The authors wish to make the following changes to their paper [1].
(1) Figure 5 should be replaced with:
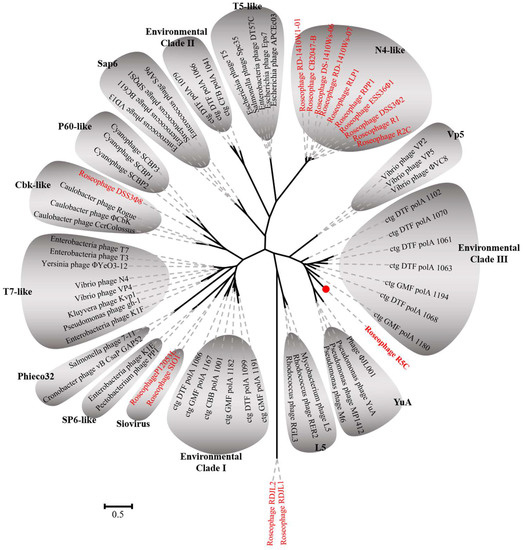
Figure 5.
Unrooted maximum likelihood phylogenetic tree of DNA polymerase I of bacteriophages. Red color represents the roseophages. The sequences were aligned by ClustalW and Bootstrap = 1000.
(2) In Section 3.4. “Phylogenetic Analyses” within Results and Discussion, the sentence of “On the DNA polymerase I phylogenetic tree, R5C was most closely related to ctg DTF polA 1086, which was an environmental DNA polymerase sequence from Dry Tortugas surface water (Figure 5)” should be corrected as “On the DNA polymerase I phylogenetic tree, R5C formed a relatively single branch (Figure 5)”.
The changes were caused by a mistake during the sequence alignment.
The authors apologize for any inconvenience this may have caused. The manuscript will be updated and the original will remain online on the article webpage, with a reference to this erratum.
References
- Yang, Y.; Cai, L.; Ma, R.; Xu, Y.; Tong, Y.; Huang, Y.; Jiao, N.; Zhang, R. A Novel Roseosiphophage Isolated from the Oligotrophic South China Sea. Viruses 2017, 9, 109. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).