RNA Phage Biology in a Metagenomic Era
Abstract
:1. Introduction
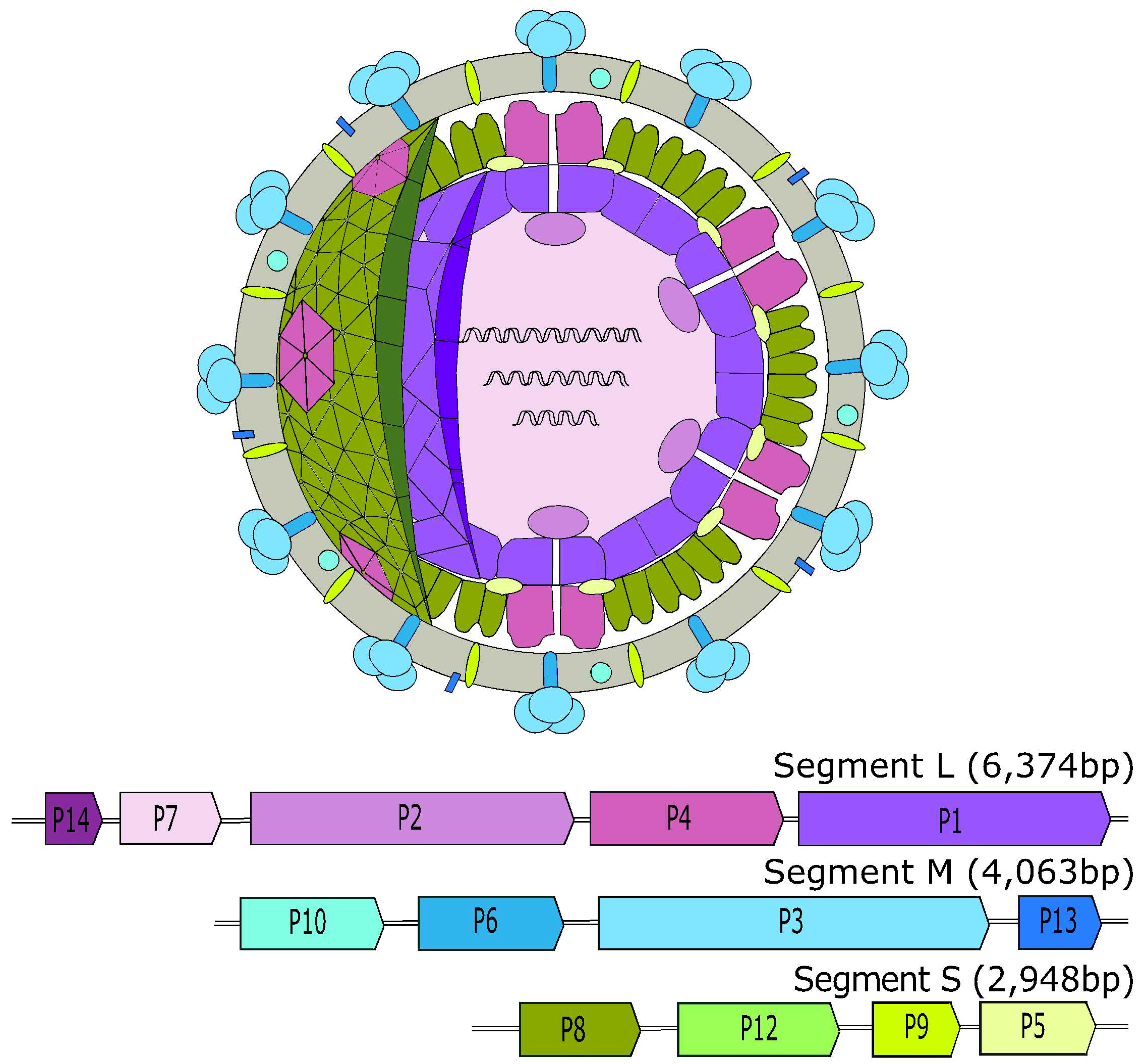
2. Cystoviridae
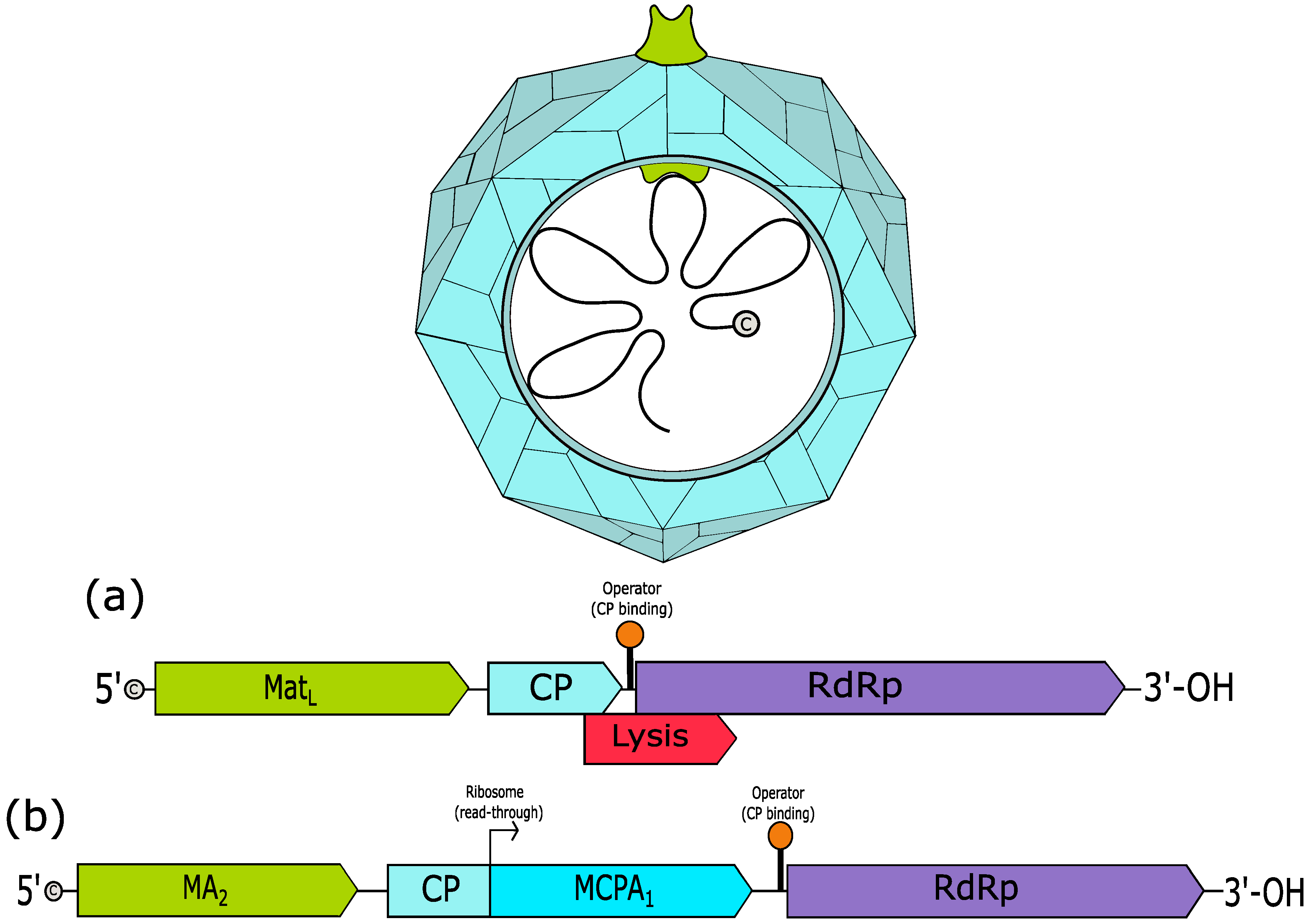
3. Leviviridae
3.1. Levivirus
3.2. Allolevirus
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hatfull, G.F. Dark Matter of the Biosphere: The Amazing World of Bacteriophage Diversity. J. Virol. 2015, 89, 8107–8110. [Google Scholar] [CrossRef] [PubMed]
- Twort, F.W. An Investigation on the nature of ultra-microscopic viruses. Lancet 1915, 186, 1241–1243. [Google Scholar] [CrossRef]
- d’Herelle, F. Sur un microbe invisible antagoniste des bacilles dysentériques. CR Acad. Sci. Paris 1917, 165, 373–375. [Google Scholar]
- Rodriguez-Valera, F.; Martin-Cuadrado, A.-B.; Rodriguez-Brito, B.; Pašić, L.; Thingstad, T.F.; Rohwer, F.; Mira, A. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 2009, 7, 828–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlton, R. Phage Therapy: Past History and Future Prospects. Arch. Immunol. Ther. Exp.-Engl. Ed. 1999, 47, 267–274. [Google Scholar]
- Summers, W.C. The strange history of phage therapy. Bacteriophage 2012, 2, 130–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loeb, T.; Zinder, N.D. A bacteriophage containing RNA. Proc. Natl. Acad. Sci. USA 1961, 47, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Weissmann, C. The making of a phage. FEBS Lett. 1974, 40, S3–S9. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.; Gold, L. RNA replication by Q beta replicase: A working model. Proc. Natl. Acad. Sci. USA 1996, 93, 11558–11562. [Google Scholar] [CrossRef] [PubMed]
- Gytz, H.; Mohr, D.; Seweryn, P.; Yoshimura, Y.; Kutlubaeva, Z.; Dolman, F.; Chelchessa, B.; Chetverin, A.B.; Mulder, F.A.A.; Brodersen, D.E.; et al. Structural basis for RNA-genome recognition during bacteriophage Qβ replication. Nucleic Acids Res. 2015, 43, 10893–10906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodish, H.F. Bacteriophage f2 RNA: Control of Translation and Gene Order. Nature 1968, 220, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Stock-Ley, P.G.; Stonehouse, N.J.; Valegård, K. Molecular mechanism of RNA phage morphogenesis. Int. J. Biochem. 1994, 26, 1249–1260. [Google Scholar] [CrossRef]
