Evasion of Influenza A Viruses from Innate and Adaptive Immune Responses
Abstract
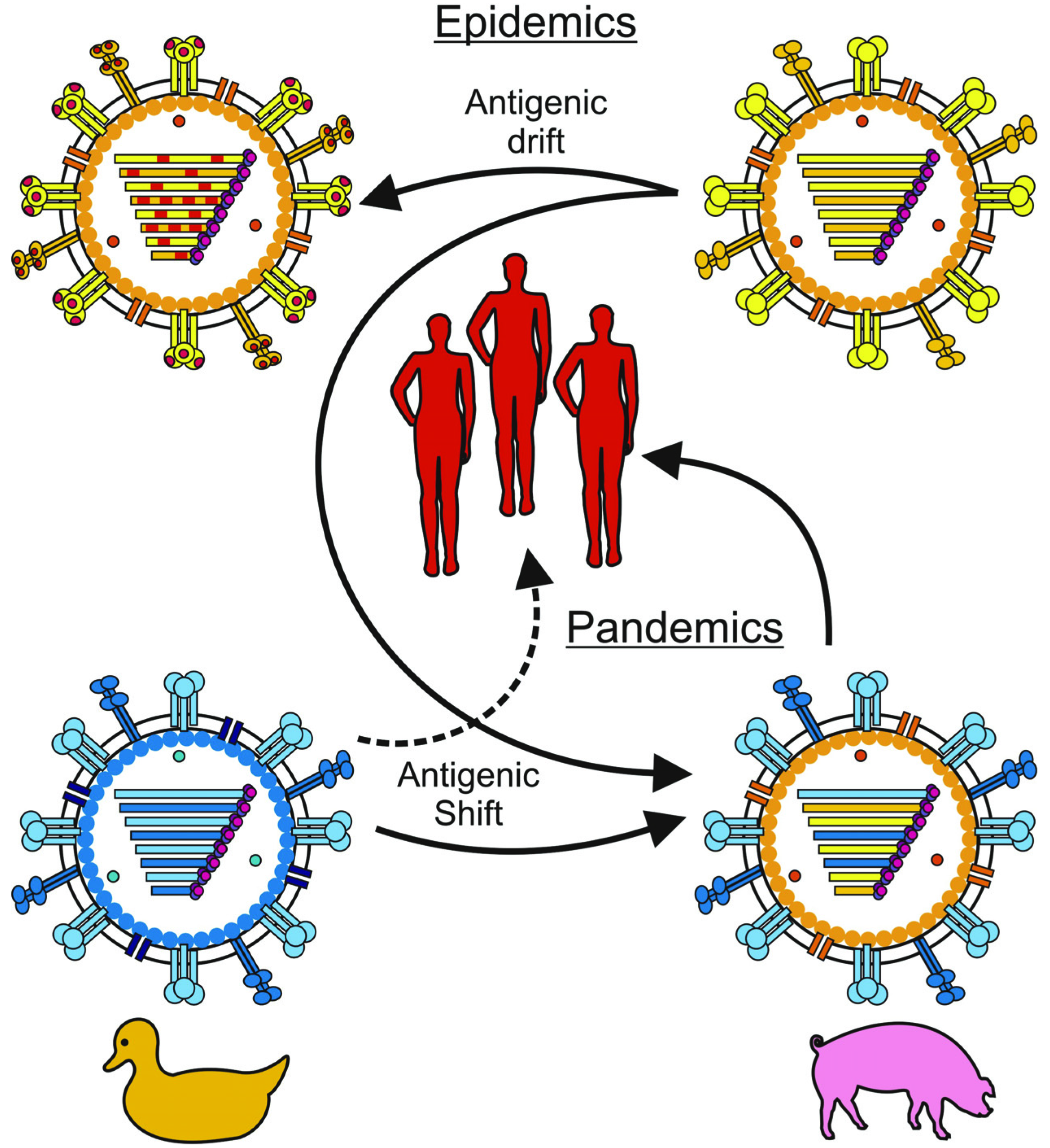
:1. Introduction
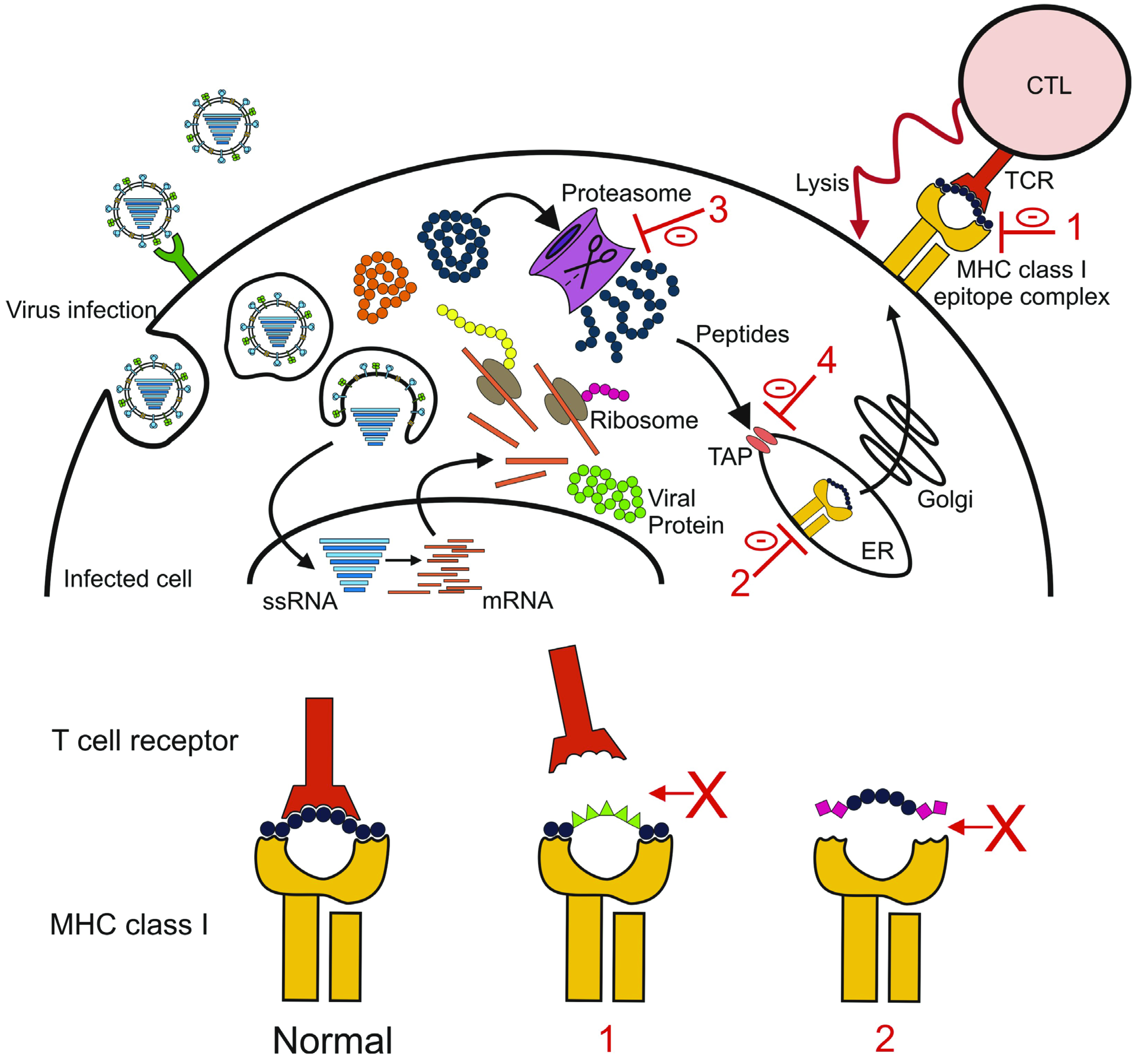
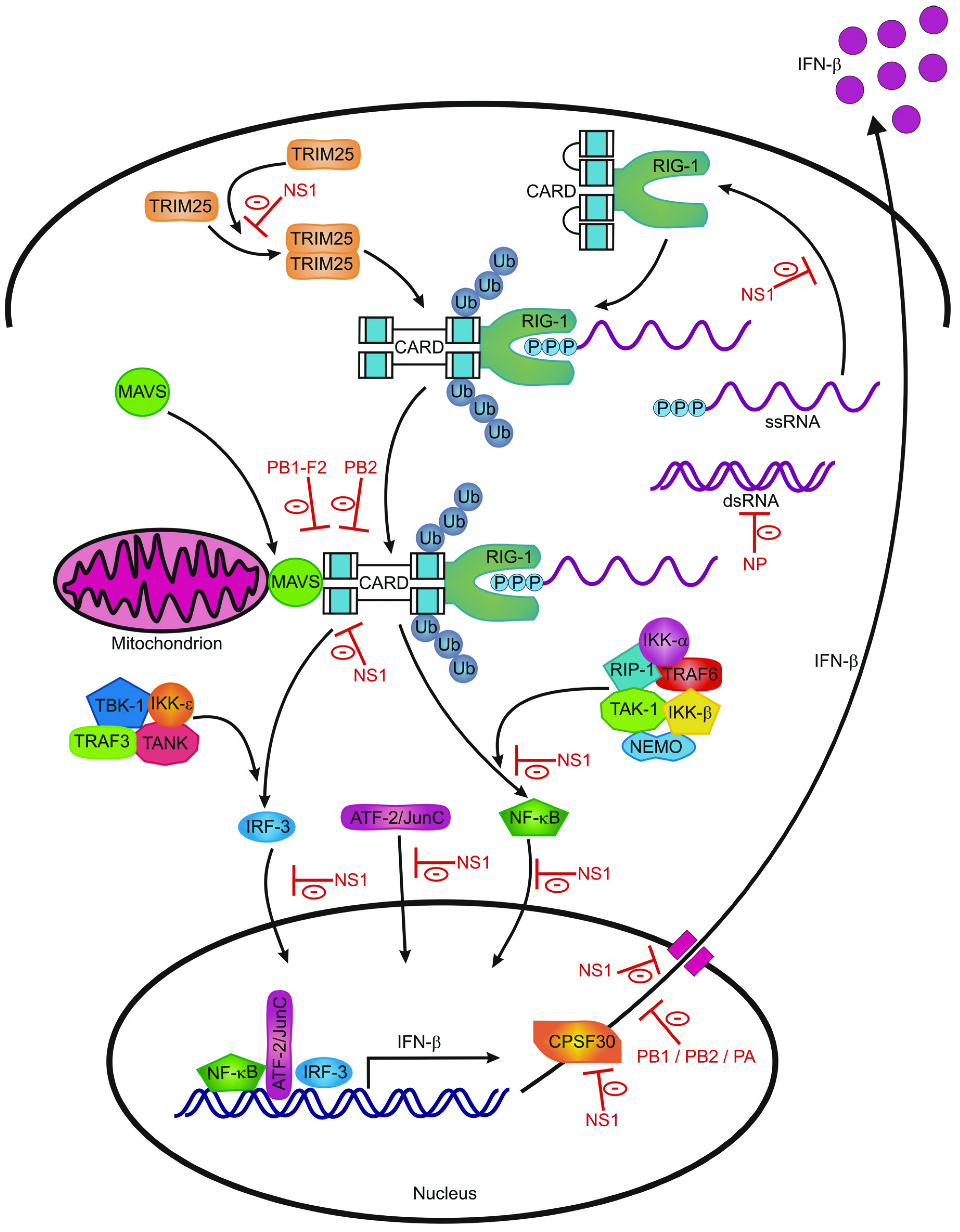
2. Innate Immunity
2.1. Sensing Of Influenza Virus Infection by Receptors of the Innate Immune System

| Protein | Function | Reference |
|---|---|---|
| MxA (Myxovirus resistance gene A) | Inhibits viral replication by interfering with the viral ribonucleoprotein structure | [54,55,56] |
| PKR (Protein kinase R) | Limits viral replication by blocking general translation | [57,58] |
| OAS (2'–5'oligoadenylate synthetase) | Stops viral replication by means of activating RNAseL which results in degradation of viral and cellular RNA and eventually apoptosis of the virus infected cell | [59,60] |
| ISG15 (IFN-stimulated gene 15) | Regulates a number of IFN-stimulated proteins | [61] |
| Viperin | Inhibits viral release by interfering with viral budding | [62] |
| Tetherin | Inhibits formation of influenza virus particles | [63,64] |
| IFITMs | Restrict viral entry | [65] |
2.2. Macrophages
2.3. Natural Killer Cells
2.4. Dendritic Cells
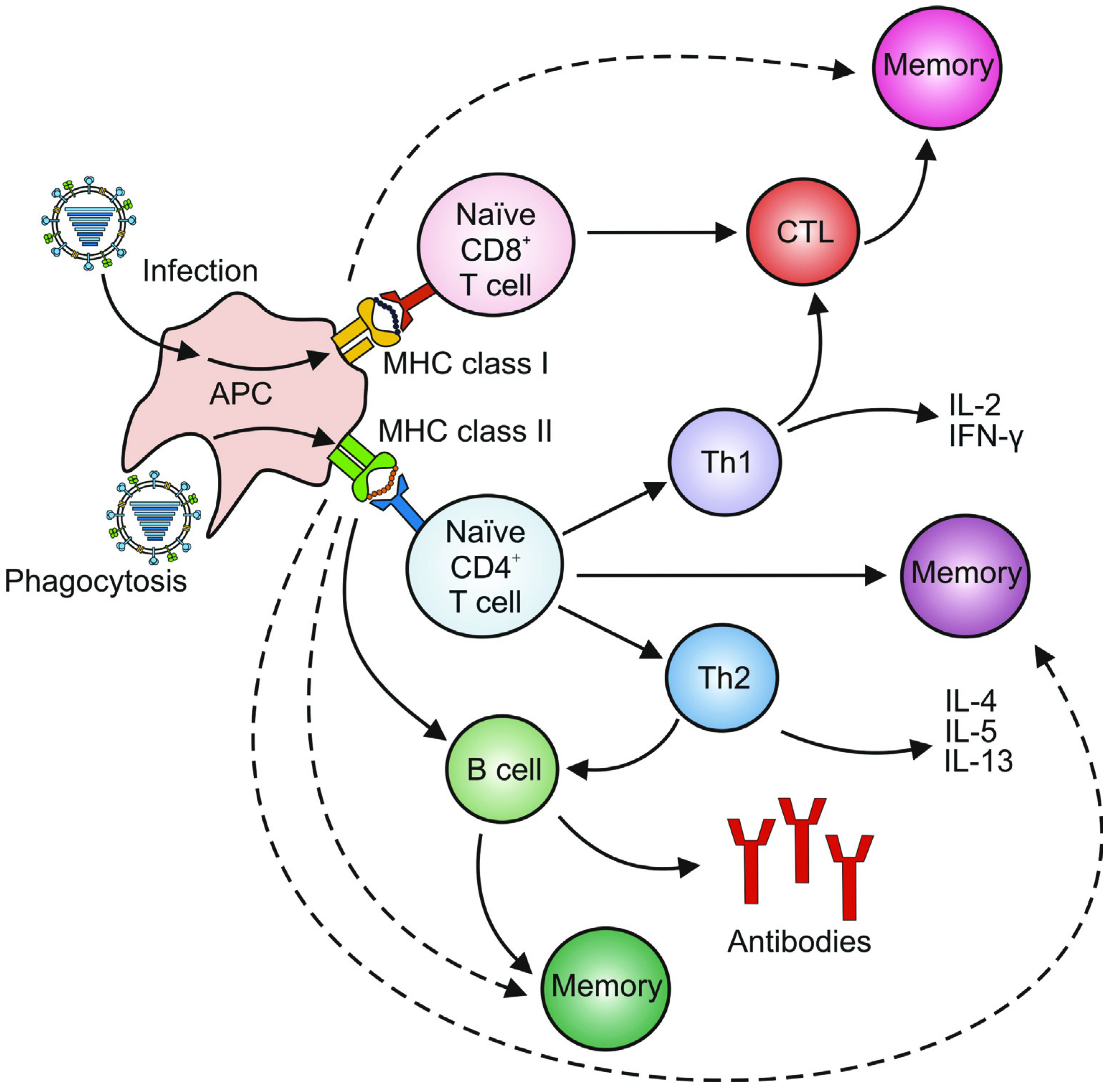
3. Adaptive Immunity
3.1. Humoral Immunity
3.2. Cellular Immunity
3.2.1. CD4+ T Cells
3.2.2. CD8+ T Cells
3.2.3. Regulatory T Cells and Th17 Cells
5. Implications for Vaccine Development
5.1. Current Influenza Vaccines
5.2. Novel Vaccines
6. Concluding Remarks
References and Notes
- Palese, P.; Shaw, M.L. Orthomyxoviridae: The viruses and their replication. In Fields Virology, 5th ed; Lippincott Williams & Wilkins, a Wolters Kluwer Business: Philadelphia, PA, USA, 2007; Volume 2, pp. 1647–1689. [Google Scholar]
- WHO. Influenza (seasonal) fact sheet No 211. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/index.html (accessed on 31 March 2012).
- Stohr, K. Influenza—WHO cares. Lancet Infect. Dis. 2002, 2, 517. [Google Scholar] [CrossRef]
- Jagger, B.W.; Wise, H.M.; Kash, J.C.; Walters, K.A.; Wills, N.M.; Xiao, Y.L.; Dunfee, R.L.; Schwartzman, L.M.; Ozinsky, A.; Bell, G.L.; et al. An overlapping protein-coding region in influenza a virus segment 3 modulates the host response. Science 2012, 337, 199–204. [Google Scholar] [CrossRef]
- Fouchier, R.A.; Munster, V.; Wallensten, A.; Bestebroer, T.M.; Herfst, S.; Smith, D.; Rimmelzwaan, G.F.; Olsen, B.; Osterhaus, A.D. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 2005, 79, 2814–2822. [Google Scholar]
- WHO. A revision of the system of nomenclature for influenza viruses: A WHO memorandum. Bull World Health Organ. 1980, 58, 585–591.
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 4269–4274. [Google Scholar]
- Smith, D.J.; Lapedes, A.S.; de Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Fouchier, R.A. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar]
- Herfst, S.; Schrauwen, E.J.A.; Linster, M.; Chutinimitkul, S.; de Wit, E.; Munster, V.J.; Sorrell, E.M.; Bestebroer, T.M.; Burke, D.F.; Smith, D.J.; et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 2012, 336, 1534–1541. [Google Scholar]
