Phylogenetic Relationships among Species of Phellinus sensu stricto, Cause of White Trunk Rot of Hardwoods, from Northern North America
Abstract
:1. Introduction
2. Experimental Section
2.1. Isolates Used, DNA Extraction, PCR, and DNA Sequencing
2.2. Sequence Alignment and Phylogenetic Analysis
| Species & Code | Host | State/Province | Country | Reference | GenBank Accession Nos. | |||
|---|---|---|---|---|---|---|---|---|
| ITS | nLSU | rpb2 | tef1 | |||||
| Phellinus alni | ||||||||
| DLL2009-140 | Acer rubrum | MN | USA | This study | KU139159 | KU139211 | KU139282 | KU139331 |
| NJB2011-SM1 | Acer rubrum | MA | USA | This study | KU139161 | KU139217 | KU139279 | KU139324 |
| NJB2011-SM4 | Acer rubrum | MA | USA | This study | KU139163 | KU139219 | KU139284 | KU139326 |
| NJB2011-SM3 | Acer saccharum | MA | USA | This study | KU139162 | KU139218 | KU139286 | KU139325 |
| FP-134638-Sp | Alnus sp. | ID | USA | This study | KU139167 | KU139213 | KU139280 | KU139330 |
| HHB-15085-Sp | Alnus sp. | AK | USA | This study | KU139160 | KU139216 | KU139285 | KU139328 |
| NJB2011-WEN2 | Betula lenta | MA | USA | This study | KU139164 | KU139220 | KU139283 | KU139327 |
| FP-125027-T | Betula papyrifera | NH | USA | This study | KU139166 | KU139212 | KU139281 | KU139329 |
| FP-70831-T | Fagus grandifolia | MI | USA | This study | KU139165 | KU139214 | KU139278 | KU139322 |
| NJB2011-GR3 | Fagus grandifolia | MA | USA | This study | KU139168 | KU139215 | KU139287 | KU139323 |
| BRNM 714864 | Alnus incana | Mikkeli, Kakriala | Finland | [12] | GQ383775 | GQ383821 | ||
| BRNM 714865 | Alnus incana | Červený Kláštor | Slovakia | [12] | GQ383730 | GQ383840 | ||
| BRNM 714891 | Betula sp. | Vyborg | Russia | [12] | GQ383770 | GQ383853 | ||
| MJ 51/96 | Fagus sylvatica | Havlíčkův Brod | Czech Republic | [12] | GQ383756 | GQ383831 | ||
| BRNM 714881 | Sorbus cf. intermedia | Uppsala | Sweden | [12] | GQ383732 | GQ383848 | ||
| TN3301 | Betula sp. | n/a | Finland | [9] | AY340040 | |||
| TW-162 | Laburnum anagyroides | n/a | Germany | [7] | AF311025 | |||
| Phellinus arctostaphyli | ||||||||
| FP-94140-R | Arctostaphylos manzanita | OR | USA | This study | KU139143 | KU139250 | KU139264 | KU139348 |
| FP-94186-R | Arctostaphylos patula | CA | USA | This study | KU139145 | KU139252 | KU139266 | KU139350 |
| M-92-2 | Arctostaphylos patula | OR | USA | This study | KU139144 | KU139251 | KU139265 | KU139349 |
| Phellinus laevigatus s.l. P. betulinus | ||||||||
