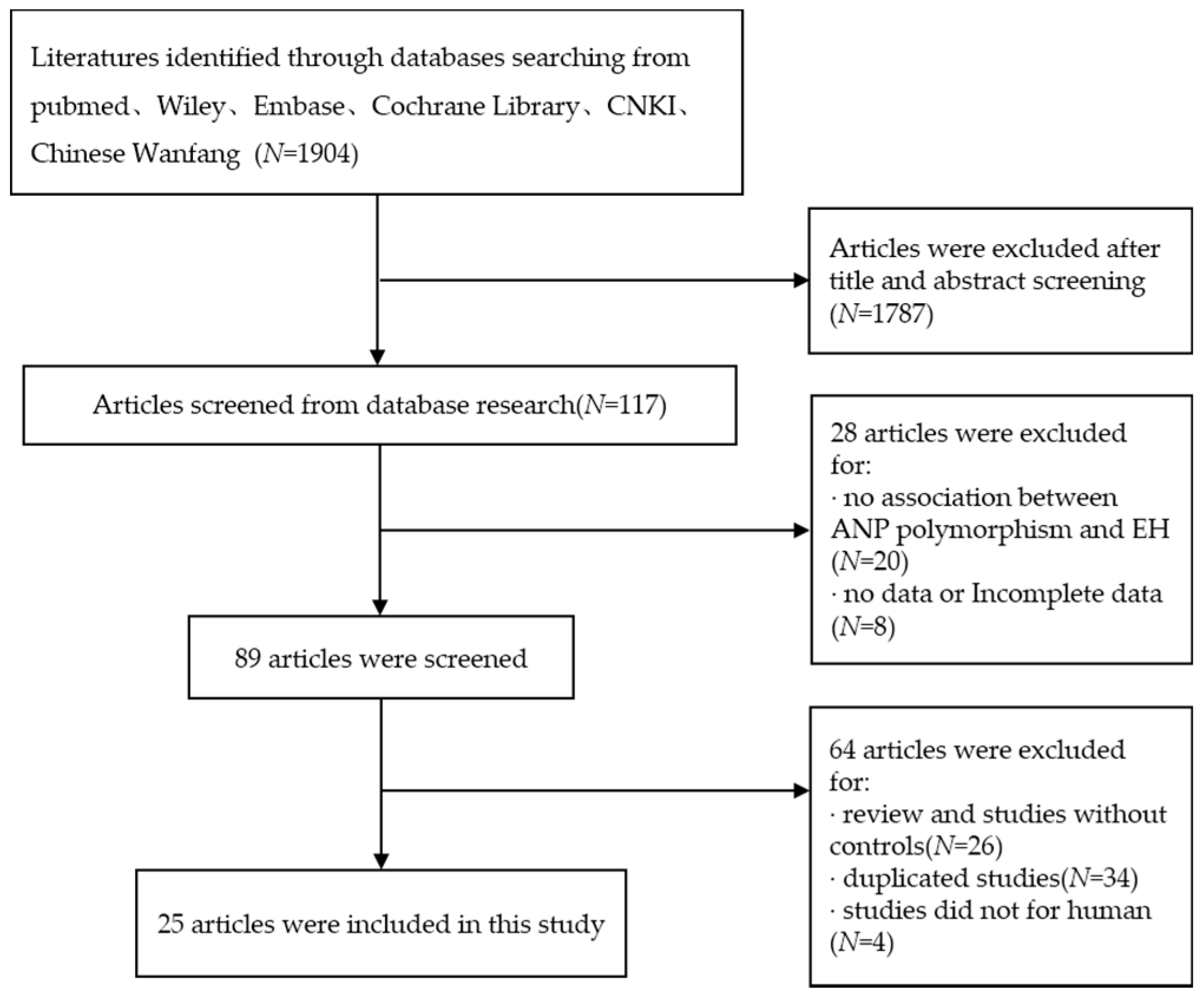
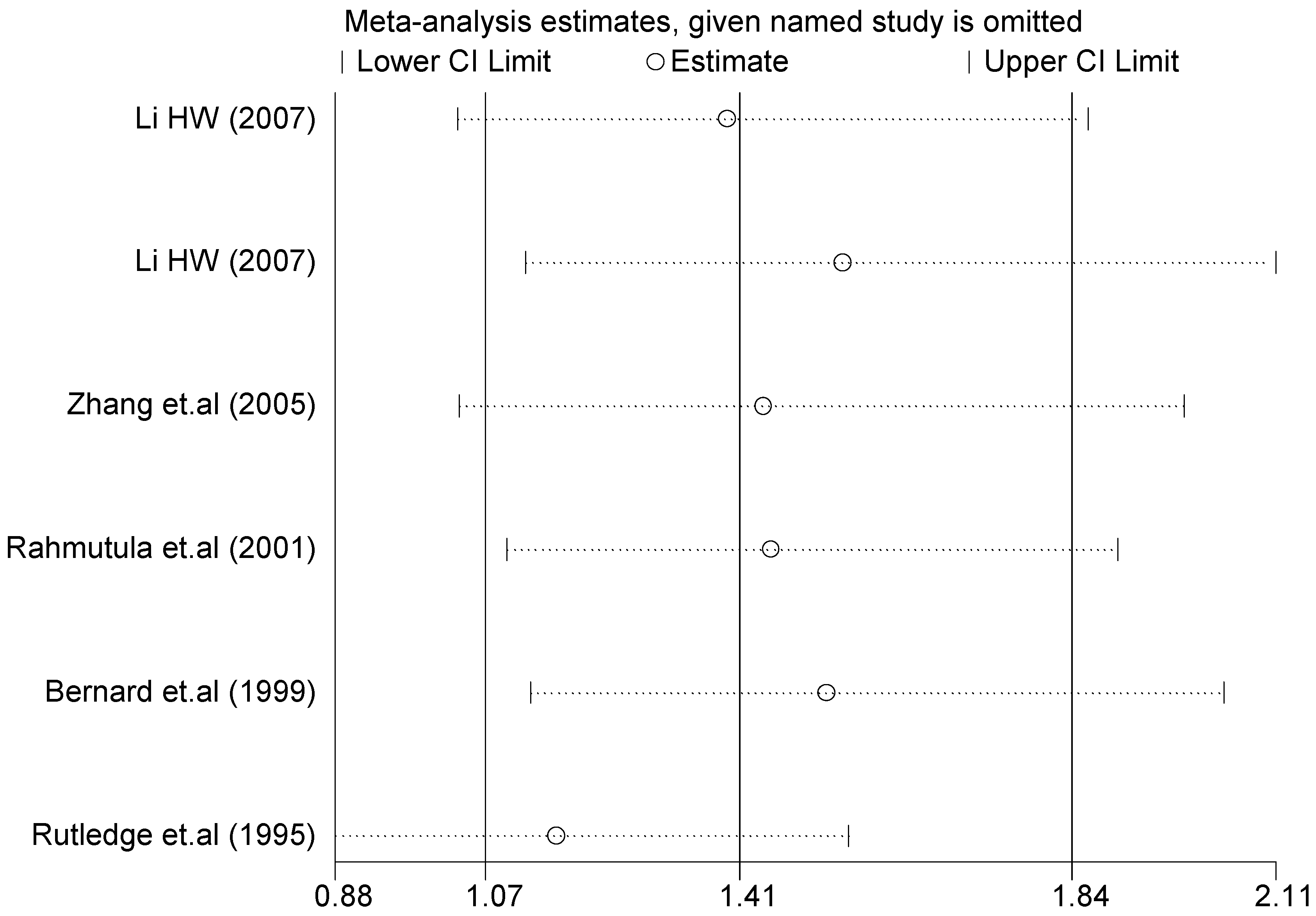Figure 1.
Flow chart of studies included in this meta-analysis.
Figure 1.
Flow chart of studies included in this meta-analysis.
Figure 2.
Forest plot of overall population of T2238C additive model (C vs. T).
Figure 2.
Forest plot of overall population of T2238C additive model (C vs. T).