- De Jong, J.C.; Rimmelzwaan, G.F.; Fouchier, R.A.; Osterhaus, A.D. Influenza virus: A master of metamorphosis. J. Infect. 2000, 40, 218–228. [Google Scholar] [CrossRef]
- Johnson, N.P.; Mueller, J. Updating the accounts: Global mortality of the 1918-1920 "Spanish" influenza pandemic. Bull. Hist. Med. 2002, 76, 105–115. [Google Scholar] [CrossRef]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201. [Google Scholar]
- de Wit, E.; Kawaoka, Y.; de Jong, M.D.; Fouchier, R.A. Pathogenicity of highly pathogenic avian influenza virus in mammals. Vaccine 2008, 26, D54–D58. [Google Scholar] [CrossRef]
- Kuiken, T.; Holmes, E.C.; McCauley, J.; Rimmelzwaan, G.F.; Williams, C.S.; Grenfell, B.T. Host species barriers to influenza virus infections. Science 2006, 312, 394–397. [Google Scholar]
- Lin, Y.P.; Shaw, M.; Gregory, V.; Cameron, K.; Lim, W.; Klimov, A.; Subbarao, K.; Guan, Y.; Krauss, S.; Shortridge, K.; et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: Relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 9654–9658. [Google Scholar]
- Fouchier, R.A.; Schneeberger, P.M.; Rozendaal, F.W.; Broekman, J.M.; Kemink, S.A.; Munster, V.; Kuiken, T.; Rimmelzwaan, G.F.; Schutten, M.; Van Doornum, G.J.; et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 1356–1361. [Google Scholar]
- Koopmans, M.; Wilbrink, B.; Conyn, M.; Natrop, G.; van der Nat, H.; Vennema, H.; Meijer, A.; van Steenbergen, J.; Fouchier, R.; Osterhaus, A.; et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 2004, 363, 587–593. [Google Scholar]
- de Jong, J.C.; Claas, E.C.; Osterhaus, A.D.; Webster, R.G.; Lim, W.L. A pandemic warning? Nature 1997, 389, 554. [Google Scholar]
- de Jong, M.D.; Bach, V.C.; Phan, T.Q.; Vo, M.H.; Tran, T.T.; Nguyen, B.H.; Beld, M.; Le, T.P.; Truong, H.K.; Nguyen, V.V.; et al. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. New Engl. J. Med. 2005, 352, 686–691. [Google Scholar]
- Gambotto, A.; Barratt-Boyes, S.M.; de Jong, M.D.; Neumann, G.; Kawaoka, Y. Human infection with highly pathogenic H5N1 influenza virus. Lancet 2008, 371, 1464–1475. [Google Scholar] [CrossRef]
- Abdel-Ghafar, A.N.; Chotpitayasunondh, T.; Gao, Z.; Hayden, F.G.; Nguyen, D.H.; de Jong, M.D.; Naghdaliyev, A.; Peiris, J.S.; Shindo, N.; Soeroso, S.; et al. Update on avian influenza A (H5N1) virus infection in humans. New Engl. J. Med. 2008, 358, 261–273. [Google Scholar] [CrossRef]
- Beigel, J.H.; Farrar, J.; Han, A.M.; Hayden, F.G.; Hyer, R.; de Jong, M.D.; Lochindarat, S.; Nguyen, T.K.; Nguyen, T.H.; Tran, T.H.; et al. Avian influenza A (H5N1) infection in humans. New Engl. J. Med. 2005, 353, 1374–1385. [Google Scholar] [CrossRef]
- WHO. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003-2012. Available online: http://www.who.int/influenza/human_animal_interface/avian_influenza/EN_GIP_20120326CumulativeNumberH5N1cases.pdf (accessed on 1 April 2012).