| FP-105325-Sp | Betula alleghaniensis | WV | USA | This study | KU139154 | KU139239 | KU139312 | KU139369 |
| DLL2009-143 | Betula alleghaniensis | MN | USA | This study | KU139146 | KU139236 | KU139308 | KU139371 |
| DVB-Betula | Betula nigra | VA | USA | This study | KU139151 | KU139246 | KU139314 | KU139365 |
| NJB2009-FpG | Betula papyrifera | MA | USA | This study | KU139153 | KU139248 | KU139311 | KU139368 |
| NJB2009-FpE | Betula papyrifera | MA | USA | This study | KU139152 | KU139247 | KU139307 | KU139370 |
| DLL2009-175 | Betula papyrifera | MN | USA | This study | KU139155 | KU139237 | KU139309 | KU139367 |
| RLG-5835-T | Betula papyrifera | MT | USA | This study | KU139150 | KU139240 | KU139306 | KU139364 |
| RLG-645-T | Betula papyrifera | MT | USA | This study | KU139147 | KU139238 | KU139313 | KU139366 |
| P. laevigatus s.s. | ||||||||
| NJB2011-PLa1-F | Betula pubescens | Hame | Finland | This study | KU139148 | KU139241 | KU139305 | KU139372 |
| NJB2011-PLa2-F | Betula pubescens | Hame | Finland | This study | KU139149 | KU139242 | KU139310 | KU139373 |
| BRNM 714875 | Betula sp. | Bohemian Forest | Czech Republic | [12] | GQ383779 | GQ383857 | ||
| BRNM 714867 | Betula sp. | Bohemian Forest | Czech Republic | [12] | GQ383778 | GQ383856 | ||
| BRNM 714877 | Betula sp. | Uppsala | Sweden | [12] | GQ383777 | GQ383855 | ||
| TN-3260 | Betula pubescens | n/a | Finland | [7] | AF311034 | |||
| P. orienticus | ||||||||
| TN-6392 | Betula sp. | Jilin | China | This study | KU139156 | KU139243 | KU139374 | |
| TN-6432 | Betula costa | Jilin | China | This study | KU139158 | KU139245 | KU139376 | |
| TN-6425 | Betula sp. | Jilin | China | This study | KU139157 | KU139244 | KU139375 | |
| Phellinus lundellii | ||||||||
| NJB2011-SM2 | Betula alleghaniensis | MA | USA | This study | KU139184 | KU139235 | KU139299 | KU139335 |
| NJB2011-WEN1 | Betula alleghaniensis | MA | USA | This study | KC551835 | KC551859 | KU139302 | KC551884 |
| DLL2011-321 | Betula alleghaniensis | MI | USA | This study | KU139181 | |||
| NJB2011-GR1 | Betula lenta | MA | USA | This study | KU139182 | KU139232 | KU139301 | KU139336 |
| JJW-694 | Betula sp. | NY | USA | This study | KU139183 | KU139233 | KU139298 | KU139334 |
| NJB2011-PLu-F | Betula pubescens | Hame | Finland | This study | KU139185 | KU139234 | KU139300 | KU139337 |
| BRNU 604719 | Betula carpatica | Jeseníky Mountains | Czech Republic | [12] | GQ383704 | |||
| Phellinus nigricans | ||||||||