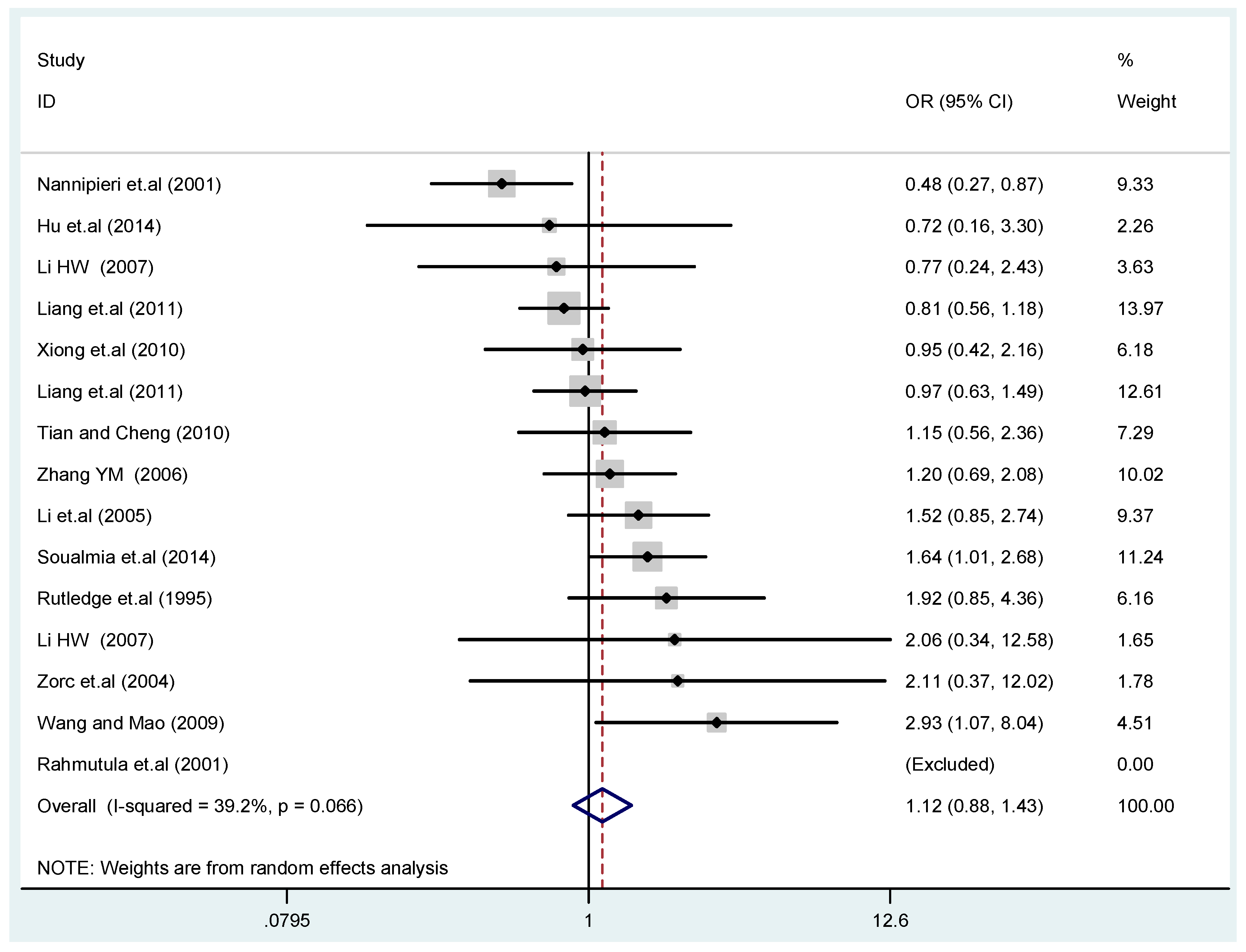
Figure 3.
Forest plot of overall population of T2238C co-dominant model-1 (TC vs. TT).
Figure 3.
Forest plot of overall population of