramoto, Y.; Noda, T.; Kawaoka, Y. The genome packaging signal of the influenza A virus genome comprises a genome incorporation signal and a genome bundling signal. J. Virol. 2013. [Google Scholar] [CrossRef]
- Gao, Q.; Chou, Y.Y.; Doganay, S.; Vafabakhsh, R.; Ha, T.; Palese, P. The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging. J. Virol. 2012, 86, 7043–7051. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; Curran, M.D.; Read, E.K.; Gog, J.R.; Digard, P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 2008, 82, 11869–11879. [Google Scholar] [CrossRef]
- Muramoto, Y.; Takada, A.; Fujii, K.; Noda, T.; Iwatsuki-Horimoto, K.; Watanabe, S.; Horimoto, T.; Kida, H.; Kawaoka, Y. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 2006, 80, 2318–2325. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; Wise, H.M.; Kudryavtseva, K.; Curran, M.D.; Digard, P. Characterisation of influenza A viruses with mutations in segment 5 packaging signals. Vaccine 2009, 27, 6270–6275. [Google Scholar] [CrossRef]
- Marsh, G.A.; Hatami, R.; Palese, P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 2007, 81, 9727–9736. [Google Scholar] [CrossRef]
- Marsh, G.A.; Rabadan, R.; Levine, A.J.; Palese, P. Highly conserved regions of influenza a virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 2008, 82, 2295–2304. [Google Scholar] [CrossRef]
- Brooke, C.B.; Ince, W.L.; Wrammert, J.; Ahmed, R.; Wilson, P.C.; Bennink, J.R.; Yewdell, J.W. Most influenza a virions fail to express at least one essential viral protein. J. Virol. 2013, 87, 3155–3162. [Google Scholar] [CrossRef]
- Takizawa, N.; Kumakura, M.; Takeuchi, K.; Kobayashi, N.; Nagata, K. Sorting of influenza A virus RNA genome segments after nuclear export. Virology 2010, 401, 248–256. [Google Scholar] [CrossRef]
- Marshall, N.; Priyamvada, L.; Ende, Z.; Steel, J.; Lowen, A.C. Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog. 2013, 9, e1003421. [Google Scholar] [CrossRef]
- Barry, R.D. The multiplication of influenza virus. II. Multiplicity reactivation of ultraviolet irradiated virus. Virology 1961, 14, 398–405. [Google Scholar]
- Sugiura, A.; Tobita, K.; Kilbourne, E.D. Isolation and preliminary characterization of temperature-sensitive mutants of influenza virus. J. Virol. 1972, 10, 639–647. [Google Scholar]
- Blumenkrantz, D.; Roberts, K.L.; Shelton, H.; Lycett, S.; Barclay, W.S. The short stalk length of HPAI H5N1 influenza neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J. Virol. 2013. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar]
- Simonsen, L.; Viboud, C.; Grenfell, B.T.; Dushoff, J.; Jennings, L.; Smit, M.; Macken, C.; Hata, M.; Gog, J.; Miller, M.A.; et al. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol. Biol. Evol. 2007, 24, 1811–1820. [Google Scholar] [CrossRef]
- Rossman, J.S.; Lamb, R.A. Influenza virus assembly and budding. Virology 2011, 411, 229–236. [Google Scholar] [CrossRef]
- Gorai, T.; Goto, H.; Noda, T.; Watanabe, T.; Kozuka-Hata, H.; Oyama, M.; Takano, R.; Neumann, G.; Watanabe, S.; Kawaoka, Y. F1Fo-ATPase, F-type proton-translocating ATPase, at the plasma membrane is critical for efficient influenza virus budding. Proc. Natl. Acad. Sci. USA 2012, 109, 4615–4620. [Google Scholar] [CrossRef]
- Hui, E.K.; Nayak, D.P. Role of ATP in influenza virus budding. Virology 2001, 290, 329–341. [Google Scholar] [CrossRef]
- Hui, E.K.; Nayak, D.P. Role of G protein and protein kinase signalling in influenza virus budding in MDCK cells. J. Gen. Virol. 2002, 83, 3055–3066. [Google Scholar]
- Gujuluva, C.N.; Kundu, A.; Murti, K.G.; Nayak, D.P. Abortive replication of influenza virus A/WSN/33 in HeLa229 cells: Defective viral entry and budding processes. Virology 1994, 204, 491–505. [Google Scholar] [CrossRef]
- Demirov, D.; Gabriel, G.; Schneider, C.; Hohenberg, H.; Ludwig, S. Interaction of influenza A virus matrix protein with RACK1 is required for virus release. Cell. Microbiol. 2012, 14, 774–789. [Google Scholar] [CrossRef]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef]
- Chu, C.M.; Dawson, I.M.; Elford, W.J. Filamentous forms associated with newly isolated influenza virus. Lancet 1949, 1, 602. [Google Scholar]