| HHB-15513-T | Betula nana | AK | USA | This study | KU139171 | KU139226 | KU139295 | KU139347 |
| FP-62186-T | Betula occidentalis | WA | USA | This study | KU139176 | KU139227 | KU139290 | KU139343 |
| FP-140068-T | Betula papyrifera | MN | USA | This study | KU139175 | KU139225 | KU139292 | KU139345 |
| DMR-94-13 | Betula papyrifera | MN | USA | This study | KU139173 | KU139221 | KU139291 | KU139344 |
| FP-135209-R | Betula papyrifera | MI | USA | This study | KU139174 | KU139224 | KU139293 | KU139346 |
| OKM-3356 | Betula papyrifera | ID | USA | This study | KU139178 | KU139228 | KU139288 | KU139342 |
| HHB-12617-T | Betula papyrifera | AK | USA | This study | KU139172 | KU139230 | KU139294 | KU139338 |
| RLG-5844-Sp | Betula sp. | MT | USA | This study | KU139177 | KU139229 | KU139289 | KU139341 |
| NJB2011-PA1-F | Betula pubescens | Hame | Finland | This study | KU139169 | KU139222 | KU139297 | KU139340 |
| NJB2011-PA2-F | Betula pubescens | Hame | Finland | This study | KU139170 | KU139223 | KU139296 | KU139339 |
| MJ 557/94 | Betula sp. | Rondane | Norway | [12] | GQ383724 | GQ383801 | ||
| BRNM 714879 | Betula sp. | Uppsala | Sweden | [12] | GQ383721 | GQ383803 | ||
| BRNM 714883 | Betula sp. | Vyborg | Russia | [12] | GQ383719 | GQ383851 | ||
| 85-917 | Betula pubescens | n/a | Germany | [7] | AF311027 | |||
| Phellinus populicola | ||||||||
| TN-526 (1) (ATCC 36122; CBS 638.75) | Populus tremula | Uusimaa | Finland | This study | KU139179 | KU139231 | KU139303 | KU139333 |
| BRNM 714885 | Populus tremula | Uppsala | Sweden | [12] | GQ383706 | GQ383785 | ||
| BRNM 714890 | Populus tremula | Uppsala | Sweden | [12] | GQ383707 | GQ383786 | ||
| MJ 92/96 | Populus alba | Lanžhot | Czech Republic | [12] | GQ383705 | GQ383787 | ||
| 84-61 | Populus alba | n/a | Germany | [7] | AF311038 | |||
| Phellinus igniarius s.s. | ||||||||
| MJ 19/94 | Populus nigra | Jihlava | Czech Republic | [12] | GQ383718 | GQ383793 | ||
| BRNM 714866 | Populus nigra | Zábřeh | Czech Republic | [12] | GQ383710 | GQ383792 | ||
| BRNM 714889 | Salix alba | Brno | Czech Republic | [12] | GQ383709 | GQ383791 | ||
| MJ 40/07 | Salix caprea | Jihlava | Czech Republic | [12] | GQ383712 | GQ383790 | ||
| BRNM 714884 | Salix caprea | Uppsala | Sweden | [12] | GQ383715 | GQ383795 | ||
| 83-1110a | Salix fragilis | n/a | Germany | [7] | AF311033 | |||
| Phellinus tremulae | ||||||||
| FP-135820-T | Populus grandidentata | WI | USA | This study | KU139136 | KU139206 | KU139274 | KU139363 |