T2238C co-dominant model-1 (TC vs. TT).
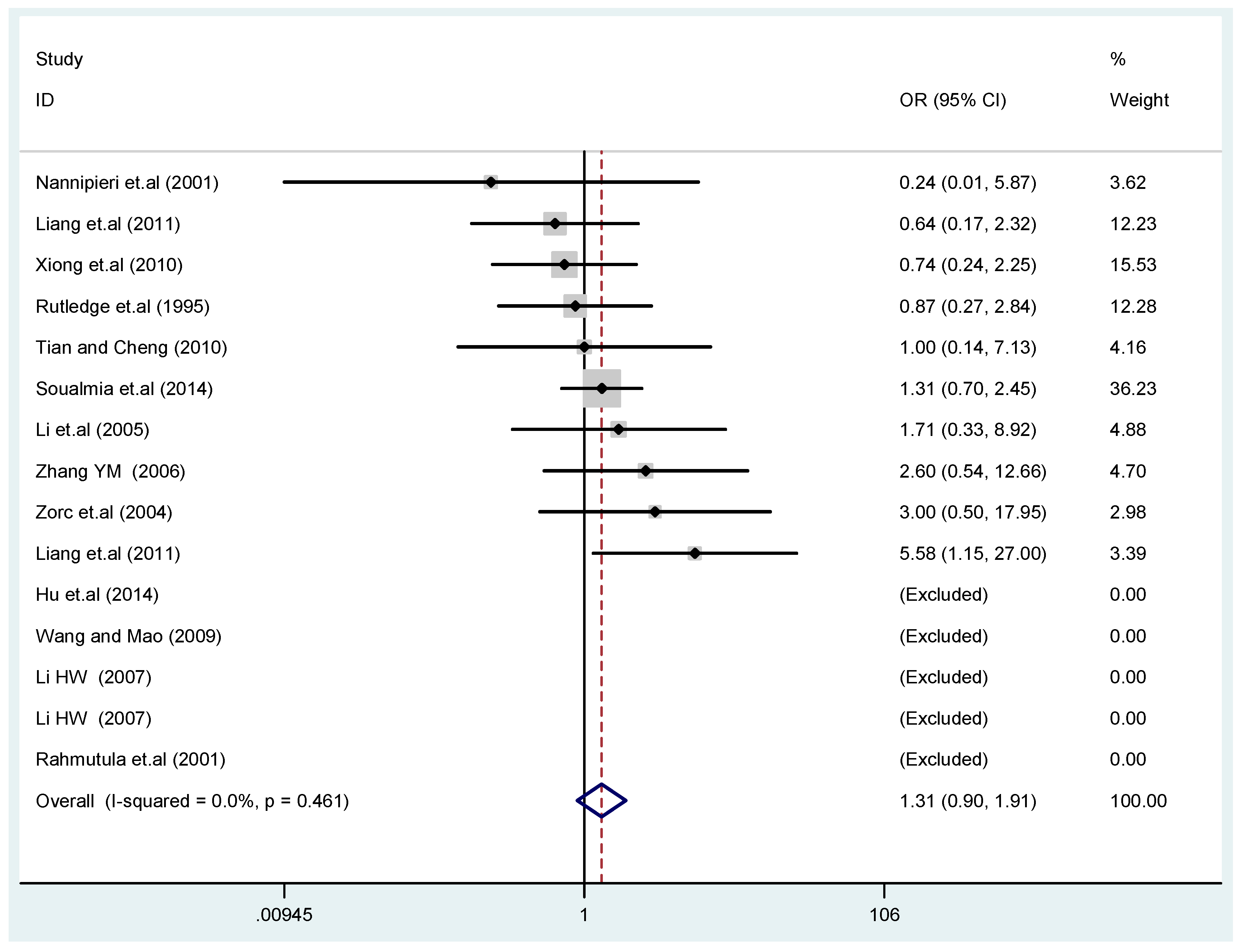
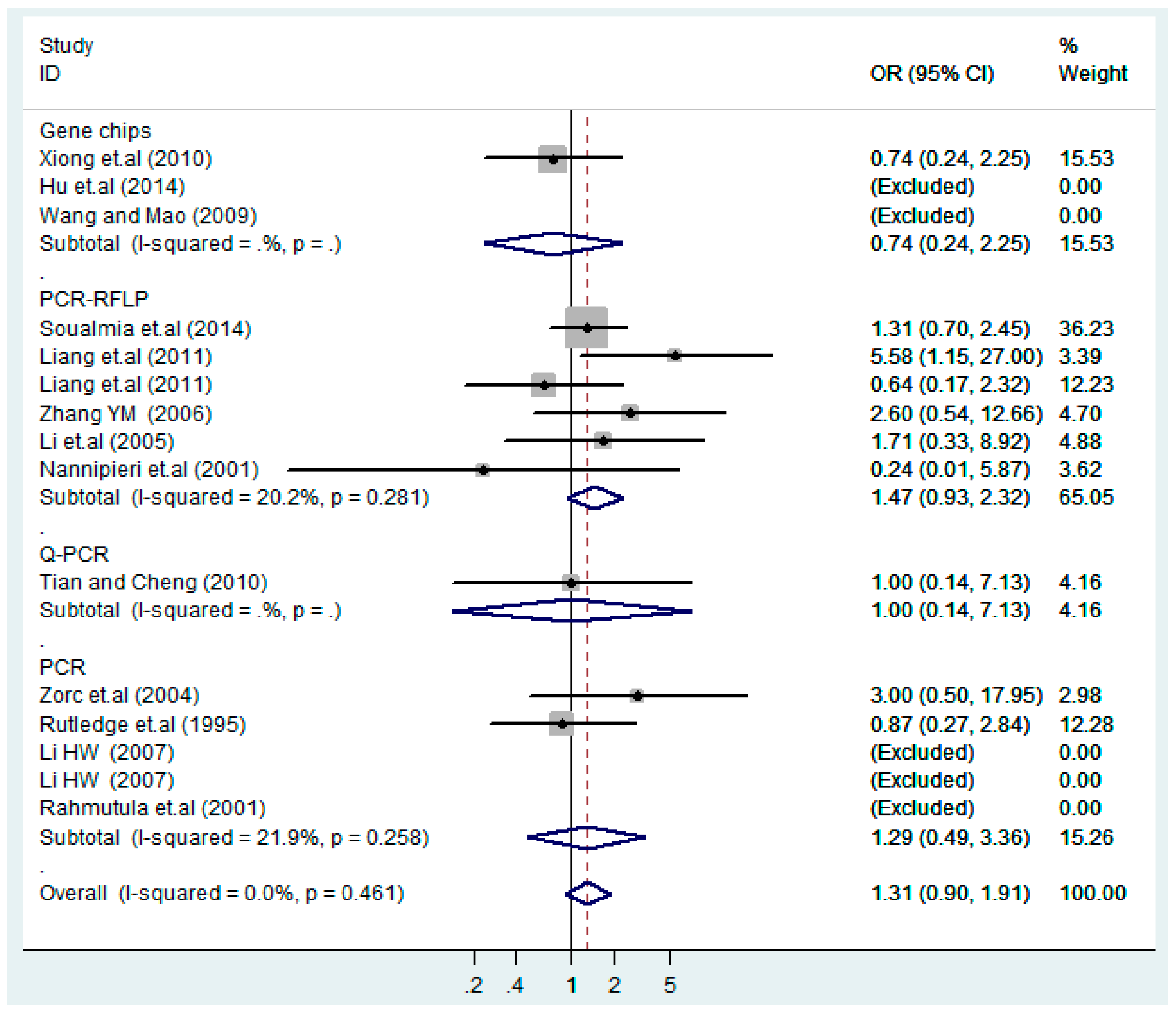
Figure 4.
Forest plot of overall population of T2238C