- Davis, J.E.; Strauss, J.H.; Sinsheimer, R.L. Bacteriophage MS2: Another RNA Phage. Science 1961, 14, 1427. [Google Scholar]
- Jou, W.M.; Haegeman, G.; Ysebaert, M.; Fiers, W. Nucleotide Sequence of the Gene Coding for the Bacteriophage MS2 Coat Protein. Nature 1972, 237, 82–88. [Google Scholar] [CrossRef]
- Fiers, W.; Contreras, R.; Duerinck, F.; Haegeman, G.; Iserentant, D.; Merregaert, J.; Min Jou, W.; Molemans, F.; Raeymaekers, A.; Van den Berghe, A.; et al. Complete nucleotide sequence of bacteriophage MS2 RNA: Primary and secondary structure of the replicase gene. Nature 1976, 260, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Air, G.M.; Barrell, B.G.; Brown, N.L.; Coulson, A.R.; Fiddes, J.C.; Iii, C.A.H.; Slocombe, P.M.; Smith, M. Nucleotide sequence of bacteriophage φX174 DNA. Nature 1977, 265, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Adcock, N.J.; Rice, E.W.; Sivaganesan, M.; Brown, J.D.; Stallknecht, D.E.; Swayne, D.E. The use of bacteriophages of the family Cystoviridae as surrogates for H5N1 highly pathogenic avian influenza viruses in persistence and inactivation studies. J. Environ. Sci. Health Part A 2009, 44, 1362–1366. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.C.; Prestwood, L.J.; Lever, A.M.L. A novel combined RNA-protein interaction analysis distinguishes HIV-1 Gag protein binding sites from structural change in the viral RNA leader. Sci. Rep. 2015, 5, 14369. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Y.; Li, D.; Zhou, T.; Gao, S.; Zha, E.; Yue, X. Preparation and evaluation of MS2 bacteriophage-like particles packaging hepatitis E virus RNA. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, J.T.; Sherer, N.M. Subcellular Localization of HIV-1 gag-pol mRNAs Regulates Sites of Virion Assembly. J. Virol. 2017, 91, e02315-16. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.; Brockhurst, M.A. Ecological and Evolutionary Benefits of Temperate Phage: What Does or Doesn’t Kill You Makes You Stronger. BioEssays 2017, 39, 1700112. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, I.; Kozlovska, T.; Cielens, I.; Strelnikova, A.; Kazaks, A.; Ose, V.; Pumpens, P. Mosaic Qβ coats as a new presentation model. FEBS Lett. 1998, 431, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Hatfull, G.F. Bacteriophage Genomics. Curr. Opin. Microbiol. 2008, 11, 447–453. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, F.; Davies, J. Horizontal gene transfer and the origin of species: Lessons from bacteria. Trends Microbiol. 2000, 8, 128–133. [Google Scholar] [CrossRef]
- Cenens, W.; Makumi, A.; Govers, S.K.; Lavigne, R.; Aertsen, A. Viral Transmission Dynamics at Single-Cell Resolution Reveal Transiently Immune Subpopulations Caused by a Carrier State Association. PLoS Genet. 2015, 11, e1005770. [Google Scholar] [CrossRef] [PubMed]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Olsthoorn, R.C.L.; Van Duin, J. Leviviridae—Positive Sense RNA Viruses—Positive Sense RNA Viruses (2011)—International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/263/leviviridae (accessed on 26 February 2018).
- Poranen, M.M.; Mäntynen, S. ICTV Virus Taxonomy Profile: Cystoviridae. J. Gen. Virol. 2017, 98, 2423–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamurthy, S.R.; Janowski, A.B.; Zhao, G.; Barouch, D.; Wang, D. Hyperexpansion of RNA Bacteriophage Diversity. PLoS Biol. 2016, 14, e1002409. [Google Scholar] [CrossRef] [PubMed]
- Saberi, A.; Gulyaeva, A.A.; Brubacher, J.; Newmark, P.A.; Gorbalenya, A. A planarian nidovirus expands the limits of RNA genome size. bioRxiv 2018. [Google Scholar] [CrossRef]
- Van Etten, J.; Lane, L.; Gonzalez, C.; Partridge, J.; Vidaver, A. Comparative Properties of Bacteriophage φ6 and φ6 Nucleocapsid. J. Virol. 1976, 18, 652–658. [Google Scholar]
- Alphonse, S.; Ghose, R. Cystoviral RNA-directed RNA polymerases: Regulation of RNA synthesis on multiple time and length scales. Virus Res. 2017, 234, 135–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viral Zone: Cystoviridae. Available online: https://viralzone.expasy.org/165?outline=all_by_species (accessed on 30 May 2018).