- Wang, T.T.; Parides, M.K.; Palese, P. Seroevidence for H5N1 influenza infections in humans: Meta-analysis. Science 2012, 335, 1463. [Google Scholar] [CrossRef]
- Kandun, I.N.; Wibisono, H.; Sedyaningsih, E.R.; Yusharmen; Hadisoedarsuno, W.; Purba, W.; Santoso, H.; Septiawati, C.; Tresnaningsih, E.; Heriyanto, B.; et al. Three Indonesian clusters of H5N1 virus infection in 2005. New Engl. J. Med. 2006, 355, 2186–2194. [Google Scholar]
- Ungchusak, K.; Auewarakul, P.; Dowell, S.F.; Kitphati, R.; Auwanit, W.; Puthavathana, P.; Uiprasertkul, M.; Boonnak, K.; Pittayawonganon, C.; Cox, N.J.; et al. Probable person-to-person transmission of avian influenza A (H5N1). New Engl. J. Med. 2005, 352, 333–340. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Z.; Shu, Y.; Yu, H.; Zhou, L.; Zu, R.; Huai, Y.; Dong, J.; Bao, C.; Wen, L.; et al. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China. Lancet 2008, 371, 1427–1434. [Google Scholar]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar]
- Russell, C.A.; Fonville, J.M.; Brown, A.E.X.; Burke, D.F.; Smith, D.L.; James, S.L.; Herfst, S.; van Boheemen, S.; Linster, M.; Schrauwen, E.J.; et al. The potential for respiratory droplet transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 2012, 336, 1541–1547. [Google Scholar]
- Holt, P.G.; Strickland, D.H.; Wikstrom, M.E.; Jahnsen, F.L. Regulation of immunological homeostasis in the respiratory tract. Nat. Rev. 2008, 8, 142–152. [Google Scholar] [CrossRef]
- Sanders, C.J.; Doherty, P.C.; Thomas, P.G. Respiratory epithelial cells in innate immunity to influenza virus infection. Cell Tissue Res. 2011, 343, 13–21. [Google Scholar]
- Blasius, A.L.; Beutler, B. Intracellular toll-like receptors. Immunity 2010, 32, 305–315. [Google Scholar] [CrossRef]
- Pang, I.K.; Iwasaki, A. Inflammasomes as mediators of immunity against influenza virus. Trends Immunol. 2011, 32, 34–41. [Google Scholar]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar]
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 5598–5603. [Google Scholar]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef]
- Guillot, L.; Le Goffic, R.; Bloch, S.; Escriou, N.; Akira, S.; Chignard, M.; Si-Tahar, M. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J. Biol. Chem. 2005, 280, 5571–5580. [Google Scholar]
- Imai, Y.; Kuba, K.; Neely, G.G.; Yaghubian-Malhami, R.; Perkmann, T.; van Loo, G.; Ermolaeva, M.; Veldhuizen, R.; Leung, Y.H.; Wang, H.; et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 2008, 133, 235–249. [Google Scholar] [CrossRef]
- Kurt-Jones, E.A.; Popova, L.; Kwinn, L.; Haynes, L.M.; Jones, L.P.; Tripp, R.A.; Walsh, E.E.; Freeman, M.W.; Golenbock, D.T.; Anderson, L.J.; et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000, 1, 398–401. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Moki, T.; Takizawa, T.; Shiratsuchi, A.; Nakanishi, Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J. Immunol. 2007, 178, 2448–2457. [Google Scholar]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzozka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.K.; Schlee, M.; et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar]
- Loo, Y.M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692. [Google Scholar]
- Kanneganti, T.D.; Body-Malapel, M.; Amer, A.; Park, J.H.; Whitfield, J.; Franchi, L.; Taraporewala, Z.F.; Miller, D.; Patton, J.T.; Inohara, N.; et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 2006, 281, 36560–36568. [Google Scholar]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef]
- Hale, B.G.; Albrecht, R.A.; Garcia-Sastre, A. Innate immune evasion strategies of influenza viruses. Future Microbiol. 2010, 5, 23–41. [Google Scholar]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasure. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar]
- Chelbi-Alix, M.K.; Wietzerbin, J. Interferon, a growing cytokine family: 50 years of interferon research. Biochimie 2007, 89, 713–718. [Google Scholar] [CrossRef]
- Van Hoeven, N.; Belser, J.A.; Szretter, K.J.; Zeng, H.; Staeheli, P.; Swayne, D.E.; Katz, J.M.; Tumpey, T.M. Pathogenesis of 1918 pandemic and H5N1 influenza virus infections in a guinea pig model: Antiviral potential of exogenous alpha interferon to reduce virus shedding. J. Virol. 2009, 83, 2851–2861. [Google Scholar]
- Szretter, K.J.; Gangappa, S.; Belser, J.A.; Zeng, H.; Chen, H.; Matsuoka, Y.; Sambhara, S.; Swayne, D.E.; Tumpey, T.M.; Katz, J.M. Early control of H5N1 influenza virus replication by the type I interferon response in mice. J. Virol. 2009, 83, 5825–5834. [Google Scholar]
- Sato, M.; Hata, N.; Asagiri, M.; Nakaya, T.; Taniguchi, T.; Tanaka, N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998, 441, 106–110. [Google Scholar]
- von der Malsburg, A.; Abutbul-Ionita, I.; Haller, O.; Kochs, G.; Danino, D. Stalk domain of the dynamin-like MxA GTPase protein mediates membrane binding and liposome tubulation via the unstructured L4 loop. J. Biol. Chem. 2011, 286, 37858–37865. [Google Scholar]
- Haller, O.; Gao, S.; von der Malsburg, A.; Daumke, O.; Kochs, G. Dynamin-like MxA GTPase: Structural insights into oligomerization and implications for antiviral activity. J. Biol. Chem. 2010, 285, 28419–28424. [Google Scholar]
- Haller, O.; Kochs, G. Interferon-induced mx proteins: Dynamin-like GTPases with antiviral activity. Traffic 2002, 3, 710–717. [Google Scholar] [CrossRef]
- Pindel, A.; Sadler, A. The role of protein kinase R in the interferon response. J. Interferon Cytokine Res. 2011, 31, 59–70. [Google Scholar] [CrossRef]
- Garcia, M.A.; Gil, J.; Ventoso, I.; Guerra, S.; Domingo, E.; Rivas, C.; Esteban, M. Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006, 70, 1032–1060. [Google Scholar]
- Chakrabarti, A.; Jha, B.K.; Silverman, R.H. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011, 31, 49–57. [Google Scholar] [CrossRef]
- Silverman, R.H. Viral encounters with 2',5'-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007, 81, 12720–12729. [Google Scholar] [CrossRef]
- Lenschow, D.J.; Lai, C.; Frias-Staheli, N.; Giannakopoulos, N.V.; Lutz, A.; Wolff, T.; Osiak, A.; Levine, B.; Schmidt, R.E.; Garcia-Sastre, A.; et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 1371–1376. [Google Scholar]
- Wang, X.; Hinson, E.R.; Cresswell, P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2007, 2, 96–105. [Google Scholar]
- Watanabe, R.; Leser, G.P.; Lamb, R.A. Influenza virus is not restricted by tetherin whereas influenza VLP production is restricted by tetherin. Virology 2011, 417, 50–56. [Google Scholar]
- Yondola, M.A.; Fernandes, F.; Belicha-Villanueva, A.; Uccelini, M.; Gao, Q.; Carter, C.; Palese, P. Budding capability of the influenza virus neuraminidase can be modulated by tetherin. J. Virol. 2011, 85, 2480–2491. [Google Scholar] [CrossRef]
- Brass, A.L.; Huang, I.C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009, 139, 1243–1254. [Google Scholar]
- Turner, S.J.; Olivas, E.; Gutierrez, A.; Diaz, G.; Doherty, P.C. Disregulated influenza A virus-specific CD8+ T cell homeostasis in the absence of IFN-gamma signaling. J. Immunol. 2007, 178, 7616–7622. [Google Scholar]
- Bot, A.; Bot, S.; Bona, C.A. Protective role of gamma interferon during the recall response to influenza virus. J. Virol. 1998, 72, 6637–6645. [Google Scholar]
- Mordstein, M.; Kochs, G.; Dumoutier, L.; Renauld, J.C.; Paludan, S.R.; Klucher, K.; Staeheli, P. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008, 4, e1000151. [Google Scholar] [CrossRef]
- McGill, J.; Heusel, J.W.; Legge, K.L. Innate immune control and regulation of influenza virus infections. J. Leukoc. Biol. 2009, 86, 803–812. [Google Scholar]
- Snelgrove, R.J.; Goulding, J.; Didierlaurent, A.M.; Lyonga, D.; Vekaria, S.; Edwards, L.; Gwyer, E.; Sedgwick, J.D.; Barclay, A.N.; Hussell, T. A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat. Immunol. 2008, 9, 1074–1083. [Google Scholar]
- Lin, K.L.; Suzuki, Y.; Nakano, H.; Ramsburg, E.; Gunn, M.D. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J. Immunol. 2008, 180, 2562–2572. [Google Scholar]
- Dawson, T.C.; Beck, M.A.; Kuziel, W.A.; Henderson, F.; Maeda, N. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am. J. Pathol. 2000, 156, 1951–1959. [Google Scholar] [CrossRef]
- Herold, S.; von Wulffen, W.; Steinmueller, M.; Pleschka, S.; Kuziel, W.A.; Mack, M.; Srivastava, M.; Seeger, W.; Maus, U.A.; Lohmeyer, J. Alveolar epithelial cells direct monocyte transepithelial migration upon influenza virus infection: Impact of chemokines and adhesion molecules. J. Immunol. 2006, 177, 1817–1824. [Google Scholar]
- van Riel, D.; Leijten, L.M.; van der Eerden, M.; Hoogsteden, H.C.; Boven, L.A.; Lambrecht, B.N.; Osterhaus, A.D.; Kuiken, T. Highly pathogenic avian influenza virus H5N1 infects alveolar macrophages without virus production or excessive TNF-alpha induction. PLoS Pathog. 2011, 7, e1002099. [Google Scholar]
- Becker, S.; Quay, J.; Soukup, J. Cytokine (tumor necrosis factor, IL-6, and IL-8) production by respiratory syncytial virus-infected human alveolar macrophage. J. Immunol. 1991, 147, 4307–4312. [Google Scholar]
- Tumpey, T.M.; Garcia-Sastre, A.; Taubenberger, J.K.; Palese, P.; Swayne, D.E.; Pantin-Jackwood, M.J.; Schultz-Cherry, S.; Solorzano, A.; van Rooijen, N.; Katz, J.M.; et al. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: Functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J. Virol. 2005, 79, 14933–14944. [Google Scholar]
- Kim, H.M.; Lee, Y.W.; Lee, K.J.; Kim, H.S.; Cho, S.W.; van Rooijen, N.; Guan, Y.; Seo, S.H. Alveolar macrophages are indispensable for controlling influenza viruses in lungs of pigs. J. Virol. 2008, 82, 4265–4274. [Google Scholar]
- Huber, V.C.; Lynch, J.M.; Bucher, D.J.; Le, J.; Metzger, D.W. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 2001, 166, 7381–7388. [Google Scholar]
- Jayasekera, J.P.; Vinuesa, C.G.; Karupiah, G.; King, N.J. Enhanced antiviral antibody secretion and attenuated immunopathology during influenza virus infection in nitric oxide synthase-2-deficient mice. J. Gen. Virol. 2006, 87, 3361–3371. [Google Scholar] [CrossRef]
- Peper, R.L.; van Campen, H. Tumor necrosis factor as a mediator of inflammation in influenza A viral pneumonia. Microb. Pathog. 1995, 19, 175–183. [Google Scholar] [CrossRef]
- Mendelson, M.; Tekoah, Y.; Zilka, A.; Gershoni-Yahalom, O.; Gazit, R.; Achdout, H.; Bovin, N.V.; Meningher, T.; Mandelboim, M.; Mandelboim, O.; et al. NKp46 O-glycan sequences that are involved in the interaction with hemagglutinin type 1 of influenza virus. J. Virol. 2010, 84, 3789–3797. [Google Scholar]
- Arnon, T.I.; Lev, M.; Katz, G.; Chernobrov, Y.; Porgador, A.; Mandelboim, O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur. J. Immunol. 2001, 31, 2680–2689. [Google Scholar]
- Mandelboim, O.; Lieberman, N.; Lev, M.; Paul, L.; Arnon, T.I.; Bushkin, Y.; Davis, D.M.; Strominger, J.L.; Yewdell, J.W.; Porgador, A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 2001, 409, 1055–1060. [Google Scholar]
- Gazit, R.; Gruda, R.; Elboim, M.; Arnon, T.I.; Katz, G.; Achdout, H.; Hanna, J.; Qimron, U.; Landau, G.; Greenbaum, E.; et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat. Immunol. 2006, 7, 517–523. [Google Scholar]
- Hashimoto, G.; Wright, P.F.; Karzon, D.T. Antibody-dependent cell-mediated cytotoxicity against influenza virus-infected cells. J. Infect. Dis. 1983, 148, 785–794. [Google Scholar]