co-dominant model-2 (CC vs. TT).
Figure 4.
Forest plot of overall population of T2238C co-dominant model-2 (CC vs. TT).
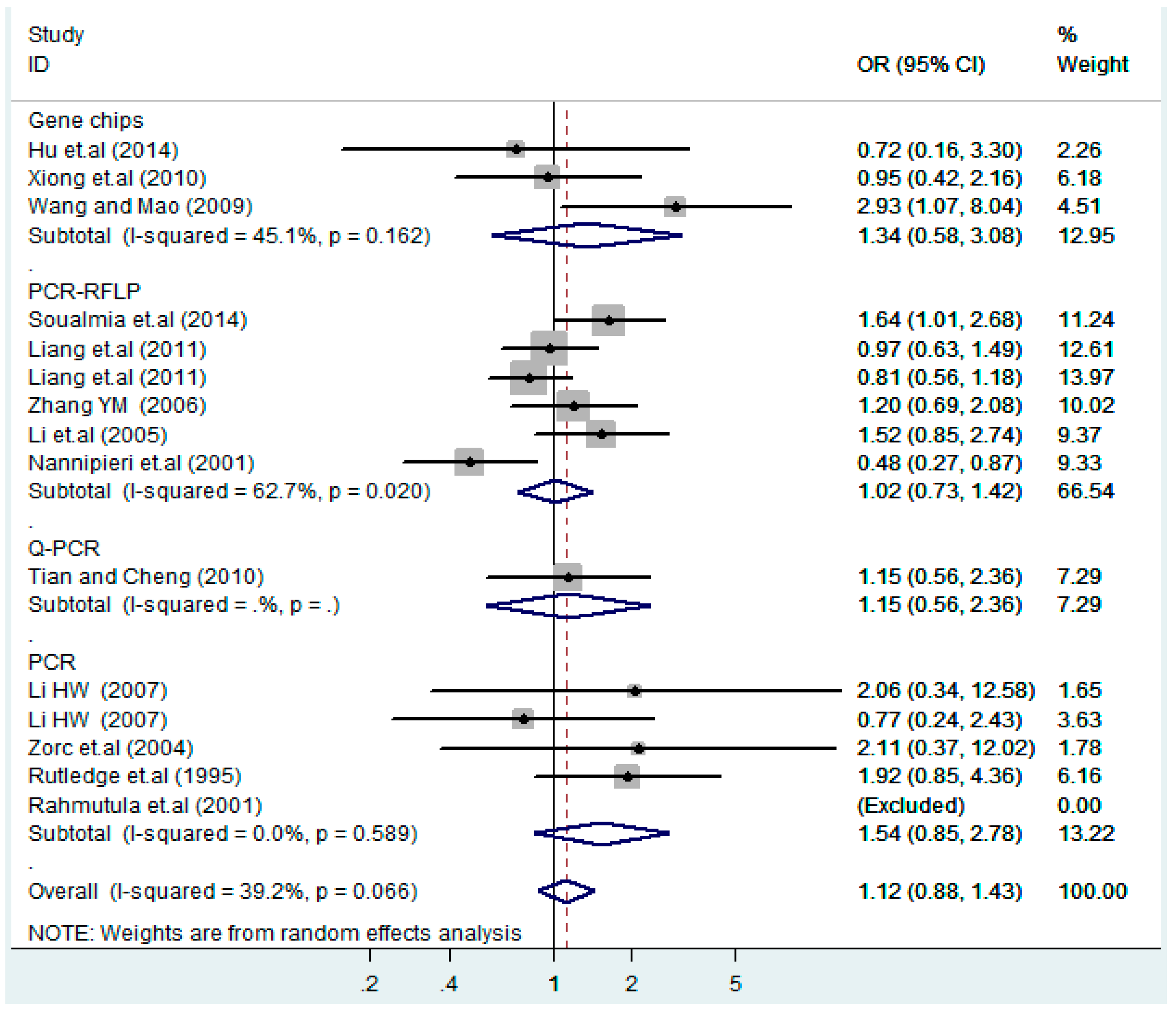
Figure 5.