| FP-59023-T | Populus tremuloides | NH | USA | This study | KU139135 | KU139205 | KU139270 | KU139358 |
| FP-135202-T | Populus tremuloides | MI | USA | This study | KU139134 | KU139207 | KU139271 | KU139359 |
| FP-140050-T | Populus tremuloides | MN | USA | This study | KU139131 | KU139204 | KU139272 | KU139362 |
| A-17 | Populus tremuloides | CO | USA | This study | KU139137 | KU139200 | KU139273 | KU139361 |
| FP-105919-R | Populus tremuloides | SD | USA | This study | KU139130 | KU139203 | KU139275 | KU139360 |
| NJB2011-PT1-F | Populus tremula | Hame | Finland | This study | KU139132 | KU139201 | KU139276 | KU139357 |
| NJB2011-PT2-F | Populus tremula | Hame | Finland | This study | KU139133 | KU139202 | KU139277 | KU139356 |
| MJ 45/07 | Populus tremula | Havlíčkův Brod | Czech Republic | [12] | GQ383782 | GQ383860 | ||
| BRNM 714886 | Populus tremula | Třeboň | Czech Republic | [12] | GQ383780 | GQ383862 | ||
| MJ 32/07 | Populus tremula | Jihlava | Czech Republic | [12] | GQ383781 | GQ383861 | ||
| Dai2352 | Populus sp. | n/a | China | [9] | AY340063 | |||
| 89-826c | Populus tremula | n/a | Estonia | [7] | AF311042 | |||
| Dai-Pt | n/a | n/a | China | Unpublished | HQ328536 | |||
| Phellinus tuberculosus | ||||||||
| TN-449 (ATCC 38666) | Prunus domestica | Nauvo | Finland | This study | KU139142 | KU139254 | KU139263 | KU139352 |
| TN-236 (ATCC 38665) | Prunus insititia | Tammisaari | Finland | This study | KU139141 | KU139253 | KU139262 | KU139351 |
| MJ 47/07 | Prunus domestica | Havlíčkův Brod | Czech Republic | [12] | GQ383784 | GQ383859 | ||
| MJ 44/07 | Prunus spinosa | Havlíčkův Brod | Czech Republic | [12] | GQ383783 | GQ383858 | ||
| Phellinus NA1 | ||||||||
| OKM-4173 | Populus tremuloides | ID | USA | This study | KU139180 | KU139249 | KU139304 | KU139332 |
| Phellinus NA2 | ||||||||
| SRM-158-Sp | Prunus americana | NE | USA | This study | KU139140 | KU139210 | KU139267 | KU139353 |
| FP-103293-R | Prunus angustifolia | SC | USA | This study | KU139138 | KU139208 | KU139268 | KU139355 |
| FP-105670-R | Prunus sp. | GA | USA | This study | KU139139 | KU139209 | KU139269 | KU139354 |
| Inonotus vaninii | ||||||||
| DMR-95-1-T | Populus tremuloides | MN | USA | This study | KU139198 | KU139258 | KU139318 | KU139380 |
| DLL2010-102 | Populus tremuloides | MN | USA | [57] | KU139197 | |||
| Inocutis dryophila | ||||||||
| DLL2012-001 | Quercus alba | WI | USA | This study | KU139186 | KU139255 | KU139317 | |