- Gottlieb, P.; Wei, H.; Potgieter, C.; Toporovsky, I. Characterization of φ12, a Bacteriophage Related to φ6: Nucleotide Sequence of the Small and Middle Double-Stranded RNA. Virology 2002, 293, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, P.; Metzger, S.; Romantschuk, M.; Carton, J.; Strassman, J.; Bamford, D.H.; Kalkkinen, N.; Mindich, L. Nucleotide sequence of the middle dsRNA segment of bacteriophage φ6: Placement of the genes of membrane-associated proteins. Virology 1988, 163, 183–190. [Google Scholar] [CrossRef]
- Hoogstraten, D.; Qiao, X.; Sun, Y.; Hu, A.; Onodera, S.; Mindich, L. Characterization of φ8, a Bacteriophage Containing Three Double-Stranded RNA Genomic Segments and Distantly Related to φ6. Virology 2000, 272, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Mäntynen, S.; Laanto, E.; Kohvakka, A.; Poranen, M.M.; Bamford, J.K.H.; Ravantti, J.J. New enveloped dsRNA phage from freshwater habitat. J. Gen. Virol. 2015, 96, 1180–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGraw, T.; Mindich, L.; Frangione, B. Nucleotide sequence of the small double-stranded RNA segment of bacteriophage phi 6: Novel mechanism of natural translational control. J. Virol. 1986, 58, 142–151. [Google Scholar] [PubMed]
- Mindich, L.; Nemhauser, I.; Gottlieb, P.; Romantschuk, M.; Carton, J.; Frucht, S.; Strassman, J.; Bamford, D.H.; Kalkkinen, N. Nucleotide sequence of the large double-stranded RNA segment of bacteriophage phi 6: Genes specifying the viral replicase and transcriptase. J. Virol. 1988, 62, 1180–1185. [Google Scholar] [PubMed]
- Qiao, X.; Sun, Y.; Qiao, J.; Sanzo, F.D.; Mindich, L. Characterization of F2954, a newly isolated bacteriophage containing three dsRNA genomic segments. BMC Microbiol. 2010, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Qiao, J.; Onodera, S.; Mindich, L. Characterization of φ13, a Bacteriophage Related to φ6 and Containing Three dsRNA Genomic Segments. Virology 2000, 275, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lu, S.; Shen, W.; Zhao, X.; Shen, M.; Tan, Y.; Li, G.; Li, M.; Wang, J.; Hu, F.; et al. Characterization of the first double-stranded RNA bacteriophage infecting Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 38795. [Google Scholar] [CrossRef] [PubMed]
- Carpino, J. Structure and Function in Bacteriophage Phi6. Ph.D. Thesis, City University of New York, New York, NY, USA, 2014. [Google Scholar]
- Mindich, L. Precise Packaging of the Three Genomic Segments of the Double-Stranded-RNA Bacteriophage φ6. Microbiol. Mol. Biol. Rev. 1999, 63, 149–160. [Google Scholar] [PubMed]
- Mäntynen, S.; Sundberg, L.-R.; Poranen, M.M. Recognition of six additional cystoviruses: Pseudomonas virus phi6 is no longer the sole species of the family Cystoviridae. Arch. Virol. 2018, 163, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Bamford, D.H.; Palva, E.T.; Lounatmaa, K. Ultrastructure and Life Cycle of the Lipid-containing Bacteriophage φ6. J. Gen. Virol. 1976, 32, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Roine, E.; Raineri, D.M.; Romantschuk, M.; Wilson, M.; Nunn, D.N. Characterization of Type IV Pilus Genes in Pseudomonas syringae pv. tomato DC3000. Mol. Plant-Microbe Interact. 1998, 11, 1048–1056. [Google Scholar] [CrossRef] [PubMed]
- Mindich, L.; Qiao, X.; Qiao, J.; Onodera, S.; Romantschuk, M.; Hoogstraten, D. Isolation of Additional Bacteriophages with Genomes of Segmented Double-Stranded RNA. J. Bacteriol. 1999, 181, 4505–4508. [Google Scholar] [PubMed]
- Mindich, L.; Lehman, J. Cell Wall Lysin as a Component of the Bacteriophage ø6 Virion. J. Virol. 1979, 30, 489–496. [Google Scholar] [PubMed]
- Caldentey, J.; Bamford, D.H. The lytic enzyme of the Pseudomonas phage φ6. Purification and biochemical characterization. Biochim. Biophys. Acta BBA—Protein Struct. Mol. Enzymol. 1992, 1159, 44–50. [Google Scholar] [CrossRef]
- Poranen, M.M.; Daugelavičius, R.; Ojala, P.M.; Hess, M.W.; Bamford, D.H. A Novel Virus–Host Cell Membrane Interaction: Membrane Voltage–Dependent Endocytic-like Entry of Bacteriophage φ6 Nucleocapsid. J. Cell Biol. 1999, 147, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Romantschuk, M.; Olkkonen, V.M.; Bamford, D.H. The nucleocapsid of bacteriophage phi 6 penetrates the host cytoplasmic membrane. EMBO J. 1988, 7, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Poranen, M.M.; Bamford, D.H. Assembly of Large Icosahedral Double-Stranded RNA Viruses. In Viral Molecular Machines; Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 2012; pp. 379–402. ISBN 978-1-4614-0979-3. [Google Scholar]