Forest plot of overall population of T2238C dominant model (CC + TC) vs. TT.
Figure 5.
Forest plot of overall population of T2238C dominant model (CC + TC) vs. TT.
Figure 6.
Forest plot of overall population of T2238C recessive model CC vs. (TT + TC).
Figure 6.
Forest plot of overall population of T2238C recessive model CC vs. (TT + TC).
Figure 7.
Forest plot of sensitivity analysis of overall population of G1837A polymorphism and EH (GA vs. GG).
Figure 7.
Forest plot of sensitivity analysis of overall population of G1837A polymorphism and EH (GA vs. GG).
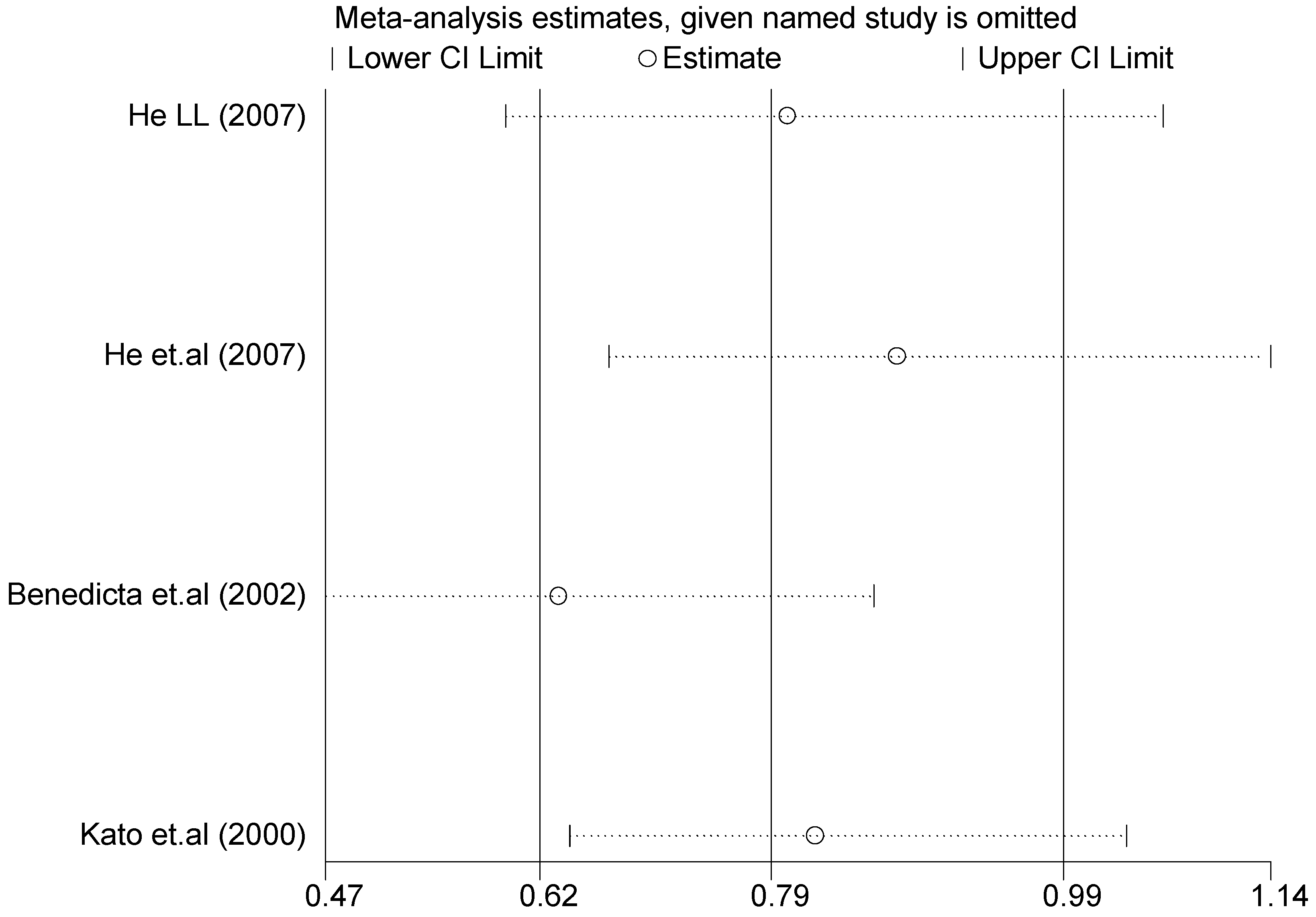
Figure 8.