| Phellinopsis conchata | ||||||||
| DLL2009-149 | Acer sp. | MN | USA | [57] | KU139187 | KU139256 | KU139316 | KU139378 |
| L-7601 | Fraxinus nigra | NY | USA | This study | KU139188 | KU139257 | KU139315 | KU139377 |
| 89-1014 | Salix sp. | — | Germany | [7] | AF311028 | |||
| Fuscoporia ferrea | ||||||||
| FP-133592-Sp | Alnus sp. | OR | USA | This study | KU139189 | KU139259 | KU139319 | KU139379 |
| DLL2009-035 | Populus tremuloides | MN | USA | [57] | KU139190 | |||
| Fuscoporia gilva | ||||||||
| DLL2011-109 | Acer saccharum | WI | USA | [36] | KU139195 | |||
| DLL2011-147 | Acer saccharum | WI | USA | [36] | KU139196 | |||
| Fuscoporia sp. 1 | ||||||||
| DLL2009-025 | Populus tremuloides | MN | USA | [57] | KU139193 | |||
| DLL2011-211 | Prunus serotina | WI | USA | [36] | KU139194 | |||
| Fuscoporia sp. 2 | ||||||||
| DLL2011-140 | Acer saccharum | WI | USA | [36] | KU139192 | |||
| DLL2011-256 | Ostrya virginiana | WI | USA | [36] | KU139191 | |||
| Fomes fomentarius | ||||||||
| NJB2011-KD3 | Fagus grandifolia | MA | USA | This study | KU139199 | KU139260 | KU139320 | KU139382 |
| NJB2011-GR2 | Populus grandidentata | MA | USA | This study | KU139261 | KU139321 | KU139381 | |
3. Results
3.1. Phylogenetic Analysis and Species Identification

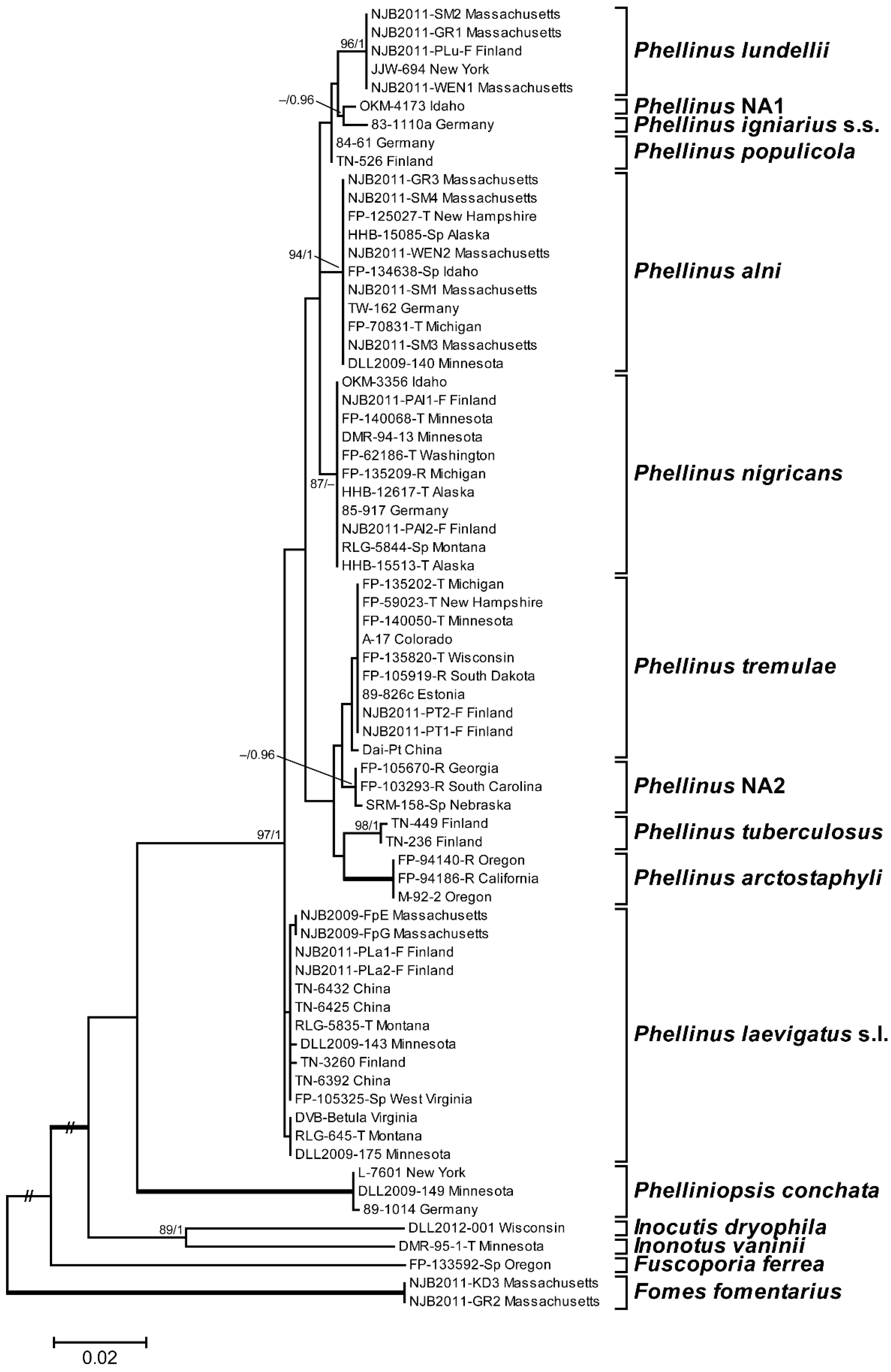

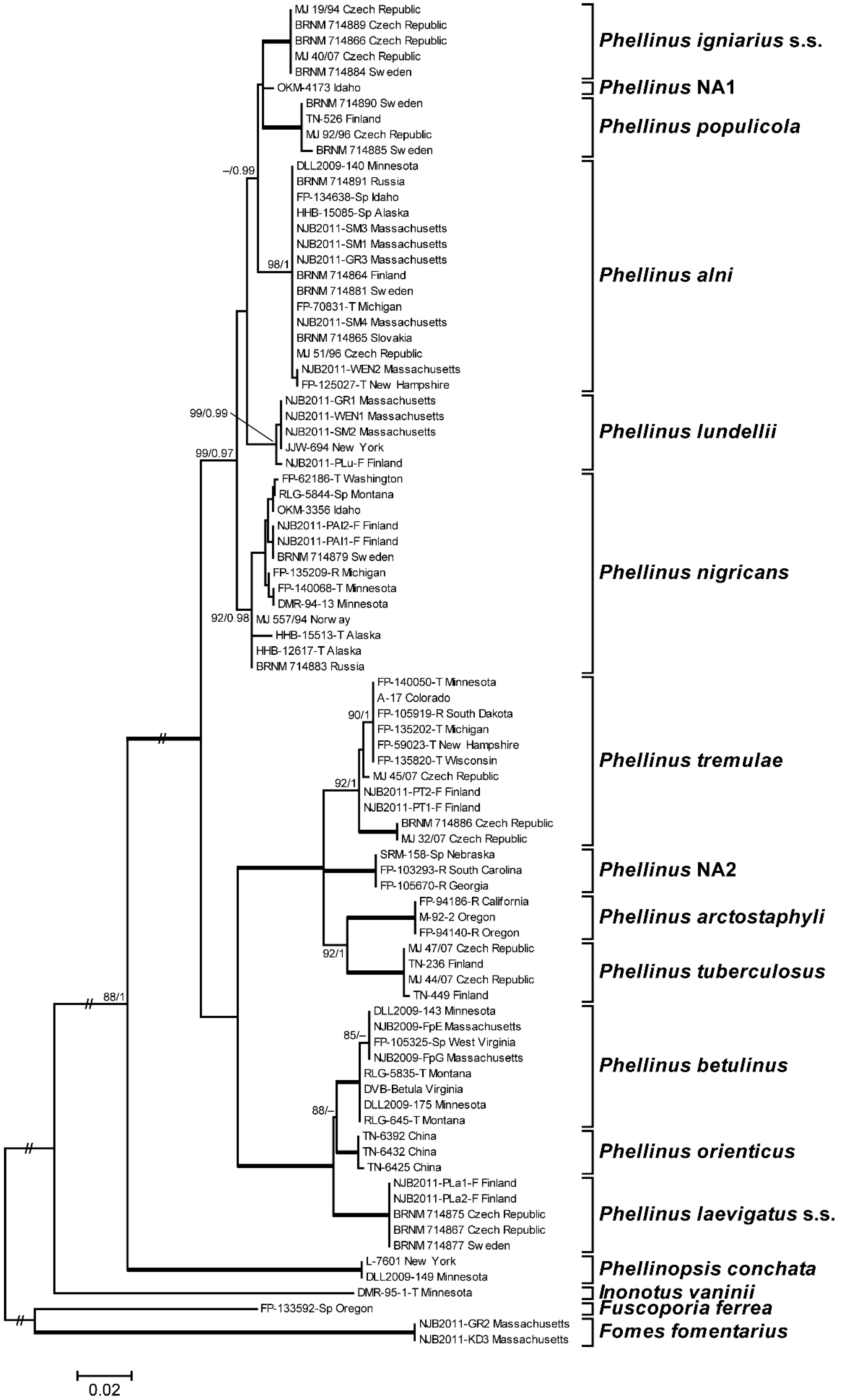
3.2. Phylogenetic Relationships by Locus
3.3. Phylogenetic Relationships by Host
| Phellinus Species | Known Range | Known Host Genera | Known Host Families |
|---|---|---|---|
| P. alni | Holarctic | Acer, Alnus, Betula, Carpinus, Corylus, Fagus, Fraxinus, Juglans, Laburnum, Malus, Padus, Prunus and Sorbus | Betulaceae, Fagaceae, Juglandaceae, Oleaceae, Rosaceae, Sapindaceae & Ulmaceae |
| P. lundellii | Holarctic | Betula | Betulaceae |
| P. nigricans | Holarctic | Betula | Betulaceae |
| P. tremulae | Holarctic | Populus | Salicaceae |
| P. arctostaphyli | North America | Arctostaphylos | Ericaceae |
| P. betulinus | North America | Betula | Betulaceae |
| P. NA1 | North America | Populus | Salicaceae |
| P. NA2 | North America | Prunus | Rosaceae |
| P. igniarius s.s. | Europe | Populus, Salix and Malus | Salicaceae and Rosaceae |
| P. populicola | Europe & Asia | Populus | Salicaceae |
| P. tuberculosus | Europe & Asia | Prunus | Rosaceae |
| P. laevigatus s.s. | Europe & West Asia | Betula | Betulaceae |
| P. orienticus | East Asia | Betula | Betulaceae |
4. Discussion
Acknowledgments
Conflicts of Interest