- Juuti, J.T.; Bamford, D.H. Protein P7 of phage phi6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro assembly of the purified protein onto deficient particles. J. Mol. Biol. 1997, 266, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Poranen, M.M.; Paatero, A.O.; Tuma, R.; Bamford, D.H. Self-Assembly of a Viral Molecular Machine from Purified Protein and RNA Constituents. Mol. Cell 2001, 7, 845–854. [Google Scholar] [CrossRef]
- Usala, S.J.; Brownstein, B.H.; Haselkorn, R. Displacement of parental RNA strands during in vitro transcription by bacteriophage φ6 nucleocapsids. Cell 1980, 19, 855–862. [Google Scholar] [CrossRef]
- Blumenthal, T. Qbeta replicase template specificity: Different templates require different GTP concentrations for initiation. Proc. Natl. Acad. Sci. USA 1980, 77, 2601–2605. [Google Scholar] [CrossRef] [PubMed]
- Silverman, P.M. Replication of RNA viruses: Specific binding of the Qβ RNA polymerase to Qβ RNA. Arch. Biochem. Biophys. 1973, 157, 222–233. [Google Scholar] [CrossRef]
- Butcher, S.J.; Grimes, J.M.; Makeyev, E.V.; Bamford, D.H.; Stuart, D.I. A mechanism for initiating RNA-dependent RNA polymerization. Nature 2001, 410, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Makeyev, E.V.; Butcher, S.J.; Gaidelyte, A.; Bamford, D.H. Two Distinct Mechanisms Ensure Transcriptional Polarity in Double-Stranded RNA Bacteriophages. J. Virol. 2003, 77, 1195–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coplin, D.L.; Etten, J.L.V.; Koski, R.K.; Vidaver, A.K. Intermediates in the biosynthesis of double-stranded ribonucleic acids of bacteriophage phi 6. Proc. Natl. Acad. Sci. USA 1975, 72, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Emori, Y.; Iba, H.; Okada, Y. Transcriptional regulation of three double-stranded RNA segments of bacteriophage phi 6 in vitro. J. Virol. 1983, 46, 196–203. [Google Scholar] [PubMed]
- Frilander, M.; Bamford, D.H. In VitroPackaging of the Single-stranded RNA Genomic Precursors of the Segmented Double-stranded RNA Bacteriophage ψ: The Three Segments Modulate Each Other’s Packaging Efficiency. J. Mol. Biol. 1995, 246, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Poranen, M.M.; Tuma, R.; Bamford, D.H. Assembly of Double-Stranded RNA Bacteriophages. In Advances in Virus Research; Virus Structure and Assembly; Academic Press: Cambridge, MA, USA, 2005; Volume 64, pp. 15–43. [Google Scholar]
- Pirttimaa, M.J.; Paatero, A.O.; Frilander, M.J.; Bamford, D.H. Nonspecific Nucleoside Triphosphatase P4 of Double-Stranded RNA Bacteriophage 6 Is Required for Single-Stranded RNA Packaging and Transcription. J. Virol. 2002, 76, 10122–10127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, J.; Qiao, X.; Sun, Y.; Mindich, L. Isolation and Analysis of Mutants of Double-Stranded-RNA Bacteriophage φ6 with Altered Packaging Specificity. J. Bacteriol. 2003, 185, 4572–4577. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Qiao, J.; Mindich, L. Analysis of Specific Binding Involved in Genomic Packaging of the Double-Stranded-RNA Bacteriophage φ6. J. Bacteriol. 2003, 185, 6409–6414. [Google Scholar] [CrossRef] [PubMed]
- Kainov, D.E.; Butcher, S.J.; Bamford, D.H.; Tuma, R. Conserved Intermediates on the Assembly Pathway of Double-stranded RNA Bacteriophages. J. Mol. Biol. 2003, 328, 791–804. [Google Scholar] [CrossRef]
- Stitt, B.L.; Mindich, L. Morphogenesis of bacteriophage phi 6: A presumptive viral membrane precursor. Virology 1983, 127, 446–458. [Google Scholar] [CrossRef]
- Onodera, S.; Olkkonen, V.M.; Gottlieb, P.; Strassman, J.; Qiao, X.Y.; Bamford, D.H.; Mindich, L. Construction of a transducing virus from double-stranded RNA bacteriophage phi6: Establishment of carrier states in host cells. J. Virol. 1992, 66, 190–196. [Google Scholar] [PubMed]
- Vidaver, A.K.; Koski, R.K.; Etten, J.L.V. Bacteriophage φ6: A Lipid-Containing Virus of Pseudomonas phaseolicola. J. Virol. 1973, 11, 799–805. [Google Scholar] [PubMed]
- O’Keefe, K.J.; Silander, O.K.; McCreery, H.; Weinreich, D.M.; Wright, K.M.; Chao, L.; Edwards, S.V.; Remold, S.K.; Turner, P.E. Geographic Differences in Sexual Reassortment in Rna Phage. Evolution 2010, 64, 3010–3023. [Google Scholar] [CrossRef] [PubMed]
- Silander, O.K.; Weinreich, D.M.; Wright, K.M.; O’Keefe, K.J.; Rang, C.U.; Turner, P.E.; Chao, L. Widespread genetic exchange among terrestrial bacteriophages. Proc. Natl. Acad. Sci. USA 2005, 102, 19009–19014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drake, J.W. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. USA 1993, 90, 4171–4175. [Google Scholar] [CrossRef] [PubMed]
- Viral Zone: Leviviridae. Available online: https://viralzone.expasy.org/163?outline=all_by_species (accessed on 30 May 2018).