- Kilbourne, E.D.; Murphy, J.S. Genetic studies of influenza viruses. I. Viral morphology and growth capacity as exchangeable genetic traits. Rapid in ovo adaptation of early passage Asian strain isolates by combination with PR8. J. Exp. Med. 1960, 111, 387–406. [Google Scholar] [CrossRef]
- Itoh, Y.; Shinya, K.; Kiso, M.; Watanabe, T.; Sakoda, Y.; Hatta, M.; Muramoto, Y.; Tamura, D.; Sakai-Tagawa, Y.; Noda, T.; et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 2009, 460, 1021–1025. [Google Scholar]
- Calder, L.J.; Wasilewski, S.; Berriman, J.A.; Rosenthal, P.B. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. USA 2010, 107, 10685–10690. [Google Scholar]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Balannik, V.; Pinto, L.H.; Lamb, R.A. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 2010, 84, 5078–5088. [Google Scholar]
- Vijayakrishnan, S.; Loney, C.; Jackson, D.; Suphamungmee, W.; Rixon, F.J.; Bhella, D. Cryotomography of budding influenza A virus reveals filaments with diverse morphologies that mostly do not bear a genome at their distal end. PLoS Pathog. 2013, 9, e1003413. [Google Scholar]
- Roberts, P.C.; Compans, R.W. Host cell dependence of viral morphology. Proc. Natl. Acad. Sci. USA 1998, 95, 5746–5751. [Google Scholar] [CrossRef]
- Simpson-Holley, M.; Ellis, D.; Fisher, D.; Elton, D.; McCauley, J.; Digard, P. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 2002, 301, 212–225. [Google Scholar]
- Choppin, P.W.; Murphy, J.S.; Tamm, I. Studies of two kinds of virus particles which comprise influenza A2 virus strains. III. Morphological characteristics: independence to morphological and functional traits. J. Exp. Med. 1960, 112, 945–952. [Google Scholar] [CrossRef]
- Ada, G.L.; Perry, B.T.; Abbot, A. Biological and physical properties of the Ryan strain of filamentous influenza virus. J. Gen. Microbiol. 1958, 19, 23–39. [Google Scholar] [CrossRef]
- Burnet, F.M.; Lind, P.E. Studies on filamentary forms of influenza virus with special reference to the use of dark-ground-microscopy. Archiv. Fur Die Gesamte Virusforsch. 1957, 7, 413–428. [Google Scholar]
- Portocala, R.; Dumitrescu, S.; Rothschild, L.; Ionescu, N.I. Morphological characteristics of type A2 (“A-Asia”) influenza virus isolated in Rumania. Acta Virol. 1959, 3, 113–114. [Google Scholar]
- Harris, A.; Cardone, G.; Winkler, D.C.; Heymann, J.B.; Brecher, M.; White, J.M.; Steven, A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA 2006, 103, 19123–19127. [Google Scholar]
- Laver, W.G.; Downie, J.C. Influenza virus recombination. I. Matrix protein markers and segregation during mixed infections. Virology 1976, 70, 105–117. [Google Scholar] [CrossRef]
- Lubeck, M.D.; Palese, P.; Schulman, J.L. Nonrandom association of parental genes in influenza A virus recombinants. Virology 1979, 95, 269–274. [Google Scholar] [CrossRef]
- Nakajima, K.; Sugiura, A. Three-Factor cross of influenza virus. Virology 1977, 81, 486–489. [Google Scholar] [CrossRef]
- Varich, N.L.; Gitelman, A.K.; Shilov, A.A.; Smirnov, Y.A.; Kaverin, N.V. Deviation from the random distribution pattern of influenza A virus gene segments in reassortants produced under non-selective conditions. Arch. Virol. 2008, 153, 1149–1154. [Google Scholar] [CrossRef]
- Chou, Y.Y.; Vafabakhsh, R.; Doganay, S.; Gao, Q.; Ha, T.; Palese, P. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc. Natl. Acad. Sci. USA 2012, 109, 9101–9106. [Google Scholar]
- Gao, Q.; Brydon, E.W.; Palese, P. A seven-segmented influenza A virus expressing the influenza C virus glycoprotein HEF. J. Virol. 2008, 82, 6419–6426. [Google Scholar]
- Moules, V.; Ferraris, O.; Terrier, O.; Giudice, E.; Yver, M.; Rolland, J.P.; Bouscambert-Duchamp, M.; Bergeron, C.; Ottmann, M.; Fournier, E.; et al. In vitro characterization of naturally occurring influenza H3NA- viruses lacking the NA gene segment: Toward a new mechanism of viral resistance? Virology 2010, 404, 215–224. [Google Scholar] [CrossRef]
- Rossman, J.S.; Leser, G.P.; Lamb, R.A. Filamentous influenza virus enters cells via macropinocytosis. J. Virol. 2012, 86, 10950–10960. [Google Scholar] [CrossRef]
- Sieczkarski, S.B.; Whittaker, G.R. Characterization of the host cell entry of filamentous influenza virus. Arch. Virol. 2005, 150, 1783–1796. [Google Scholar] [CrossRef]