References
- Niemelä, T. On Fennoscandian Polypores. II. Phellinus laevigatus (Fr.) Bourd. & Galz. and P. lundellii Niemelä, n. sp. Ann. Bot. Fennici 1972, 9, 41–59. [Google Scholar]
- Niemelä, T. On Fennoscandian Polypores. III. Phellinus tremulae (Bond.) Bond. & Borisov. Ann. Bot. Fennici 1974, 11, 202–215. [Google Scholar]
- Niemelä, T. On Fennoscandian Polypores. IV. Phellinus igniarius, P. nigricans and P. populicola, n. sp. Ann. Bot. Fennici 1975, 12, 93–122. [Google Scholar]
- Fiasson, J.; Niemelä, T. The Hymenochaetales: A revision of the European poroid taxa. Karstenia 1984, 24, 14–28. [Google Scholar]
- Gilbertson, R.L.; Ryvarden, L. North American Polypores; Fungiflora: Oslo, Norway, 1987. [Google Scholar]
- Fischer, M. Phellinus igniarius and its closest relatives in Europe. Mycol. Res. 1995, 99, 735–744. [Google Scholar] [CrossRef]
- Wagner, T.; Fischer, M. Natural groups and a revised system for the European poroid Hymenochaetales (Basidiomycota) supported by nLSU rDNA sequence data. Mycol. Res. 2001, 105, 773–782. [Google Scholar] [CrossRef]
- Wagner, T.; Fischer, M. Proceedings towards a natural classification of the worldwide taxa Phellinus s.l. and Inonotus s.l., and phylogenetic relationships of allied genera. Mycologia 2002, 94, 998–1016. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Binder, M. Species recognition, geographic distribution and host-pathogen relationships: A case study in a group of lignicolous basidiomycetes, Phellinus s.l. Mycologia 2004, 96, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Larsson, K.H.; Parmasto, E.; Fischer, M.; Langer, E.; Nakasone, K.K.; Redhead, S.A. Hymenochaetales: A molecular phylogeny for the hymenochaetoid clade. Mycologia 2006, 98, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Sell, I. Taxonomy of the species in the Phellinus igniarius group. Mycotaxon 2008, 104, 337–347. [Google Scholar]
- Tomšovský, M.; Vampola, P.; Sedlák, P.; Byrtusová, Z.; Jankovský, L. Delimitation of central and northern European species of the Phellinus igniarius group (Basidiomycota, Hymenochaetales) based on analysis of ITS and translation elongation factor 1 alpha DNA sequences. Mycol. Prog. 2010, 9, 431–445. [Google Scholar] [CrossRef]
- Larsen, M.J.; Cobb-Poulle, L.A. Phellinus (Hymenochaetaceae): A Survey of the World Taxa; Fungiflora: Oslo, Norway, 1990. [Google Scholar]
- Ohman, J.H.; Kessler, K.J. White Trunk Rot of Hardwoods; USDA Forest Service: Fort Collins, CO, USA, 1964.