- Olsthoorn, R.; Duin, J. van Bacteriophages with ssRNA. In eLS; American Cancer Society: Atlanta, GA, USA, 2011; ISBN 978-0-470-01590-2. [Google Scholar]
- Zinder, N.D. RNA phages. Annu. Rev. Microbiol. 1965, 19, 455–473. [Google Scholar] [CrossRef] [PubMed]
- Dryden, S.K.; Ramaswami, B.; Yuan, Z.; Giammar, D.E.; Angenent, L.T. A rapid reverse transcription-PCR assay for F+ RNA coliphages to trace fecal pollution in Table Rock Lake on the Arkansas–Missouri border. Water Res. 2006, 40, 3719–3724. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.; Long, S.C.; Sobsey, M.D. Evaluation of F+ RNA and DNA Coliphages as Source-Specific Indicators of Fecal Contamination in Surface Waters. Appl. Environ. Microbiol. 2003, 69, 6507–6514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnik, S.; Billeter, M. The lysis function of RNA bacteriophage Qβ is mediated by the maturation (A2) protein. EMBO J. 1983, 2, 1521–1526. [Google Scholar] [PubMed]
- Young, R. Bacteriophage Lysis: Mechanism and Regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [PubMed]
- Chamakura, K.R.; Edwards, G.B.; Young, R. Mutational analysis of the MS2 lysis protein L. Microbiology 2017, 163, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.W.; Steitz, J.E. The reconstitution of infective bacteriophage R17. Proc. Natl. Acad. Sci. USA 1967, 58, 1416–1421. [Google Scholar] [CrossRef] [PubMed]
- Krahn, P.M.; O’Callaghan, R.J.; Paranchych, W. Stages in phage R17 infection. VI. Injection of A protein and RNA into the host cell. Virology 1972, 47, 628–637. [Google Scholar] [CrossRef]
- Shiba, T.; Suzuki, Y. Localization of A protein in the RNA-A protein complex of RNA phage MS2. Biochim. Biophys. Acta BBA-Nucleic Acids Protein Synth. 1981, 654, 249–255. [Google Scholar] [CrossRef]
- Zechner, E.L.; Lang, S.; Schildbach, J.F. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1073–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eigen, M.; Biebricher, C.K.; Gebinoga, M.; Gardiner, W.C. The hypercycle. Coupling of RNA and protein biosynthesis in the infection cycle of an RNA bacteriophage. Biochemistry 1991, 30, 11005–11018. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.D.; Genthner, F.J.; Gentry, J.; Sobsey, M.D.; Vinjé, J. Gene Mapping and Phylogenetic Analysis of the Complete Genome from 30 Single-Stranded RNA Male-Specific Coliphages (Family Leviviridae). J. Virol. 2009, 83, 11233–11243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahba, A.J.; Miller, M.J.; Niveleau, A.; Landers, T.A.; Carmichael, G.G.; Weber, K.; Hawley, D.A.; Slobin, L.I. Subunit I of Qβ Replicase and 30 S Ribosomal Protein Sl of Escherichia coli EVIDENCE FOR THE IDENTITY OF THE TWO PROTEINS. J. Biol. Chem. 1974, 249, 3314–3316. [Google Scholar] [PubMed]
- Blumenthal, T.; Landers, T.A.; Weber, K. Bacteriophage Qβ Replicase Contains the Protein Biosynthesis Elongation Factors EF Tu and EF Ts. Proc. Natl. Acad. Sci. USA 1972, 69, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Agirrezabala, X.; Frank, J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 2009, 42, 159–200. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Voorhees, R.M.; Kelley, A.C.; Gao, Y.-G.; Murphy, F.V.; Weir, J.R.; Ramakrishnan, V. The Crystal Structure of the Ribosome Bound to EF-Tu and Aminoacyl-tRNA. Science 2009, 326, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Schuette, J.-C.; Murphy, F.V.; Kelley, A.C.; Weir, J.R.; Giesebrecht, J.; Connell, S.R.; Loerke, J.; Mielke, T.; Zhang, W.; Penczek, P.A.; et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009, 28, 755–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamen, R.; Kondo, M.; Römer, W.; Weissmann, C. Reconstitution of Qβ Replicase Lacking Subunit α with Protein-Synthesis-Interference Factor i. Eur. J. Biochem. 1972, 31, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Weber, H.; Weissmann, C. Interactions of Qβ replicase with Qβ RNA. J. Mol. Biol. 1981, 153, 631–660. [Google Scholar] [CrossRef]