- Sun, P.D. Structure and function of natural-killer-cell receptors. Immunol. Res. 2003, 27, 539–548. [Google Scholar] [CrossRef]
- Jegerlehner, A.; Schmitz, N.; Storni, T.; Bachmann, M.F. Influenza A vaccine based on the extracellular domain of M2: Weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 2004, 172, 5598–5605. [Google Scholar]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Guermonprez, P.; Valladeau, J.; Zitvogel, L.; Thery, C.; Amigorena, S. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 2002, 20, 621–667. [Google Scholar] [CrossRef]
- GeurtsvanKessel, C.H.; Lambrecht, B.N. Division of labor between dendritic cell subsets of the lung. Mucosal Immunol. 2008, 1, 442–450. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bender, A.; Gonzalez, N.; Bui, L.K.; Garrett, M.C.; Steinman, R.M. Influenza virus-infected dendritic cells stimulate strong proliferative and cytolytic responses from human CD8+ T cells. J. Clin. Investig. 1994, 94, 797–807. [Google Scholar]
- Hamilton-Easton, A.; Eichelberger, M. Virus-specific antigen presentation by different subsets of cells from lung and mediastinal lymph node tissues of influenza virus-infected mice. J. Virol. 1995, 69, 6359–6366. [Google Scholar]
- Yewdell, J.W.; Reits, E.; Neefjes, J. Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat. Rev. 2003, 3, 952–961. [Google Scholar]
- Mori, I.; Komatsu, T.; Takeuchi, K.; Nakakuki, K.; Sudo, M.; Kimura, Y. In vivo induction of apoptosis by influenza virus. J. Gen. Virol. 1995, 76, 2869–2873. [Google Scholar] [CrossRef]
- Braciale, T.J.; Sun, J.; Kim, T.S. Regulating the adaptive immune response to respiratory virus infection. Nat. Rev. 2012, 12, 295–305. [Google Scholar] [CrossRef]
- Kim, T.S.; Braciale, T.J. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS One 2009, 4, e4204. [Google Scholar]
- Norbury, C.C.; Malide, D.; Gibbs, J.S.; Bennink, J.R.; Yewdell, J.W. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat. Immunol. 2002, 3, 265–271. [Google Scholar] [CrossRef]
- Heer, A.K.; Harris, N.L.; Kopf, M.; Marsland, B.J. CD4+ and CD8+ T cells exhibit differential requirements for CCR7-mediated antigen transport during influenza infection. J. Immunol. 2008, 181, 6984–6994. [Google Scholar]
- Legge, K.L.; Braciale, T.J. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity 2003, 18, 265–277. [Google Scholar] [CrossRef]
- Baumgarth, N.; Herman, O.C.; Jager, G.C.; Brown, L.E.; Herzenberg, L.A.; Chen, J. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. J. Exp. Med. 2000, 192, 271–280. [Google Scholar]
- Baumgarth, N.; Tung, J.W.; Herzenberg, L.A. Inherent specificities in natural antibodies: A key to immune defense against pathogen invasion. Springer Semin. Immunopathol. 2005, 26, 347–362. [Google Scholar] [CrossRef]
- Potter, C.W.; Oxford, J.S. Determinants of immunity to influenza infection in man. Br. Med. Bull. 1979, 35, 69–75. [Google Scholar]
- Waffarn, E.E.; Baumgarth, N. Protective B cell responses to flu—No fluke! J. Immunol. 2011, 186, 3823–3829. [Google Scholar] [CrossRef]
- de Jong, J.C.; Palache, A.M.; Beyer, W.E.; Rimmelzwaan, G.F.; Boon, A.C.; Osterhaus, A.D. Haemagglutination-inhibiting antibody to influenza virus. Dev. Biologicals 2003, 115, 63–73. [Google Scholar]
- Knossow, M.; Skehel, J.J. Variation and infectivity neutralization in influenza. Immunology 2006, 119, 1–7. [Google Scholar] [CrossRef]
- Wilson, I.A.; Cox, N.J. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 1990, 8, 737–771. [Google Scholar] [CrossRef]
- Kilbourne, E.D. Influenza as a problem in immunology. J. Immunol. 1978, 120, 1447–1452. [Google Scholar]
- Potter, C.W.; Oxford, J.S.; Shore, S.L.; McLaren, C.; Stuart-Harris, C. Immunity to influenza in ferrets. I. Response to live and killed virus. Br. J. Exp. Pathol. 1972, 53, 153–167. [Google Scholar]
- Yu, X.; Tsibane, T.; McGraw, P.A.; House, F.S.; Keefer, C.J.; Hicar, M.D.; Tumpey, T.M.; Pappas, C.; Perrone, L.A.; Martinez, O.; et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 2008, 455, 532–536. [Google Scholar]
- Ikonen, N.; Strengell, M.; Kinnunen, L.; Osterlund, P.; Pirhonen, J.; Broman, M.; Davidkin, I.; Ziegler, T.; Julkunen, I. High frequency of cross-reacting antibodies against 2009 pandemic influenza A(H1N1) virus among the elderly in Finland. Euro Surveill. 2010, 15, pii=19478. [Google Scholar]
- Reed, C.; Katz, J.M. Serological surveys for 2009 pandemic influenza A H1N1. Lancet 2010, 375, 1062–1063. [Google Scholar] [CrossRef]
- Hancock, K.; Veguilla, V.; Lu, X.; Zhong, W.; Butler, E.N.; Sun, H.; Liu, F.; Dong, L.; DeVos, J.R.; Gargiullo, P.M.; et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. New Engl. J. Med. 2009, 361, 1945–1952. [Google Scholar]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.A.; Friesen, R.H.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope. Science 2009, 324, 246–251. [Google Scholar]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B.; et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011, 333, 843–850. [Google Scholar]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273. [Google Scholar]
- Wang, T.T.; Tan, G.S.; Hai, R.; Pica, N.; Petersen, E.; Moran, T.M.; Palese, P. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010, 6, e1000796. [Google Scholar] [CrossRef]
- Steel, J.; Lowen, A.C.; Wang, T.T.; Yondola, M.; Gao, Q.; Haye, K.; Garcia-Sastre, A.; Palese, P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 2010, 1, e00018–10. [Google Scholar]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 2008, 3, e3942. [Google Scholar]
- Wang, T.T.; Tan, G.S.; Hai, R.; Pica, N.; Ngai, L.; Ekiert, D.C.; Wilson, I.A.; Garcia-Sastre, A.; Moran, T.M.; Palese, P. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 18979–18984. [Google Scholar]
- Okuno, Y.; Isegawa, Y.; Sasao, F.; Ueda, S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993, 67, 2552–2558. [Google Scholar]
- Bosch, B.J.; Bodewes, R.; de Vries, R.P.; Kreijtz, J.H.; Bartelink, W.; van Amerongen, G.; Rimmelzwaan, G.F.; de Haan, C.A.; Osterhaus, A.D.; Rottier, P.J. Recombinant soluble, multimeric HA and NA exhibit distinctive types of protection against pandemic swine-origin 2009 A(H1N1) influenza virus infection in ferrets. J. Virol. 2010, 84, 10366–10374. [Google Scholar]
- Johansson, B.E.; Bucher, D.J.; Kilbourne, E.D. Purified influenza virus hemagglutinin and neuraminidase are equivalent in stimulation of antibody response but induce contrasting types of immunity to infection. J. Virol. 1989, 63, 1239–1246. [Google Scholar]
- Johansson, B.E.; Grajower, B.; Kilbourne, E.D. Infection-permissive immunization with influenza virus neuraminidase prevents weight loss in infected mice. Vaccine 1993, 11, 1037–1039. [Google Scholar] [CrossRef]
- Kilbourne, E.D.; Pokorny, B.A.; Johansson, B.; Brett, I.; Milev, Y.; Matthews, J.T. Protection of mice with recombinant influenza virus neuraminidase. J. Infect. Dis. 2004, 189, 459–461. [Google Scholar] [CrossRef]
- Schulman, J.L.; Khakpour, M.; Kilbourne, E.D. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J. Virol. 1968, 2, 778–786. [Google Scholar]
- Treanor, J.J.; Tierney, E.L.; Zebedee, S.L.; Lamb, R.A.; Murphy, B.R. Passively transferred monoclonal antibody to the M2 protein inhibits influenza A virus replication in mice. J. Virol. 1990, 64, 1375–1377. [Google Scholar]
- Zebedee, S.L.; Lamb, R.A. Influenza A virus M2 protein: Monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 1988, 62, 2762–2772. [Google Scholar]
- Mozdzanowska, K.; Maiese, K.; Furchner, M.; Gerhard, W. Treatment of influenza virus-infected SCID mice with nonneutralizing antibodies specific for the transmembrane proteins matrix 2 and neuraminidase reduces the pulmonary virus titer but fails to clear the infection. Virology 1999, 254, 138–146. [Google Scholar] [CrossRef]
- El Bakkouri, K.; Descamps, F.; de Filette, M.; Smet, A.; Festjens, E.; Birkett, A.; van Rooijen, N.; Verbeek, S.; Fiers, W.; Saelens, X. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J. Immunol. 2011, 186, 1022–1031. [Google Scholar] [CrossRef]
- Ebrahimi, S.M.; Tebianian, M. Influenza A viruses: Why focusing on M2e-based universal vaccines. Virus Genes 2011, 42, 1–8. [Google Scholar] [CrossRef]
- Fiers, W.; de Filette, M.; Birkett, A.; Neirynck, S.; Min Jou, W. A "universal" human influenza A vaccine. Virus Res. 2004, 103, 173–176. [Google Scholar]
- Fiers, W.; de Filette, M.; El Bakkouri, K.; Schepens, B.; Roose, K.; Schotsaert, M.; Birkett, A.; Saelens, X. M2e-based universal influenza A vaccine. Vaccine 2009, 27, 6280–6283. [Google Scholar] [CrossRef]
- Neirynck, S.; Deroo, T.; Saelens, X.; Vanlandschoot, P.; Jou, W.M.; Fiers, W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999, 5, 1157–1163. [Google Scholar]
- Wang, Y.; Zhou, L.; Shi, H.; Xu, H.; Yao, H.; Xi, X.G.; Toyoda, T.; Wang, X.; Wang, T. Monoclonal antibody recognizing SLLTEVET epitope of M2 protein potently inhibited the replication of influenza A viruses in MDCK cells. Biochem. Biophys. Res. Comm. 2009, 385, 118–122. [Google Scholar] [CrossRef]
- Fu, T.M.; Freed, D.C.; Horton, M.S.; Fan, J.; Citron, M.P.; Joyce, J.G.; Garsky, V.M.; Casimiro, D.R.; Zhao, Q.; Shiver, J.W.; et al. Characterizations of four monoclonal antibodies against M2 protein ectodomain of influenza A virus. Virology 2009, 385, 218–226. [Google Scholar]
- Schotsaert, M.; de Filette, M.; Fiers, W.; Saelens, X. Universal M2 ectodomain-based influenza A vaccines: Preclinical and clinical developments. Expert Rev. Vaccine 2009, 8, 499–508. [Google Scholar] [CrossRef]
- Tompkins, S.M.; Zhao, Z.S.; Lo, C.Y.; Misplon, J.A.; Liu, T.; Ye, Z.; Hogan, R.J.; Wu, Z.; Benton, K.A.; Tumpey, T.M.; et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg. Infect. Dis. 2007, 13, 426–435. [Google Scholar] [CrossRef]
- Sukeno, N.; Otsuki, Y.; Konno, J.; Yamane, N.; Odagiri, T.; Arikawa, J.; Ishida, N. Anti-nucleoprotein antibody response in influenza A infection. Tohoku J. Exp. Med. 1979, 128, 241–249. [Google Scholar] [CrossRef]
- Lamere, M.W.; Moquin, A.; Lee, F.E.; Misra, R.S.; Blair, P.J.; Haynes, L.; Randall, T.D.; Lund, F.E.; Kaminski, D.A. Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance. J. Virol. 2011, 85, 5027–5035. [Google Scholar] [CrossRef]
- Carragher, D.M.; Kaminski, D.A.; Moquin, A.; Hartson, L.; Randall, T.D. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J. Immunol. 2008, 181, 4168–4176. [Google Scholar]
- Bodewes, R.; Osterhaus, A.D.; Rimmelzwaan, G.F. Targets for the induction of protective immunity against influenza a viruses. Viruses 2010, 2, 166–188. [Google Scholar] [CrossRef]
- Sambhara, S.; Kurichh, A.; Miranda, R.; Tumpey, T.; Rowe, T.; Renshaw, M.; Arpino, R.; Tamane, A.; Kandil, A.; James, O.; et al. Heterosubtypic immunity against human influenza A viruses, including recently emerged avian H5 and H9 viruses, induced by FLU-ISCOM vaccine in mice requires both cytotoxic T-lymphocyte and macrophage function. Cell. Immunol. 2001, 211, 143–153. [Google Scholar]
- Rimmelzwaan, G.F.; Baars, M.; van Beek, R.; van Amerongen, G.; Lovgren-Bengtsson, K.; Claas, E.C.; Osterhaus, A.D. Induction of protective immunity against influenza virus in a macaque model: Comparison of conventional and iscom vaccines. J. Gen. Virol. 1997, 78, 757–765. [Google Scholar]
- Fernandez Gonzalez, S.; Jayasekera, J.P.; Carroll, M.C. Complement and natural antibody are required in the long-term memory response to influenza virus. Vaccine 2008, 26, I86–I93. [Google Scholar]