- De Vries, E.; Tscherne, D.M.; Wienholts, M.J.; Cobos-Jimenez, V.; Scholte, F.; Garcia-Sastre, A.; Rottier, P.J.; de Haan, C.A. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011, 7, e1001329. [Google Scholar] [CrossRef]
- Chen, C.; Zhuang, X. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc. Natl. Acad. Sci. USA 2008, 105, 11790–11795. [Google Scholar] [CrossRef]
- Lakadamyali, M.; Rust, M.J.; Zhuang, X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell 2006, 124, 997–1009. [Google Scholar] [CrossRef]
- Eierhoff, T.; Hrincius, E.R.; Rescher, U.; Ludwig, S.; Ehrhardt, C. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 2010, 6, e1001099. [Google Scholar] [CrossRef]
- De Conto, F.; Covan, S.; Arcangeletti, M.C.; Orlandini, G.; Gatti, R.; Dettori, G.; Chezzi, C. Differential infectious entry of human influenza A/NWS/33 virus (H1N1) in mammalian kidney cells. Virus Res. 2011, 155, 221–230. [Google Scholar] [CrossRef]
- Roy, A.M.; Parker, J.S.; Parrish, C.R.; Whittaker, G.R. Early stages of influenza virus entry into Mv-1 lung cells: Involvement of dynamin. Virology 2000, 267, 17–28. [Google Scholar]
- Yoshimura, A.; Ohnishi, S. Uncoating of influenza virus in endosomes. J. Virol. 1984, 51, 497–504. [Google Scholar]
- Pinto, L.H.; Lamb, R.A. The M2 proton channels of influenza A and B viruses. J. Biol. Chem. 2006, 281, 8997–9000. [Google Scholar] [CrossRef]
- Bui, M.; Whittaker, G.; Helenius, A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 1996, 70, 8391–8401. [Google Scholar]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef]
- Martin, K.; Helenius, A. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 1991, 65, 232–244. [Google Scholar]
- Koff, W.C.; Knight, V. Inhibition of influenza virus uncoating by rimantadine hydrochloride. J. Virol. 1979, 31, 261–263. [Google Scholar]
- Nevalainen, M.; Nissinen, M.; Kaakinen, M.; Metsikko, K. Influenza virus infection in multinucleated skeletal myofibers. Exp. Cell Res. 2010, 316, 1784–1794. [Google Scholar] [CrossRef]
- Martin, K.; Helenius, A. Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell 1991, 67, 117–130. [Google Scholar] [CrossRef]
- Whittaker, G.; Bui, M.; Helenius, A. Nuclear trafficking of influenza virus ribonuleoproteins in heterokaryons. J. Virol. 1996, 70, 2743–2756. [Google Scholar]
- Wu, W.W.; Pante, N. The directionality of the nuclear transport of the influenza A genome is driven by selective exposure of nuclear localization sequences on nucleoprotein. Virol. J. 2009, 6, 68. [Google Scholar] [CrossRef]
- O’Neill, R.E.; Jaskunas, R.; Blobel, G.; Palese, P.; Moroianu, J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J. Biol. Chem. 1995, 270, 22701–22704. [Google Scholar]
- Wu, W.W.; Sun, Y.H.; Pante, N. Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol. J. 2007, 4, 49. [Google Scholar] [CrossRef]
- Wu, W.W.; Weaver, L.L.; Pante, N. Ultrastructural analysis of the nuclear localization sequences on influenza A ribonucleoprotein complexes. J. Mol. Biol. 2007, 374, 910–916. [Google Scholar] [CrossRef]
- Cros, J.F.; Garcia-Sastre, A.; Palese, P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 2005, 6, 205–213. [Google Scholar] [CrossRef]
- Ozawa, M.; Fujii, K.; Muramoto, Y.; Yamada, S.; Yamayoshi, S.; Takada, A.; Goto, H.; Horimoto, T.; Kawaoka, Y. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 2007, 81, 30–41. [Google Scholar] [CrossRef]
- Hudjetz, B.; Gabriel, G. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-alpha1 and -alpha7. PLoS Pathog. 2012, 8, e1002488. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hutchinson, E.C.; Fodor, E. Transport of the Influenza Virus Genome from Nucleus to Nucleus. Viruses 2013, 5, 2424-2446. https://doi.org/10.3390/v5102424
Hutchinson EC, Fodor E. Transport of the Influenza Virus Genome from Nucleus to Nucleus. Viruses. 2013; 5(10):2424-2446. https://doi.org/10.3390/v5102424
Chicago/Turabian StyleHutchinson, Edward C., and Ervin Fodor. 2013. "Transport of the Influenza Virus Genome from Nucleus to Nucleus" Viruses 5, no. 10: 2424-2446. https://doi.org/10.3390/v5102424
APA StyleHutchinson, E. C., & Fodor, E. (2013). Transport of the Influenza Virus Genome from Nucleus to Nucleus. Viruses, 5(10), 2424-2446. https://doi.org/10.3390/v5102424