- Schuppli, D.; Miranda, G.; Qiu, S.; Weber, H. A branched stem-loop structure in the M-site of bacteriophage Qβ RNA is important for template recognition by Qβ replicase holoenzyme. J. Mol. Biol. 1998, 283, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Miranda, G.; Schuppli, D.; Barrera, I.; Hausherr, C.; Sogo, J.M.; Weber, H. Recognition of bacteriophage Qβ plus strand RNA as a template by Qβ replicase: Role of RNA interactions mediated by ribosomal proteins S1 and host factor1. J. Mol. Biol. 1997, 267, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Schuppli, D.; Georgijevic, J.; Weber, H. Synergism of mutations in bacteriophage qβ RNA affecting host factor dependence of qβ replicase1. J. Mol. Biol. 2000, 295, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.; Webster, R.E.; Zinder, N.D. Bacteriophage Coat Protein as Repressor. Nature 1968, 218, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Valegrad, K.; Murray, J.B.; Stockley, P.G.; Stonehouse, N.J.; Liljas, L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature 1994, 371, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, T.G.; Wang, I.-N.; Struck, D.K.; Young, R. Breaking free: “Protein antibiotics” and phage lysis. Res. Microbiol. 2002, 153, 493–501. [Google Scholar] [CrossRef]
- Walderich, B.; Höltje, J.V. Specific localization of the lysis protein of bacteriophage MS2 in membrane adhesion sites of Escherichia coli. J. Bacteriol. 1989, 171, 3331–3336. [Google Scholar] [CrossRef] [PubMed]
- Beremand, M.N.; Blumenthal, T. Overlapping genes in RNA phage: A new protein implicated in lysis. Cell 1979, 18, 257–266. [Google Scholar] [CrossRef]
- Goessens, W.H.; Driessen, A.J.; Wilschut, J.; van Duin, J. A synthetic peptide corresponding to the C-terminal 25 residues of phage MS2 coded lysis protein dissipates the protonmotive force in Escherichia coli membrane vesicles by generating hydrophilic pores. EMBO J. 1988, 7, 867–873. [Google Scholar] [PubMed]
- Walderich, B.; Ursinus-Wössner, A.; van Duin, J.; Höltje, J.V. Induction of the autolytic system of Escherichia coli by specific insertion of bacteriophage MS2 lysis protein into the bacterial cell envelope. J. Bacteriol. 1988, 170, 5027–5033. [Google Scholar] [CrossRef] [PubMed]
- Chamakura, K.R.; Tran, J.S.; Young, R. MS2 Lysis of Escherichia coli Depends on Host Chaperone DnaJ. J. Bacteriol. 2017, 199, e00058-17. [Google Scholar] [CrossRef] [PubMed]
- Lodish, H.F. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein biosynthesis. J. Mol. Biol. 1970, 50, 689–702. [Google Scholar] [CrossRef]
- Kozak, M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 1983, 47, 1–45. [Google Scholar] [PubMed]
- Poot, R.A.; Tsareva, N.V.; Boni, I.V.; van Duin, J. RNA folding kinetics regulates translation of phage MS2 maturation gene. Proc. Natl. Acad. Sci. USA 1997, 94, 10110–10115. [Google Scholar] [CrossRef] [PubMed]
- Weber, H. The binding site for coat protein on bacteriophage Qβ RNA. Biochim. Biophys. Acta BBA-Nucleic Acids Protein Synth. 1976, 418, 175–183. [Google Scholar] [CrossRef]
- Groeneveld, H.; Thimon, K.; van Duin, J. Translational control of maturation-protein synthesis in phage MS2: A role for the kinetics of RNA folding? RNA 1995, 1, 79–88. [Google Scholar] [PubMed]
- Reed, C.A.; Langlais, C.; Wang, I.-N.; Young, R. A2 expression and assembly regulates lysis in Qβ infections. Microbiology 2013, 159, 507–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumnieks, J.; Tars, K. Crystal Structure of the Maturation Protein from Bacteriophage Qβ. J. Mol. Biol. 2017, 429, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Atkins, J.F.; Steitz, J.A.; Anderson, C.W.; Model, P. Binding of mammalian ribosomes to ms2 phage RNA reveals an overlapping gene encoding a lysis function. Cell 1979, 18, 247–256. [Google Scholar] [CrossRef]
- Berkhout, B.; Schmidt, B.F.; van Strien, A.; van Boom, J.; van Westrenen, J.; van Duin, J. Lysis gene of bacteriophage MS2 is activated by translation termination at the overlapping coat gene. J. Mol. Biol. 1987, 195, 517–524. [Google Scholar] [CrossRef]
- Schmidt, B.F.; Berkhout, B.; Overbeek, G.P.; van Strien, A.; van Duin, J. Determination of the RNA secondary structure that regulates lysis gene expression in bacteriophage MS2. J. Mol. Biol. 1987, 195, 505–516. [Google Scholar] [CrossRef]
- Khazaie, K.; Buchanan, J.H.; Rosenberger, R.F. The accuracy of Qbeta RNA translation. 1. Errors during the synthesis of Qbeta proteins by intact Escherichia coli cells. Eur. J. Biochem. 1984, 144, 485–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiner, A.M.; Weber, K. Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat. New Biol. 1971, 234, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.D.; Lodish, H.F. Messenger Characteristics of Nascent Bacteriophage RNA. Proc. Natl. Acad. Sci. USA 1970, 67, 710–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klovins, J.; Overbeek, G.P.; van den Worm, S.H.E.; Ackermann, H.-W.; van Duin, J. Nucleotide sequence of a ssRNA phage from Acinetobacter: Kinship to coliphages. J. Gen. Virol. 2002, 83, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.E. The Structure and Infective Process of a Pseudomonas Aeruginosa Bacteriophage Containing Ribonucleic Acid. J. Gen. Microbiol. 1966, 45, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Olsen, R.H.; Thomas, D.D. Characteristics and Purification of PRR1, an RNA Phage Specific for the Broad Host Range Pseudomonas R1822 Drug Resistance Plasmid. J. Virol. 1973, 12, 1560–1567. [Google Scholar] [PubMed]