- Jayasekera, J.P.; Moseman, E.A.; Carroll, M.C. Natural antibody and complement mediate neutralization of influenza virus in the absence of prior immunity. J. Virol. 2007, 81, 3487–3494. [Google Scholar]
- Voeten, J.T.; Groen, J.; van Alphen, D.; Claas, E.C.; de Groot, R.; Osterhaus, A.D.; Rimmelzwaan, G.F. Use of recombinant nucleoproteins in enzyme-linked immunosorbent assays for detection of virus-specific immunoglobulin A (IgA) and IgG antibodies in influenza virus A- or B-infected patients. J. Clin. Microbiol. 1998, 36, 3527–3531. [Google Scholar]
- Rothbarth, P.H.; Groen, J.; Bohnen, A.M.; de Groot, R.; Osterhaus, A.D. Influenza virus serology—A comparative study. J. Virol. Meth. 1999, 78, 163–169. [Google Scholar] [CrossRef]
- Koutsonanos, D.G.; del Pilar Martin, M.; Zarnitsyn, V.G.; Jacob, J.; Prausnitz, M.R.; Compans, R.W.; Skountzou, I. Serological memory and long-term protection to novel H1N1 influenza virus after skin vaccination. J. Infect. Dis. 2011, 204, 582–591. [Google Scholar] [CrossRef]
- Onodera, T.; Takahashi, Y.; Yokoi, Y.; Ato, M.; Kodama, Y.; Hachimura, S.; Kurosaki, T.; Kobayashi, K. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 2485–2490. [Google Scholar]
- Jones, P.D.; Ada, G.L. Persistence of influenza virus-specific antibody-secreting cells and B-cell memory after primary murine influenza virus infection. Cell. Immunol. 1987, 109, 53–64. [Google Scholar] [CrossRef]
- Armstrong, S.J.; Dimmock, N.J. Neutralization of influenza virus by low concentrations of hemagglutinin-specific polymeric immunoglobulin A inhibits viral fusion activity, but activation of the ribonucleoprotein is also inhibited. J. Virol. 1992, 66, 3823–3832. [Google Scholar]
- Mazanec, M.B.; Coudret, C.L.; Fletcher, D.R. Intracellular neutralization of influenza virus by immunoglobulin A anti-hemagglutinin monoclonal antibodies. J. Virol. 1995, 69, 1339–1343. [Google Scholar]
- Zuccotti, G.; Pogliani, L.; Pariani, E.; Amendola, A.; Zanetti, A. Transplacental antibody transfer following maternal immunization with a pandemic 2009 influenza A(H1N1) MF59-adjuvanted vaccine. JAMA 2010, 304, 2360–2361. [Google Scholar]
- Bodewes, R.; de Mutsert, G.; van der Klis, F.R.; Ventresca, M.; Wilks, S.; Smith, D.J.; Koopmans, M.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin. Vaccine Immunol. 2011, 18, 469–476. [Google Scholar]
- Mbawuike, I.N.; Six, H.R.; Cate, T.R.; Couch, R.B. Vaccination with inactivated influenza A virus during pregnancy protects neonatal mice against lethal challenge by influenza A viruses representing three subtypes. J. Virol. 1990, 64, 1370–1374. [Google Scholar]
- Hwang, S.D.; Shin, J.S.; Ku, K.B.; Kim, H.S.; Cho, S.W.; Seo, S.H. Protection of pregnant mice, fetuses and neonates from lethality of H5N1 influenza viruses by maternal vaccination. Vaccine 2010, 28, 2957–2964. [Google Scholar]
- Steinhoff, M.C.; Omer, S.B.; Roy, E.; Arifeen, S.E.; Raqib, R.; Altaye, M.; Breiman, R.F.; M, B.B.S.K. Influenza immunization in pregnancy—Antibody responses in mothers and infants. New Engl. J. Med. 2010, 362, 1644–1646. [Google Scholar]
- Zhu, J.; Paul, W.E. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 2010, 238, 247–262. [Google Scholar]
- Lamb, J.R.; Woody, J.N.; Hartzman, R.J.; Eckels, D.D. In vitro influenza virus-specific antibody production in man: Antigen-specific and HLA-restricted induction of helper activity mediated by cloned human T lymphocytes. J. Immunol. 1982, 129, 1465–1470. [Google Scholar]
- Okoye, I.S.; Wilson, M.S. CD4+ T helper 2 cells—Microbial triggers, differentiation requirements and effector functions. Immunology 2011, 134, 368–377. [Google Scholar] [CrossRef]
- Eichelberger, M.C.; Wang, M.L.; Allan, W.; Webster, R.G.; Doherty, P.C. Influenza virus RNA in the lung and lymphoid tissue of immunologically intact and CD4-depleted mice. J. Gen. Virol. 1991, 72, 1695–1698. [Google Scholar] [CrossRef]
- Justewicz, D.M.; Doherty, P.C.; Webster, R.G. The B-cell response in lymphoid tissue of mice immunized with various antigenic forms of the influenza virus hemagglutinin. J. Virol. 1995, 69, 5414–5421. [Google Scholar]
- Kamperschroer, C.; Dibble, J.P.; Meents, D.L.; Schwartzberg, P.L.; Swain, S.L. SAP is required for Th cell function and for immunity to influenza. J. Immunol. 2006, 177, 5317–5327. [Google Scholar]
- Scherle, P.A.; Gerhard, W. Functional analysis of influenza-specific helper T cell clones in vivo. T cells specific for internal viral proteins provide cognate help for B cell responses to hemagglutinin. J. Exp. Med. 1986, 164, 1114–1128. [Google Scholar] [CrossRef]
- Schonbeck, U.; Libby, P. The CD40/CD154 receptor/ligand dyad. Cell. Mol. Life Sci. 2001, 58, 4–43. [Google Scholar] [CrossRef]
- Zhu, J.; Paul, W.E. Heterogeneity and plasticity of T helper cells. Cell Res. 2010, 20, 4–12. [Google Scholar]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986, 136, 2348–2357. [Google Scholar]
- Belz, G.T.; Wodarz, D.; Diaz, G.; Nowak, M.A.; Doherty, P.C. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J. Virol. 2002, 76, 12388–12393. [Google Scholar] [CrossRef]
- Deliyannis, G.; Jackson, D.C.; Ede, N.J.; Zeng, W.; Hourdakis, I.; Sakabetis, E.; Brown, L.E. Induction of long-term memory CD8(+) T cells for recall of viral clearing responses against influenza virus. J. Virol. 2002, 76, 4212–4221. [Google Scholar]
- Riberdy, J.M.; Christensen, J.P.; Branum, K.; Doherty, P.C. Diminished primary and secondary influenza virus-specific CD8(+) T-cell responses in CD4-depleted Ig(-/-) mice. J. Virol. 2000, 74, 9762–9765. [Google Scholar]
- Strutt, T.M.; McKinstry, K.K.; Dibble, J.P.; Winchell, C.; Kuang, Y.; Curtis, J.D.; Huston, G.; Dutton, R.W.; Swain, S.L. Memory CD4+ T cells induce innate responses independently of pathogen. Nat. Med. 2010, 16 551p following 564, 558–564. [Google Scholar]
- Teijaro, J.R.; Turner, D.; Pham, Q.; Wherry, E.J.; Lefrancois, L.; Farber, D.L. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 2011, 187, 5510–5514. [Google Scholar]
- Brown, D.M.; Dilzer, A.M.; Meents, D.L.; Swain, S.L. CD4 T cell-mediated protection from lethal influenza: Perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 2006, 177, 2888–2898. [Google Scholar]
- Graham, M.B.; Braciale, V.L.; Braciale, T.J. Influenza virus-specific CD4+ T helper type 2 T lymphocytes do not promote recovery from experimental virus infection. J. Exp. Med. 1994, 180, 1273–1282. [Google Scholar]
- Wilkinson, T.M.; Li, C.K.; Chui, C.S.; Huang, A.K.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar]
- McKinstry, K.K.; Strutt, T.M.; Swain, S.L. Hallmarks of CD4 T cell immunity against influenza. J. Intern. Med. 2011, 269, 507–518. [Google Scholar] [CrossRef]
- Nakanishi, Y.; Lu, B.; Gerard, C.; Iwasaki, A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature 2009, 462, 510–513. [Google Scholar]
- Assarsson, E.; Bui, H.H.; Sidney, J.; Zhang, Q.; Glenn, J.; Oseroff, C.; Mbawuike, I.N.; Alexander, J.; Newman, M.J.; Grey, H.; et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 2008, 82, 12241–12251. [Google Scholar]
- Bednarek, M.A.; Sauma, S.Y.; Gammon, M.C.; Porter, G.; Tamhankar, S.; Williamson, A.R.; Zweerink, H.J. The minimum peptide epitope from the influenza virus matrix protein. Extra and intracellular loading of HLA-A2. J. Immunol. 1991, 147, 4047–4053. [Google Scholar]
- Boon, A.C.; de Mutsert, G.; Graus, Y.M.; Fouchier, R.A.; Sintnicolaas, K.; Osterhaus, A.D.; Rimmelzwaan, G.F. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 2002, 76, 582–590. [Google Scholar] [CrossRef]
- Gotch, F.; McMichael, A.; Smith, G.; Moss, B. Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J. Exp. Med. 1987, 165, 408–416. [Google Scholar]
- Jameson, J.; Cruz, J.; Ennis, F.A. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 1998, 72, 8682–8689. [Google Scholar]
- Kreijtz, J.H.; de Mutsert, G.; van Baalen, C.A.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J. Virol. 2008, 82, 5161–5166. [Google Scholar]
- Lee, L.Y.; Ha do, L.A.; Simmons, C.; de Jong, M.D.; Chau, N.V.; Schumacher, R.; Peng, Y.C.; McMichael, A.J.; Farrar, J.J.; Smith, G.L.; et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Investig. 2008, 118, 3478–3490. [Google Scholar]
- Andrade, F. Non-cytotoxic antiviral activities of granzymes in the context of the immune antiviral state. Immunol. Rev. 2010, 235, 128–146. [Google Scholar]
- Moffat, J.M.; Gebhardt, T.; Doherty, P.C.; Turner, S.J.; Mintern, J.D. Granzyme A expression reveals distinct cytolytic CTL subsets following influenza A virus infection. Eur. J. Immunol. 2009, 39, 1203–1210. [Google Scholar] [CrossRef]
- Topham, D.J.; Tripp, R.A.; Doherty, P.C. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J. Immunol. 1997, 159, 5197–5200. [Google Scholar]
- La Gruta, N.L.; Turner, S.J.; Doherty, P.C. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: Correlation of cytokine profile and TCR avidity. J. Immunol. 2004, 172, 5553–5560. [Google Scholar]
- Metkar, S.S.; Menaa, C.; Pardo, J.; Wang, B.; Wallich, R.; Freudenberg, M.; Kim, S.; Raja, S.M.; Shi, L.; Simon, M.M.; et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity 2008, 29, 720–733. [Google Scholar]
- van Domselaar, R.; Bovenschen, N. Cell death-independent functions of granzymes: Hit viruses where it hurts. Rev. Med. Virol. 2011, 21, 301–314. [Google Scholar]
- La Gruta, N.L.; Kedzierska, K.; Stambas, J.; Doherty, P.C. A question of self-preservation: Immunopathology in influenza virus infection. Immunol. Cell Biol. 2007, 85, 85–92. [Google Scholar]
- Woodland, D.L.; Hogan, R.J.; Zhong, W. Cellular immunity and memory to respiratory virus infections. Immunol. Res. 2001, 24, 53–67. [Google Scholar] [CrossRef]
- Kedl, R.M.; Mescher, M.F. Qualitative differences between naive and memory T cells make a major contribution to the more rapid and efficient memory CD8+ T cell response. J. Immunol. 1998, 161, 674–683. [Google Scholar]
- Lalvani, A.; Brookes, R.; Hambleton, S.; Britton, W.J.; Hill, A.V.; McMichael, A.J. Rapid effector function in CD8+ memory T cells. J. Exp. Med. 1997, 186, 859–865. [Google Scholar] [CrossRef]
- Zimmermann, C.; Prevost-Blondel, A.; Blaser, C.; Pircher, H. Kinetics of the response of naive and memory CD8 T cells to antigen: Similarities and differences. Eur. J. Immunol. 1999, 29, 284–290. [Google Scholar] [CrossRef]
- DiSpirito, J.R.; Shen, H. Quick to remember, slow to forget: Rapid recall responses of memory CD8+ T cells. Cell Res. 2010, 20, 13–23. [Google Scholar]
- Chang, J.T.; Palanivel, V.R.; Kinjyo, I.; Schambach, F.; Intlekofer, A.M.; Banerjee, A.; Longworth, S.A.; Vinup, K.E.; Mrass, P.; Oliaro, J.; et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 2007, 315, 1687–1691. [Google Scholar] [CrossRef]
- Hikono, H.; Kohlmeier, J.E.; Takamura, S.; Wittmer, S.T.; Roberts, A.D.; Woodland, D.L. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cell. J. Exp. Med. 2007, 204, 1625–1636. [Google Scholar]
- Sallusto, F.; Lenig, D.; Forster, R.; Lipp, M.; Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999, 401, 708–712. [Google Scholar]
- van Gisbergen, K.P.; Klarenbeek, P.L.; Kragten, N.A.; Unger, P.P.; Nieuwenhuis, M.B.; Wensveen, F.M.; ten Brinke, A.; Tak, P.P.; Eldering, E.; Nolte, M.A.; et al. The costimulatory molecule CD27 maintains clonally diverse CD8(+) T cell responses of low antigen affinity to protect against viral variants. Immunity 2011, 35, 97–108. [Google Scholar]