- Boyce, J.S. Forest Pathology, 3rd ed.; John Wiley and Sons Inc.: New York, NY, USA, 1961. [Google Scholar]
- Sinclair, W.A.; Lyon, H.H. Diseases of Trees and Shrubs, 2nd ed.; Cornell University Press: Ithaca, NY, USA, 2005; pp. 306–308. [Google Scholar]
- Silverborg, S. Northern Hardwoods Cull Manual; Bulletin No. 31; State University of New York: Syracuse, NY, USA, 1954; pp. 4–5. [Google Scholar]
- Bondartsev, A.S. The Polyporaceae of the European USSR and Caucasia; Israel Program for Scientific Translations: Jerusalem, Israel, 1953; p. 358. [Google Scholar]
- Jones, A.C.; Ostry, M.E. Estimating white trunk rot in aspen stands. North. J. Appl. For. 1998, 15, 33–36. [Google Scholar]
- Worrall, J.J.; Fairweather, M.L. Decay and Discoloration of Aspen; USDA Forest Service: Fort Collins, CO, USA, 2009.
- Verrall, A.F. Variation in Fomes igniarius (L.) Gill; Minnesota Agricultural Experiment Station: MN, USA, 1937. [Google Scholar]
- Overholts, L.O. The Polyporaceae of the United States, Alaska, and Canada; University of Michigan Press: Ann Arbor, MI, USA, 1953. [Google Scholar]
- Worrall, J.J.; Lee, T.D.; Harrington, T.C. Forest dynamics and agents that initiate and expand canopy gaps in Picea—Abies forests of Crawford Notch, New Hampshire, USA. J. Ecol. 2005, 93, 178–190. [Google Scholar] [CrossRef]
- Lindner, D.L.; Burdsall, H.H., Jr.; Stanosz, G.R. Species diversity of polyporoid and corticioid fungi in northern hardwood forests with differing management histories. Mycologia 2006, 98, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Brazee, N.J.; Lindner, D.L. Unraveling the Phellinus pini s.l. complex in North America: A multilocus phylogeny and differentiation analysis of Porodaedalea. For. Pathol. 2013, 43, 132–143. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.; Samuels, G. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Hopple, J.S.; Vilgalys, R. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: Divergent domains, outgroups and monophyly. Mol. Phylogenet. Evol. 1999, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kauserud, H.; Schumacher, T. Outcrossing or inbreeding: DNA markers provide evidence for type of reproductive mode in Phellinus nigrolimitatus. Mycol. Res. 2001, 105, 676–683. [Google Scholar] [CrossRef]
- Matheny, P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol. Phylogenet. Evol. 2005, 35, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Tavaré, S. Some probabilistic and statistical problems on the analysis of DNA sequences. Lect. Math. Life Sci. 1986, 17, 57–86. [Google Scholar]
- Brazee, N.J.; Lindner, D.L.; D’Amato, A.W.; Fraver, S.; Forrester, J.A.; Mladenoff, D.J. Disturbance and diversity of wood-inhabiting fungi: Effects of canopy gaps and downed woody debris. Biodivers. Conserv. 2014, 23, 2155–2172. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Gadagkar, S.R. Disparity Index: A simple statistic to measure and test the homogeneity of substitution patterns between molecular sequences. Genetics 2001, 158, 1321–1327. [Google Scholar] [PubMed]
- Parmasto, E. Phellinus laevigatus s. l. (Hymenochaetales): A ring species. Folia Cryptogam. Estonica 2007, 43, 39–49. [Google Scholar]
- Eyre, F.H. Forest Cover Types of the United States and Canada; Society of American Foresters: Washington, DC, USA, 1980. [Google Scholar]
- Brazee, N.J.; Hulvey, J.P.; Wick, R.L. Evaluation of partial tef1, rpb2, and nLSU sequences for identification of isolates representing Armillaria calvescens and Armillaria gallica from northeastern North America. Fungal Biol. 2011, 115, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Ross-Davis, A.L.; Hanna, J.W.; Kim, M.-S.; Klopfenstein, N.B. Advances toward DNA-based identification and phylogeny of North American Armillaria species using elongation factor-1 alpha gene. Mycoscience 2012, 53, 161–165. [Google Scholar] [CrossRef]