- Olsthoorn, R.C.L.; Garde, G.; Dayhuff, T.; Atkins, J.F.; Van Duin, J. Nucleotide sequence of a single-stranded RNA phage from Pseudomonas aeruginosa: Kinship to coliphages and conservation of regulatory RNA structures. Virology 1995, 206, 611–625. [Google Scholar] [CrossRef]
- Ruokoranta, T.M.; Grahn, A.M.; Ravantti, J.J.; Poranen, M.M.; Bamford, D.H. Complete Genome Sequence of the Broad Host Range Single-Stranded RNA Phage PRR1 Places It in the Levivirus Genus with Characteristics Shared with Alloleviviruses. J. Virol. 2006, 80, 9326–9330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weissmann, C.; Billeter, M.A.; Goodman, H.M.; Hindley, J.; Weber, H. Structure and Function of Phage RNA. Annu. Rev. Biochem. 1973, 42, 303–328. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Storz, G. RNA reflections: Converging on Hfq. RNA 2015, 21, 511–512. [Google Scholar] [CrossRef] [PubMed]
- Schuppli, D.; Miranda, G.; Tsui, H.-C.T.; Winkler, M.E.; Sogo, J.M.; Weber, H. Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc. Natl. Acad. Sci. USA 1997, 94, 10239–10242. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Weissmann, C. The 3’-Termini of Bacteriophage Qβ Plus and Minus Strands. J. Mol. Biol. 1970, 215–224. [Google Scholar] [CrossRef]
- Blumenthal, T.; Carmichael, G.G. RNA replication: Function and structure of Qbeta-replicase. Annu. Rev. Biochem. 1979, 48, 525–548. [Google Scholar] [CrossRef] [PubMed]
- Bausch, J.N.; Kramer, F.R.; Miele, E.A.; Dobkin, C.; Mills, D.R. Terminal Adenylation in the Synthesis of RNA by Qβ Replicase. J. Biol. Chem. 1983, 258, 1978–1984. [Google Scholar] [PubMed]
- Beekwilder, J.; Nieuwenhuizen, R.; Poot, R. Secondary Structure Model for the First Three Domains of Qβ RNA. Control of A-protein synthesis. J. Mol. Biol. 1996, 256, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Staples, D.H.; Hindley, J.; Billeter, M.A.; Weissmann, C. Localization of Qβ Maturation Cistron Ribosome Binding Site. Nat. New Biol. 1971, 234, 202–204. [Google Scholar] [CrossRef] [PubMed]
- Rumnieks, J.; Tars, K. Crystal structure of the read-through domain from bacteriophage Qβ A1 protein. Protein Sci. 2011, 20, 1707–1712. [Google Scholar] [CrossRef] [PubMed]
- Kastelein, R.A.; Remaut, E.; Fiers, W.; Duin, J. van Lysis gene expression of RNA phage MS2 depends on a frameshift during translation of the overlapping coat protein gene. Nature 1982, 295, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, T.G.; Wang, I.-N.; Struck, D.K.; Young, R. A Protein Antibiotic in the Phage Qβ Virion: Diversity in Lysis Targets. Science 2001, 292, 2326–2329. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.D.; Vivas, E.I.; Walsh, C.T.; Kolter, R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 1995, 177, 4194–4197. [Google Scholar] [CrossRef] [PubMed]
- Grasis, J.A. Host-Associated Bacteriophage Isolation and Preparation for Viral Metagenomics. In Viral Metagenomics; Pantaleo, V., Chiumenti, M., Eds.; Springer: New York, NY, USA, 2018; Volume 1746, pp. 1–25. ISBN 978-1-4939-7682-9. [Google Scholar]
- Krishnamurthy, S.R.; Wang, D. Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology 2018, 516, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.; Farkas, K.; Harrison, C.; Jones, D.; Allison, H.E.; McCarthy, A.J. Viromic analysis of wastewater input to a river catchment reveals a diverse assemblage of RNA viruses. MSystems 2018, 3, e00025-18. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.S.; Bae, H.-W.; Cho, Y.-H. A Pilin Region Affecting Host Range of the Pseudomonas aeruginosa RNA Phage, PP7. Front. Microbiol. 2018, 9, 247. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Callanan, J.; Stockdale, S.R.; Shkoporov, A.; Draper, L.A.; Ross, R.P.; Hill, C. RNA Phage Biology in a Metagenomic Era. Viruses 2018, 10, 386. https://doi.org/10.3390/v10070386
Callanan J, Stockdale SR, Shkoporov A, Draper LA, Ross RP, Hill C. RNA Phage Biology in a Metagenomic Era. Viruses. 2018; 10(7):386. https://doi.org/10.3390/v10070386
Chicago/Turabian StyleCallanan, Julie, Stephen R. Stockdale, Andrey Shkoporov, Lorraine A. Draper, R. Paul Ross, and Colin Hill. 2018. "RNA Phage Biology in a Metagenomic Era" Viruses 10, no. 7: 386. https://doi.org/10.3390/v10070386