- Wherry, E.J.; Teichgraber, V.; Becker, T.C.; Masopust, D.; Kaech, S.M.; Antia, R.; von Andrian, U.H.; Ahmed, R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 2003, 4, 225–234. [Google Scholar]
- Bender, B.S.; Croghan, T.; Zhang, L.; Small, P.A., Jr. Transgenic mice lacking class I major histocompatibility complex-restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. J. Exp. Med. 1992, 175, 1143–1145. [Google Scholar]
- Graham, M.B.; Braciale, T.J. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. J. Exp. Med. 1997, 186, 2063–2068. [Google Scholar]
- Hillaire, M.L.; van Trierum, S.E.; Kreijtz, J.H.; Bodewes, R.; Geelhoed-Mieras, M.M.; Nieuwkoop, N.J.; Fouchier, R.A.; Kuiken, T.; Osterhaus, A.D.; Rimmelzwaan, G.F. Cross-protective immunity against influenza pH1N1 2009 viruses induced by seasonal influenza A (H3N2) virus is mediated by virus-specific T-cells. J. Gen. Virol. 2011, 92, 2339–2349. [Google Scholar]
- Kreijtz, J.H.; Bodewes, R.; van Amerongen, G.; Kuiken, T.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 2007, 25, 612–620. [Google Scholar]
- Kreijtz, J.H.; Bodewes, R.; van den Brand, J.M.; de Mutsert, G.; Baas, C.; van Amerongen, G.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Infection of mice with a human influenza A/H3N2 virus induces protective immunity against lethal infection with influenza A/H5N1 virus. Vaccine 2009, 27, 4983–4989. [Google Scholar]
- Taylor, P.M.; Askonas, B.A. Influenza nucleoprotein-specific cytotoxic T-cell clones are protective in vivo. Immunology 1986, 58, 417–420. [Google Scholar]
- Hillaire, M.L.; Osterhaus, A.D.; Rimmelzwaan, G.F. Induction of virus-specific cytotoxic T lymphocytes as a basis for the development of broadly protective influenza vaccines. J. Biomed. Biotechnol. 2011, 2011, 939860. [Google Scholar]
- Grebe, K.M.; Yewdell, J.W.; Bennink, J.R. Heterosubtypic immunity to influenza A virus: Where do we stand? Microb. Infect. 2008, 10, 1024–1029. [Google Scholar] [CrossRef]
- Sridhar, S.; Begom, S.; Bermingham, A.; Ziegler, T.; Roberts, K.L.; Barclay, W.S.; Openshaw, P.; Lalvani, A. Predominance of heterosubtypic IFN-gamma-only-secreting effector memory T cells in pandemic H1N1 naive adults. Eur. J. Immunol. 2012. [Google Scholar]
- McMichael, A.J.; Gotch, F.M.; Noble, G.R.; Beare, P.A. Cytotoxic T-cell immunity to influenza. New Engl. J. Med. 1983, 309, 13–17. [Google Scholar]
- Epstein, S.L. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: An experiment of nature. J. Infect. Dis. 2006, 193, 49–53. [Google Scholar]
- Slepushkin, A.N. The effect of a previous attack of A1 influenza on susceptibility to A2 virus during the 1957 outbreak. Bull. World Health Organ. 1959, 20, 297–301. [Google Scholar]
- Smallman-Raynor, M.; Cliff, A.D. Avian influenza A (H5N1) age distribution in humans. Emerg. Infect. Dis. 2007, 13, 510–512. [Google Scholar] [CrossRef]
- Campbell, D.J.; Koch, M.A. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. 2011, 11, 119–130. [Google Scholar] [CrossRef]
- Surls, J.; Nazarov-Stoica, C.; Kehl, M.; Casares, S.; Brumeanu, T.D. Differential effect of CD4+Foxp3+ T-regulatory cells on the B and T helper cell responses to influenza virus vaccination. Vaccine 2010, 28, 7319–7330. [Google Scholar] [CrossRef]
- Kudva, A.; Scheller, E.V.; Robinson, K.M.; Crowe, C.R.; Choi, S.M.; Slight, S.R.; Khader, S.A.; Dubin, P.J.; Enelow, R.I.; Kolls, J.K.; et al. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J. Immunol. 2011, 186, 1666–1674. [Google Scholar] [CrossRef]
- Garcia-Sastre, A.; Egorov, A.; Matassov, D.; Brandt, S.; Levy, D.E.; Durbin, J.E.; Palese, P.; Muster, T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 1998, 252, 324–330. [Google Scholar] [CrossRef]
- Talon, J.; Salvatore, M.; O'Neill, R.E.; Nakaya, Y.; Zheng, H.; Muster, T.; Garcia-Sastre, A.; Palese, P. Influenza A and B viruses expressing altered NS1 proteins: A vaccine approach. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 4309–4314. [Google Scholar]
- Falcon, A.M.; Fernandez-Sesma, A.; Nakaya, Y.; Moran, T.M.; Ortin, J.; Garcia-Sastre, A. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 2005, 86, 2817–2821. [Google Scholar] [CrossRef]
- Donelan, N.R.; Basler, C.F.; Garcia-Sastre, A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 2003, 77, 13257–13266. [Google Scholar] [CrossRef]
- Solorzano, A.; Webby, R.J.; Lager, K.M.; Janke, B.H.; Garcia-Sastre, A.; Richt, J.A. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 2005, 79, 7535–7543. [Google Scholar]
- Richt, J.A.; Garcia-Sastre, A. Attenuated influenza virus vaccines with modified NS1 proteins. Curr. Top. Microbiol. Immunol. 2009, 333, 177–195. [Google Scholar]
- Park, H.J.; Ferko, B.; Byun, Y.H.; Song, J.H.; Han, G.Y.; Roethl, E.; Egorov, A.; Muster, T.; Seong, B.; Kweon, M.N.; et al. Sublingual immunization with a live attenuated influenza a virus lacking the nonstructural protein 1 induces broad protective immunity in mice. PLoS One 2012, 7, e39921. [Google Scholar]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.S.; Huang, I.C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; Garcia-Sastre, A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar]
- Ludwig, S.; Wang, X.; Ehrhardt, C.; Zheng, H.; Donelan, N.; Planz, O.; Pleschka, S.; Garcia-Sastre, A.; Heins, G.; Wolff, T. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 2002, 76, 11166–11171. [Google Scholar]
- Talon, J.; Horvath, C.M.; Polley, R.; Basler, C.F.; Muster, T.; Palese, P.; Garcia-Sastre, A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 2000, 74, 7989–7996. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.; Zheng, H.; Muster, T.; Palese, P.; Beg, A.A.; Garcia-Sastre, A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J. Virol. 2000, 74, 11566–11573. [Google Scholar]
- Ruckle, A.; Haasbach, E.; Julkunen, I.; Planz, O.; Ehrhardt, C.; Ludwig, S. The NS1 protein of influenza A virus blocks RIG-I mediated activation of the noncanonical NF-kappaB pathway and p52/RelB dependent gene expression in lung epithelial cells. J. Virol. 2012, 86, 10211–7. [Google Scholar] [CrossRef]
- Mibayashi, M.; Martinez-Sobrido, L.; Loo, Y.M.; Cardenas, W.B.; Gale, M., Jr.; Garcia-Sastre, A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 2007, 81, 514–524. [Google Scholar]
- Opitz, B.; Rejaibi, A.; Dauber, B.; Eckhard, J.; Vinzing, M.; Schmeck, B.; Hippenstiel, S.; Suttorp, N.; Wolff, T. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 2007, 9, 930–938. [Google Scholar]
- Nemeroff, M.E.; Barabino, S.M.; Li, Y.; Keller, W.; Krug, R.M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3'end formation of cellular pre-mRNAs. Mol. Cell 1998, 1, 991–1000. [Google Scholar] [CrossRef]
- Noah, D.L.; Twu, K.Y.; Krug, R.M. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3' end processing of cellular pre-mRNAS. Virology 2003, 307, 386–395. [Google Scholar]
- Das, K.; Ma, L.C.; Xiao, R.; Radvansky, B.; Aramini, J.; Zhao, L.; Marklund, J.; Kuo, R.L.; Twu, K.Y.; Arnold, E.; et al. Structural basis for suppression of a host antiviral response by influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 13093–13098. [Google Scholar]
- Qian, X.Y.; Alonso-Caplen, F.; Krug, R.M. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 1994, 68, 2433–2441. [Google Scholar]
- Satterly, N.; Tsai, P.L.; van Deursen, J.; Nussenzveig, D.R.; Wang, Y.; Faria, P.A.; Levay, A.; Levy, D.E.; Fontoura, B.M. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 1853–1858. [Google Scholar]
- Varga, Z.T.; Ramos, I.; Hai, R.; Schmolke, M.; Garcia-Sastre, A.; Fernandez-Sesma, A.; Palese, P. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 2011, 7, e1002067. [Google Scholar] [CrossRef]
- Conenello, G.M.; Tisoncik, J.R.; Rosenzweig, E.; Varga, Z.T.; Palese, P.; Katze, M.G. A single N66S mutation in the PB1-F2 protein of influenza A virus increases virulence by inhibiting the early interferon response in vivo. J. Virol. 2011, 85, 652–662. [Google Scholar]
- Dudek, S.E.; Wixler, L.; Nordhoff, C.; Nordmann, A.; Anhlan, D.; Wixler, V.; Ludwig, S. The influenza virus PB1-F2 protein has interferon antagonistic activity. Biol. Chem. 2011, 392, 1135–1144. [Google Scholar]
- Graef, K.M.; Vreede, F.T.; Lau, Y.F.; McCall, A.W.; Carr, S.M.; Subbarao, K.; Fodor, E. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J. Virol. 2010, 84, 8433–8445. [Google Scholar] [CrossRef]
- Iwai, A.; Shiozaki, T.; Kawai, T.; Akira, S.; Kawaoka, Y.; Takada, A.; Kida, H.; Miyazaki, T. Influenza A virus polymerase inhibits type I interferon induction by binding to interferon beta promoter stimulator 1. J. Biol. Chem. 2010, 285, 32064–32074. [Google Scholar]
- Guilligay, D.; Tarendeau, F.; Resa-Infante, P.; Coloma, R.; Crepin, T.; Sehr, P.; Lewis, J.; Ruigrok, R.W.; Ortin, J.; Hart, D.J.; et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 2008, 15, 500–506. [Google Scholar]
- Sugiyama, K.; Obayashi, E.; Kawaguchi, A.; Suzuki, Y.; Tame, J.R.; Nagata, K.; Park, S.Y. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 2009, 28, 1803–1811. [Google Scholar] [CrossRef]
- Dias, A.; Bouvier, D.; Crepin, T.; McCarthy, A.A.; Hart, D.J.; Baudin, F.; Cusack, S.; Ruigrok, R.W. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009, 458, 914–918. [Google Scholar]
- Plotch, S.J.; Bouloy, M.; Ulmanen, I.; Krug, R.M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 1981, 23, 847–858. [Google Scholar] [CrossRef]
- Yuan, P.; Bartlam, M.; Lou, Z.; Chen, S.; Zhou, J.; He, X.; Lv, Z.; Ge, R.; Li, X.; Deng, T.; et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 2009, 458, 909–913. [Google Scholar]
- Goodman, A.G.; Smith, J.A.; Balachandran, S.; Perwitasari, O.; Proll, S.C.; Thomas, M.J.; Korth, M.J.; Barber, G.N.; Schiff, L.A.; Katze, M.G. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J. Virol. 2007, 81, 2221–2230. [Google Scholar]
- Melville, M.W.; Hansen, W.J.; Freeman, B.C.; Welch, W.J.; Katze, M.G. The molecular chaperone hsp40 regulates the activity of P58IPK, the cellular inhibitor of PKR. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 97–102. [Google Scholar]
- Sharma, K.; Tripathi, S.; Ranjan, P.; Kumar, P.; Garten, R.; Deyde, V.; Katz, J.M.; Cox, N.J.; Lal, R.B.; Sambhara, S.; et al. Influenza A virus nucleoprotein exploits Hsp40 to inhibit PKR activation. PLoS One 2011, 6, e20215. [Google Scholar]
- Guan, Z.; Liu, D.; Mi, S.; Zhang, J.; Ye, Q.; Wang, M.; Gao, G.F.; Yan, J. Interaction of Hsp40 with influenza virus M2 protein: Implications for PKR signaling pathway. Protein Cell 2010, 1, 944–955. [Google Scholar] [CrossRef]
- Pauli, E.K.; Schmolke, M.; Wolff, T.; Viemann, D.; Roth, J.; Bode, J.G.; Ludwig, S. Influenza A virus inhibits type I IFN signaling via NF-kappaB-dependent induction of SOCS-3 expression. PLoS Pathog. 2008, 4, e1000196. [Google Scholar] [CrossRef]
- Pothlichet, J.; Chignard, M.; Si-Tahar, M. Cutting edge: Innate immune response triggered by influenza A virus is negatively regulated by SOCS1 and SOCS3 through a RIG-I/IFNAR1-dependent pathway. J. Immunol. 2008, 180, 2034–2038. [Google Scholar]
- Boliar, S.; Chambers, T.M. A new strategy of immune evasion by influenza A virus: Inhibition of monocyte differentiation into dendritic cells. Vet. Immunol. Immunopathol. 2010, 136, 201–210. [Google Scholar] [CrossRef]