- Carlson, A.L.; Justo, A.; Hibbett, D.S. Species delimitation in Trametes: A comparison of ITS, RPB1, RPB2 and TEF1 gene phylogenies. Mycologia 2014, 106, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Ming-Xiao, D.; Yu-Lian, W.; Yue, G. Three species of Phellinus (Basidiomycota, Hymenochaetaceae) new to China. Mycosystema 2012, 31, 940–946. [Google Scholar]
- Niemelä, T. On Fennoscandian polypores 5, Phellinus pomaceus. Karstenia 1977, 17, 77–86. [Google Scholar]
- Baumgarten, J.C.G. Flora Lipsiensis; Leipzig: Saxony, Germany, 1790; p. 635. [Google Scholar]
- Persoon, C.H. Observationes Mycologicae; Leipzig: Saxony, Germany, 1799; Volume 2, p. 5. [Google Scholar]
- Nobles, M.K. Studies in forest pathology VI. Identification of cultures of wood-rotting fungi. Can. J. Res. 1948, 26, 281–431. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.L. The genus Phellinus (Aphyllophorales: Hymenochaetaceae) in western North America. Mycotaxon 1979, 9, 51–89. [Google Scholar]
- Niemelä, T. Taxonomic notes on the polypore genera Antrodiella, Daedaleopsis, Fibuloporia and Phellinus. Karstenia 1982, 22, 11–12. [Google Scholar]
- Riffle, J.W.; Conway, K.E. Phellinus stem decays of hardwoods. In Diseases of Trees in the Great Plants; Riffle, J.W., Peterson, G.W., Eds.; USDA Forest Service: Fort Collins, CO, USA, 1986; pp. 83–85. [Google Scholar]
- Ryvarden, L.; Gilbertson, R.L. European Polypores, part 2; Fungiflora: Oslo, Norway, 1994; p. 507. [Google Scholar]
- Nam, B.-H.; Lee, J.-Y.; Kim, G.-Y.; Jung, H.-H.; Park, H.-S.; Kim, C.-Y.; Jo, W.-S.; Jeong, S.-J.; Lee, T.-H.; Lee, J.-D. Phylogenetic analysis and rapid detection of genus Phellinus using the nucleotide sequences of 18S ribosomal RNA. Mycobiology 2003, 31, 133–138. [Google Scholar] [CrossRef]
- Schmidt, O.; Gaiser, O.; Dujesiefken, D. Molecular identification of decay fungi in the wood of urban trees. Eur. J. For. Res. 2012, 131, 885–891. [Google Scholar] [CrossRef]
- Overholts, L.O. Diagnoses of American polypores—III. Some additional brown species, with a key to the common brown species of the United States and Canada. Mycologia 1931, 23, 117–129. [Google Scholar] [CrossRef]
- Brazee, N.J.; Lindner, D.L.; Fraver, S.; D’Amato, A.W.; Milo, A.M. Wood-inhabiting, polyporoid fungi in aspen-dominated forests managed for biomass in the U.S. Lake States. Fungal Ecol. 2012, 5, 600–609. [Google Scholar] [CrossRef]
© 2015 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brazee, N.J. Phylogenetic Relationships among Species of Phellinus sensu stricto, Cause of White Trunk Rot of Hardwoods, from Northern North America. Forests 2015, 6, 4191-4211. https://doi.org/10.3390/f6114191
Brazee NJ. Phylogenetic Relationships among Species of Phellinus sensu stricto, Cause of White Trunk Rot of Hardwoods, from Northern North America. Forests. 2015; 6(11):4191-4211. https://doi.org/10.3390/f6114191
Chicago/Turabian StyleBrazee, Nicholas J. 2015. "Phylogenetic Relationships among Species of Phellinus sensu stricto, Cause of White Trunk Rot of Hardwoods, from Northern North America" Forests 6, no. 11: 4191-4211. https://doi.org/10.3390/f6114191
APA StyleBrazee, N. J. (2015). Phylogenetic Relationships among Species of Phellinus sensu stricto, Cause of White Trunk Rot of Hardwoods, from Northern North America. Forests, 6(11), 4191-4211. https://doi.org/10.3390/f6114191