- Fernandez-Sesma, A.; Marukian, S.; Ebersole, B.J.; Kaminski, D.; Park, M.S.; Yuen, T.; Sealfon, S.C.; Garcia-Sastre, A.; Moran, T.M. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol. 2006, 80, 6295–6304. [Google Scholar]
- Guo, H.; Kumar, P.; Malarkannan, S. Evasion of natural killer cells by influenza virus. J. Leukoc. Biol. 2011, 89, 189–194. [Google Scholar] [CrossRef]
- Owen, R.E.; Yamada, E.; Thompson, C.I.; Phillipson, L.J.; Thompson, C.; Taylor, E.; Zambon, M.; Osborn, H.M.; Barclay, W.S.; Borrow, P. Alterations in receptor binding properties of recent human influenza H3N2 viruses are associated with reduced natural killer cell lysis of infected cells. J. Virol. 2007, 81, 11170–11178. [Google Scholar]
- Mao, H.; Tu, W.; Liu, Y.; Qin, G.; Zheng, J.; Chan, P.L.; Lam, K.T.; Peiris, J.S.; Lau, Y.L. Inhibition of human natural killer cell activity by influenza virions and hemagglutinin. J. Virol. 2010, 84, 4148–4157. [Google Scholar] [CrossRef]
- Mao, H.; Tu, W.; Qin, G.; Law, H.K.; Sia, S.F.; Chan, P.L.; Liu, Y.; Lam, K.T.; Zheng, J.; Peiris, M.; et al. Influenza virus directly infects human natural killer cells and induces cell apoptosis. J. Virol. 2009, 83, 9215–9222. [Google Scholar]
- Rambaut, A.; Pybus, O.G.; Nelson, M.I.; Viboud, C.; Taubenberger, J.K.; Holmes, E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature 2008, 453, 615–619. [Google Scholar]
- McHardy, A.C.; Adams, B. The role of genomics in tracking the evolution of influenza A virus. PLoS Pathog. 2009, 5, e1000566. [Google Scholar]
- Sorrell, E.M.; Schrauwen, E.J.; Linster, M.; De Graaf, M.; Herfst, S.; Fouchier, R.A. Predicting 'airborne' influenza viruses: (Trans-) mission impossible? Curr. Opin. Virol. 2011, 1, 635–642. [Google Scholar] [CrossRef]
- Bodewes, R.; Nieuwkoop, N.J.; Verburgh, R.J.; Fouchier, R.A.; Osterhaus, A.; Rimmelzwaan, G.F. The use of influenza A viruses expressing reporter genes to assess the frequency of double infections in vitro. J. Gen. Virol. 2012, 93, 1645–1648. [Google Scholar]
- Ito, T.; Couceiro, J.N.; Kelm, S.; Baum, L.G.; Krauss, S.; Castrucci, M.R.; Donatelli, I.; Kida, H.; Paulson, J.C.; Webster, R.G.; et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998, 72, 7367–7373. [Google Scholar]
- Ma, W.; Lager, K.M.; Vincent, A.L.; Janke, B.H.; Gramer, M.R.; Richt, J.A. The role of swine in the generation of novel influenza viruses. Zoonoses Public Health 2009, 56, 326–337. [Google Scholar] [CrossRef]
- Suzuki, Y.; Ito, T.; Suzuki, T.; Holland, R.E., Jr.; Chambers, T.M.; Kiso, M.; Ishida, H.; Kawaoka, Y. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 2000, 74, 11825–11831. [Google Scholar] [CrossRef]
- Claas, E.C.; Kawaoka, Y.; de Jong, J.C.; Masurel, N.; Webster, R.G. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology 1994, 204, 453–457. [Google Scholar]
- Pensaert, M.; Ottis, K.; Vandeputte, J.; Kaplan, M.M.; Bachmann, P.A. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull. World Health Organ. 1981, 59, 75–78. [Google Scholar]
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006, 12, 9–14. [Google Scholar] [CrossRef]
- Smith, G.J.; Bahl, J.; Vijaykrishna, D.; Zhang, J.; Poon, L.L.; Chen, H.; Webster, R.G.; Peiris, J.S.; Guan, Y. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 11709–11712. [Google Scholar]
- Scholtissek, C.; Rohde, W.; Von Hoyningen, V.; Rott, R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 1978, 87, 13–20. [Google Scholar] [CrossRef]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar]
- Thontiravong, A.; Kitikoon, P.; Wannaratana, S.; Tantilertcharoen, R.; Tuanudom, R.; Pakpinyo, S.; Sasipreeyajan, J.; Oraveerakul, K.; Amonsin, A. Quail as a potential mixing vessel for the generation of new reassortant influenza A viruses. Vet. Microbiol. 2012, in press. [Google Scholar]
- Han, T.; Marasco, W.A. Structural basis of influenza virus neutralization. Ann. New York Acad. Sci. 2011, 1217, 178–190. [Google Scholar]
- Kwong, P.D.; Doyle, M.L.; Casper, D.J.; Cicala, C.; Leavitt, S.A.; Majeed, S.; Steenbeke, T.D.; Venturi, M.; Chaiken, I.; Fung, M.; et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 2002, 420, 678–682. [Google Scholar]
- Wyatt, R.; Kwong, P.D.; Desjardins, E.; Sweet, R.W.; Robinson, J.; Hendrickson, W.A.; Sodroski, J.G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 1998, 393, 705–711. [Google Scholar]
- Horst, D.; Verweij, M.C.; Davison, A.J.; Ressing, M.E.; Wiertz, E.J. Viral evasion of T cell immunity: Ancient mechanisms offering new applications. Curr. Opin. Immunol. 2011, 23, 96–103. [Google Scholar]
- Berkhoff, E.G.; de Wit, E.; Geelhoed-Mieras, M.M.; Boon, A.C.; Symons, J.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes. J. Virol. 2005, 79, 11239–11246. [Google Scholar]
- Del Val, M.; Schlicht, H.J.; Ruppert, T.; Reddehase, M.J.; Koszinowski, U.H. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell 1991, 66, 1145–1153. [Google Scholar] [CrossRef]
- Eisenlohr, L.C.; Yewdell, J.W.; Bennink, J.R. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J. Exp. Med. 1992, 175, 481–487. [Google Scholar]
- Neisig, A.; Roelse, J.; Sijts, A.J.; Ossendorp, F.; Feltkamp, M.C.; Kast, W.M.; Melief, C.J.; Neefjes, J.J. Major differences in transporter associated with antigen presentation (TAP)-dependent translocation of MHC class I-presentable peptides and the effect of flanking sequences. J. Immunol. 1995, 154, 1273–1279. [Google Scholar]
- Milicic, A.; Price, D.A.; Zimbwa, P.; Booth, B.L.; Brown, H.L.; Easterbrook, P.J.; Olsen, K.; Robinson, N.; Gileadi, U.; Sewell, A.K.; et al. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J. Immunol. 2005, 175, 4618–4626. [Google Scholar]
- Berkhoff, E.G.; Boon, A.C.; Nieuwkoop, N.J.; Fouchier, R.A.; Sintnicolaas, K.; Osterhaus, A.D.; Rimmelzwaan, G.F. A mutation in the HLA-B*2705-restricted NP383-391 epitope affects the human influenza A virus-specific cytotoxic T-lymphocyte response in vitro. J. Virol. 2004, 78, 5216–5222. [Google Scholar]
- Berkhoff, E.G.; Geelhoed-Mieras, M.M.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Assessment of the extent of variation in influenza A virus cytotoxic T-lymphocyte epitopes by using virus-specific CD8+ T-cell clones. J. Gen. Virol. 2007, 88, 530–535. [Google Scholar] [CrossRef]
- Price, G.E.; Ou, R.; Jiang, H.; Huang, L.; Moskophidis, D. Viral escape by selection of cytotoxic T cell-resistant variants in influenza A virus pneumonia. J. Exp. Med. 2000, 191, 1853–1867. [Google Scholar] [CrossRef]
- Rimmelzwaan, G.F.; Boon, A.C.; Voeten, J.T.; Berkhoff, E.G.; Fouchier, R.A.; Osterhaus, A.D. Sequence variation in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. Virus Res. 2004, 103, 97–100. [Google Scholar] [CrossRef]
- Voeten, J.T.; Bestebroer, T.M.; Nieuwkoop, N.J.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J. Virol. 2000, 74, 6800–6807. [Google Scholar] [CrossRef]
- Berkhoff, E.G.; Geelhoed-Mieras, M.M.; Verschuren, E.J.; van Baalen, C.A.; Gruters, R.A.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. The loss of immunodominant epitopes affects interferon-gamma production and lytic activity of the human influenza virus-specific cytotoxic T lymphocyte response in vitro. Clin. Exp. Immunol. 2007, 148, 296–306. [Google Scholar] [CrossRef]
- Boon, A.C.; de Mutsert, G.; Graus, Y.M.; Fouchier, R.A.; Sintnicolaas, K.; Osterhaus, A.D.; Rimmelzwaan, G.F. Sequence variation in a newly identified HLA-B35-restricted epitope in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. J. Virol. 2002, 76, 2567–2572. [Google Scholar]
- Cale, E.M.; Bazick, H.S.; Rianprakaisang, T.A.; Alam, S.M.; Letvin, N.L. Mutations in a dominant Nef epitope of simian immunodeficiency virus diminish TCR: Epitope peptide affinity but not epitope peptide:MHC class I binding. J. Immunol. 2011, 187, 3300–3313. [Google Scholar] [CrossRef]
- Huet, S.; Nixon, D.F.; Rothbard, J.B.; Townsend, A.; Ellis, S.A.; McMichael, A.J. Structural homologies between two HLA B27-restricted peptides suggest residues important for interaction with HLA B27. Int. Immunol. 1990, 2, 311–316. [Google Scholar] [CrossRef]
- Gog, J.R.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Grenfell, B.T. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 11143–11147. [Google Scholar]
- Boon, A.C.; de Mutsert, G.; van Baarle, D.; Smith, D.J.; Lapedes, A.S.; Fouchier, R.A.; Sintnicolaas, K.; Osterhaus, A.D.; Rimmelzwaan, G.F. Recognition of homo- and heterosubtypic variants of influenza A viruses by human CD8+ T lymphocytes. J. Immunol. 2004, 172, 2453–2460. [Google Scholar]
- Gras, S.; Kedzierski, L.; Valkenburg, S.A.; Laurie, K.; Liu, Y.C.; Denholm, J.T.; Richards, M.J.; Rimmelzwaan, G.F.; Kelso, A.; Doherty, P.C.; et al. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 12599–12604. [Google Scholar]
- Berkhoff, E.G.; de Wit, E.; Geelhoed-Mieras, M.M.; Boon, A.C.; Symons, J.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Fitness costs limit escape from cytotoxic T lymphocytes by influenza A viruses. Vaccine 2006, 24, 6594–6596. [Google Scholar] [CrossRef]
- Rimmelzwaan, G.F.; Berkhoff, E.G.; Nieuwkoop, N.J.; Fouchier, R.A.; Osterhaus, A.D. Functional compensation of a detrimental amino acid substitution in a cytotoxic-T-lymphocyte epitope of influenza a viruses by comutations. J. Virol. 2004, 78, 8946–8949. [Google Scholar] [CrossRef]
- Rimmelzwaan, G.F.; Berkhoff, E.G.; Nieuwkoop, N.J.; Smith, D.J.; Fouchier, R.A.; Osterhaus, A.D. Full restoration of viral fitness by multiple compensatory co-mutations in the nucleoprotein of influenza A virus cytotoxic T-lymphocyte escape mutants. J. Gen. Virol. 2005, 86, 1801–1805. [Google Scholar] [CrossRef]
- Berkhoff, E.G.; Geelhoed-Mieras, M.M.; Jonges, M.; Smith, D.J.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. An amino acid substitution in the influenza A virus hemagglutinin associated with escape from recognition by human virus-specific CD4+ T-cells. Virus Res. 2007, 126, 282–287. [Google Scholar] [CrossRef]
- Cox, R.J.; Brokstad, K.A.; Ogra, P. Influenza virus: Immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand. J. Immunol. 2004, 59, 1–15. [Google Scholar] [CrossRef]
- Thomas, P.G.; Keating, R.; Hulse-Post, D.J.; Doherty, P.C. Cell-mediated protection in influenza infection. Emerg. Infect. Dis. 2006, 12, 48–54. [Google Scholar] [CrossRef]
- Kreijtz, J.H.; Osterhaus, A.D.; Rimmelzwaan, G.F. Vaccination strategies and vaccine formulations for epidemic and pandemic influenza control. Hum. Vaccine. 2009, 5, 126–135. [Google Scholar] [CrossRef]
- Russell, C.A.; Jones, T.C.; Barr, I.G.; Cox, N.J.; Garten, R.J.; Gregory, V.; Gust, I.D.; Hampson, A.W.; Hay, A.J.; Hurt, A.C.; et al. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 2008, 26, D31–D34. [Google Scholar]
- de Jong, J.C.; Beyer, W.E.; Palache, A.M.; Rimmelzwaan, G.F.; Osterhaus, A.D. Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J. Med. Virol. 2000, 61, 94–99. [Google Scholar] [CrossRef]
- Salzberg, S. The contents of the syringe. Nature 2008, 454, 160–161. [Google Scholar]
- Jin, H.; Zhou, H.; Liu, H.; Chan, W.; Adhikary, L.; Mahmood, K.; Lee, M.S.; Kemble, G. Two residues in the hemagglutinin of A/Fujian/411/02-like influenza viruses are responsible for antigenic drift from A/Panama/2007/99. Virology 2005, 336, 113–119. [Google Scholar] [CrossRef]
- WHO. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 2005, 11, 1515–1521.
- Pada, S.; Tambyah, P.A. Overview/reflections on the 2009 H1N1 pandemic. Microb. Infect. 2011, 13, 470–478. [Google Scholar] [CrossRef]
- Rockman, S.; Brown, L. Pre-pandemic and pandemic influenza vaccines. Hum. Vaccine. 2010, 6, 792–801. [Google Scholar] [CrossRef]
- Belshe, R.B.; Nichol, K.L.; Black, S.B.; Shinefield, H.; Cordova, J.; Walker, R.; Hessel, C.; Cho, I.; Mendelman, P.M. Safety, efficacy, and effectiveness of live, attenuated, cold-adapted influenza vaccine in an indicated population aged 5-49 years. Clin. Infect. Dis. 2004, 39, 920–927. [Google Scholar] [CrossRef]
- Mossad, S.B. Demystifying FluMist, a new intranasal, live influenza vaccine. Cleve. Clin. J. Med. 2003, 70, 801–806. [Google Scholar] [CrossRef]
- Beyer, W.E.; Palache, A.M.; de Jong, J.C.; Osterhaus, A.D. Cold-adapted live influenza vaccine versus inactivated vaccine: Systemic vaccine reactions, local and systemic antibody response, and vaccine efficacy. A meta-analysis. Vaccine 2002, 20, 1340–1353. [Google Scholar] [CrossRef]
- Rudenko, L.; van den Bosch, H.; Kiseleva, I.; Mironov, A.; Naikhin, A.; Larionova, N.; Bushmenkov, D. Live attenuated pandemic influenza vaccine: Clinical studies on A/17/California/2009/38 (H1N1) and licensing of the Russian-developed technology to WHO for pandemic influenza preparedness in developing countries. Vaccine 2011, 29, A40–A44. [Google Scholar]
- Hoft, D.F.; Babusis, E.; Worku, S.; Spencer, C.T.; Lottenbach, K.; Truscott, S.M.; Abate, G.; Sakala, I.G.; Edwards, K.M.; Creech, C.B.; et al. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J. Infect. Dis. 2011, 204, 845–853. [Google Scholar] [CrossRef]
- He, Q.; Martinez-Sobrido, L.; Eko, F.O.; Palese, P.; Garcia-Sastre, A.; Lyn, D.; Okenu, D.; Bandea, C.; Ananaba, G.A.; Black, C.M.; et al. Live-attenuated influenza viruses as delivery vectors for Chlamydia vaccines. Immunology 2007, 122, 28–37. [Google Scholar] [CrossRef]
- Pica, N.; Langlois, R.A.; Krammer, F.; Margine, I.; Palese, P. NS1-truncated live attenuated virus vaccine provides robust protection to aged mice from viral challenge. J. Virol. 2012. [Google Scholar]
- Bodewes, R.; Kreijtz, J.H.; Rimmelzwaan, G.F. Yearly influenza vaccinations: A double-edged sword? Lancet Infect. Dis. 2009, 9, 784–788. [Google Scholar] [CrossRef]
- Bodewes, R.; Fraaij, P.L.; Geelhoed-Mieras, M.M.; van Baalen, C.A.; Tiddens, H.A.; van Rossum, A.M.; van der Klis, F.R.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Annual vaccination against influenza virus hampers development of virus-specific CD8(+) T cell immunity in children. J. Virol. 2011, 85, 11995–12000. [Google Scholar]
- Bodewes, R.; Kreijtz, J.H.; Baas, C.; Geelhoed-Mieras, M.M.; de Mutsert, G.; van Amerongen, G.; van den Brand, J.M.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Vaccination against human influenza A/H3N2 virus prevents the induction of heterosubtypic immunity against lethal infection with avian influenza A/H5N1 virus. PLoS One 2009, 4, e5538. [Google Scholar]
- Bodewes, R.; Kreijtz, J.H.; Geelhoed-Mieras, M.M.; van Amerongen, G.; Verburgh, R.J.; van Trierum, S.E.; Kuiken, T.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Vaccination against seasonal influenza A/H3N2 virus reduces the induction of heterosubtypic immunity against influenza A/H5N1 virus infection in ferrets. J. Virol. 2011, 85, 2695–2702. [Google Scholar]
- Bodewes, R.; Kreijtz, J.H.; Hillaire, M.L.; Geelhoed-Mieras, M.M.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Vaccination with whole inactivated virus vaccine affects the induction of heterosubtypic immunity against influenza virus A/H5N1 and immunodominance of virus-specific CD8+ T-cell responses in mice. J. Gen. Virol. 2010, 91, 1743–1753. [Google Scholar]
- Heiny, A.T.; Miotto, O.; Srinivasan, K.N.; Khan, A.M.; Zhang, G.L.; Brusic, V.; Tan, T.W.; August, J.T. Evolutionarily conserved protein sequences of influenza a viruses, avian and human, as vaccine targets. PLoS One 2007, 2, e1190. [Google Scholar]
- Rimmelzwaan, G.F.; Fouchier, R.A.; Osterhaus, A.D. Influenza virus-specific cytotoxic T lymphocytes: A correlate of protection and a basis for vaccine development. Curr. Opin. Biotechnol. 2007, 18, 529–536. [Google Scholar]
- Daemen, T.; de Mare, A.; Bungener, L.; de Jonge, J.; Huckriede, A.; Wilschut, J. Virosomes for antigen and DNA delivery. Adv. Drug Deliv. Rev. 2005, 57, 451–463. [Google Scholar] [CrossRef]
- Berthoud, T.K.; Hamill, M.; Lillie, P.J.; Hwenda, L.; Collins, K.A.; Ewer, K.J.; Milicic, A.; Poyntz, H.C.; Lambe, T.; Fletcher, H.A.; et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin. Infect. Dis. 2011, 52, 1–7. [Google Scholar]
- Liniger, M.; Zuniga, A.; Naim, H.Y. Use of viral vectors for the development of vaccines. Expet Rev. Vaccine 2007, 6, 255–266. [Google Scholar] [CrossRef]
- Ulmer, J.B. Influenza DNA vaccines. Vaccine 2002, 20, S74–S76. [Google Scholar] [CrossRef]
- Rimmelzwaan, G.F.; Nieuwkoop, N.; Brandenburg, A.; Sutter, G.; Beyer, W.E.; Maher, D.; Bates, J.; Osterhaus, A.D. A randomized, double blind study in young healthy adults comparing cell mediated and humoral immune responses induced by influenza ISCOM vaccines and conventional vaccines. Vaccine 2000, 19, 1180–1187. [Google Scholar] [CrossRef]
- Sambhara, S.; Woods, S.; Arpino, R.; Kurichh, A.; Tamane, A.; Underdown, B.; Klein, M.; Lovgren Bengtsson, K.; Morein, B.; Burt, D. Heterotypic protection against influenza by immunostimulating complexes is associated with the induction of cross-reactive cytotoxic T lymphocytes. J. Infect. Dis. 1998, 177, 1266–1274. [Google Scholar]
- Lipatov, A.S.; Gitelman, A.K.; Smirnov Yu, A. Prevention and treatment of lethal influenza A virus bronchopneumonia in mice by monoclonal antibody against haemagglutinin stem region. Acta Virol. 1997, 41, 337–340. [Google Scholar]
- Fan, J.; Liang, X.; Horton, M.S.; Perry, H.C.; Citron, M.P.; Heidecker, G.J.; Fu, T.M.; Joyce, J.; Przysiecki, C.T.; Keller, P.M.; et al. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine 2004, 22, 2993–3003. [Google Scholar]
- Song, J.M.; Wang, B.Z.; Park, K.M.; Van Rooijen, N.; Quan, F.S.; Kim, M.C.; Jin, H.T.; Pekosz, A.; Compans, R.W.; Kang, S.M. Influenza virus-like particles containing M2 induce broadly cross protective immunity. PLoS One 2011, 6, e14538. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Van de Sandt, C.E.; Kreijtz, J.H.C.M.; Rimmelzwaan, G.F. Evasion of Influenza A Viruses from Innate and Adaptive Immune Responses. Viruses 2012, 4, 1438-1476. https://doi.org/10.3390/v4091438
Van de Sandt CE, Kreijtz JHCM, Rimmelzwaan GF. Evasion of Influenza A Viruses from Innate and Adaptive Immune Responses. Viruses. 2012; 4(9):1438-1476. https://doi.org/10.3390/v4091438
Chicago/Turabian StyleVan de Sandt, Carolien E., Joost H. C. M. Kreijtz, and Guus F. Rimmelzwaan. 2012. "Evasion of Influenza A Viruses from Innate and Adaptive Immune Responses" Viruses 4, no. 9: 1438-1476. https://doi.org/10.3390/v4091438