Forest plot of sensitivity analysis of overall population of T1766C polymorphism and EH (TC vs. TT).
Figure 8.
Forest plot of sensitivity analysis of overall population of T1766C polymorphism and EH (TC vs. TT).
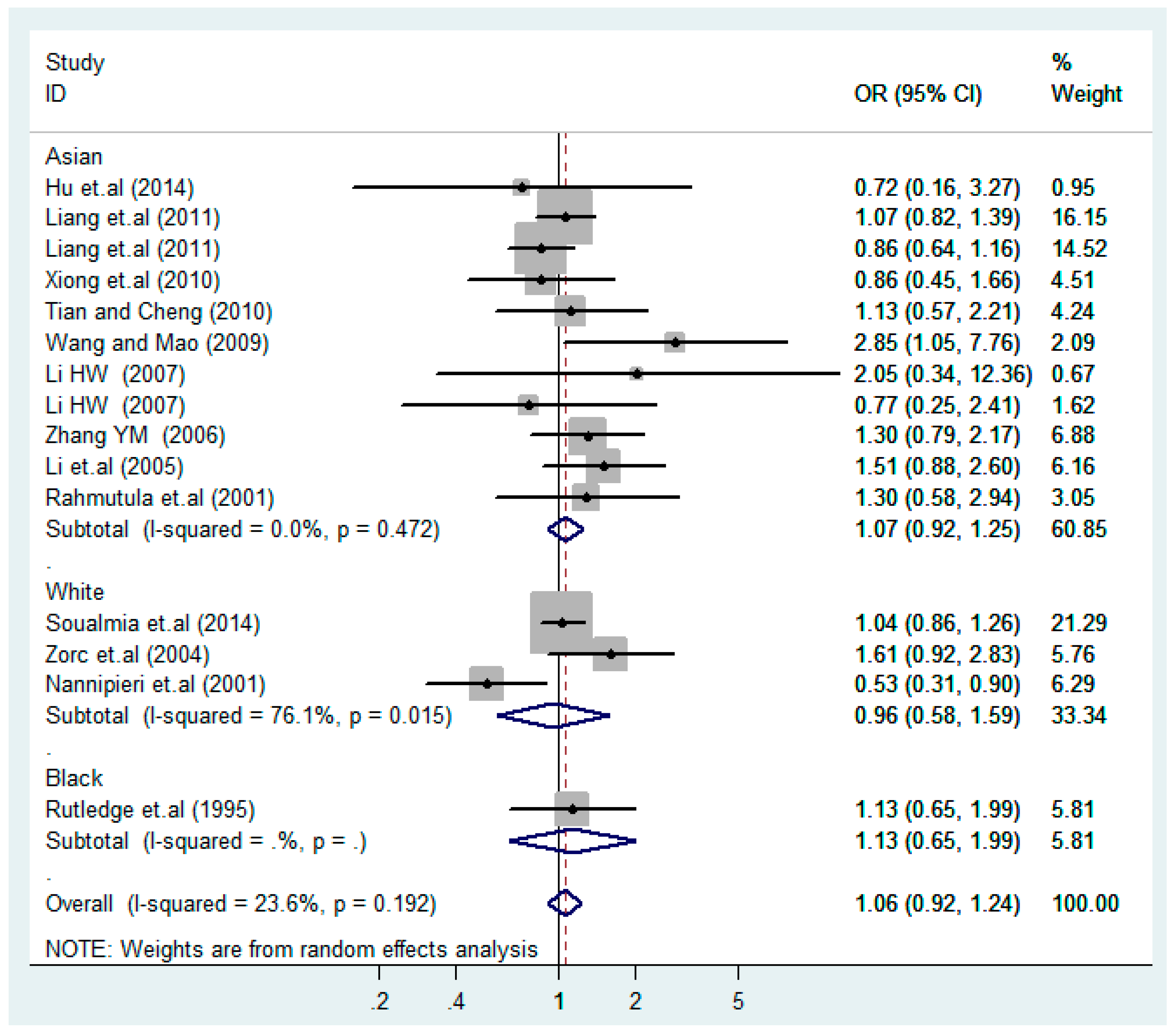
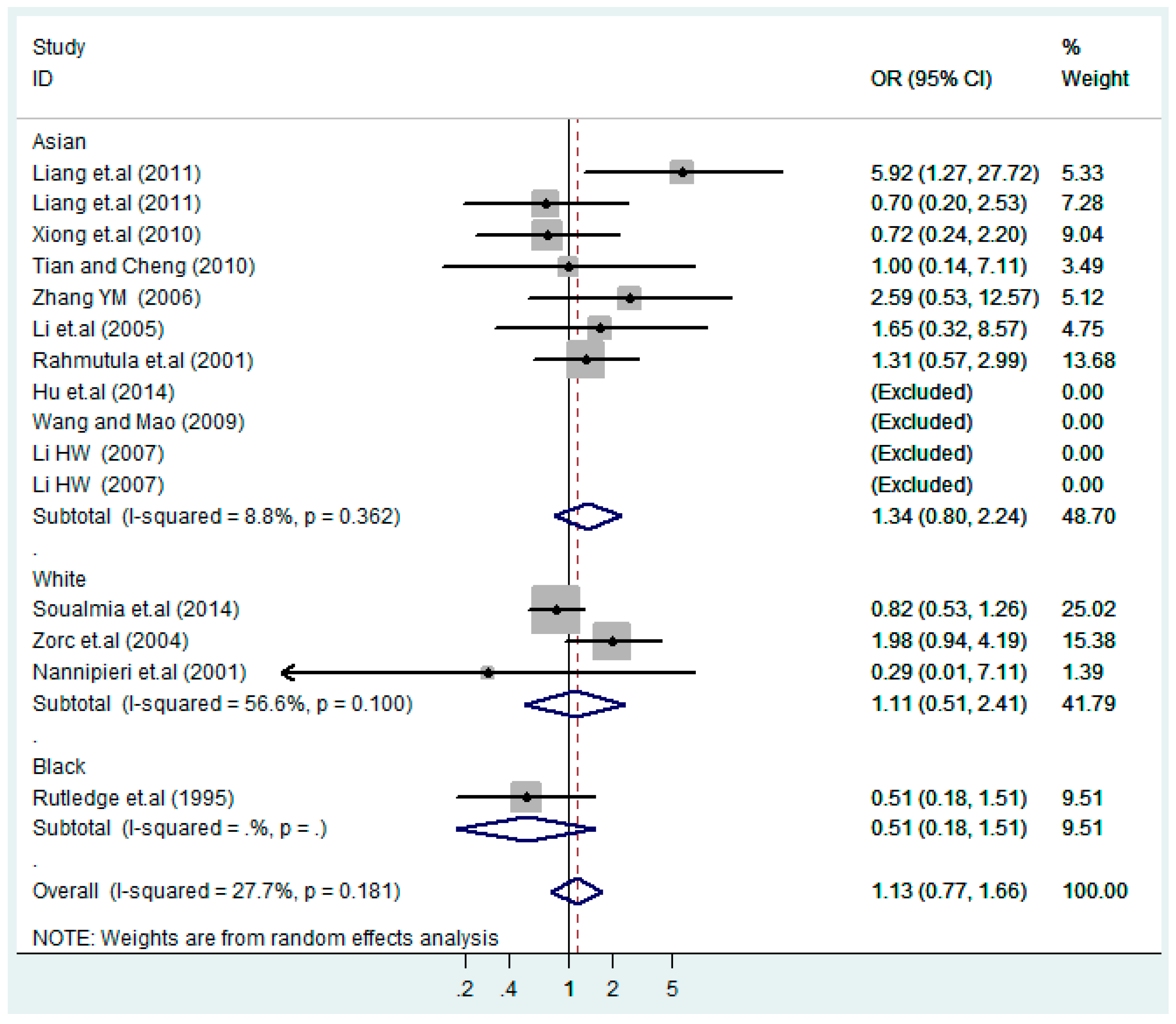
Figure 9.
Ethnic subgroups analysis of T2238C additive model (C vs. T).
Figure 9.
Ethnic subgroups analysis of T2238C additive model (C vs. T).
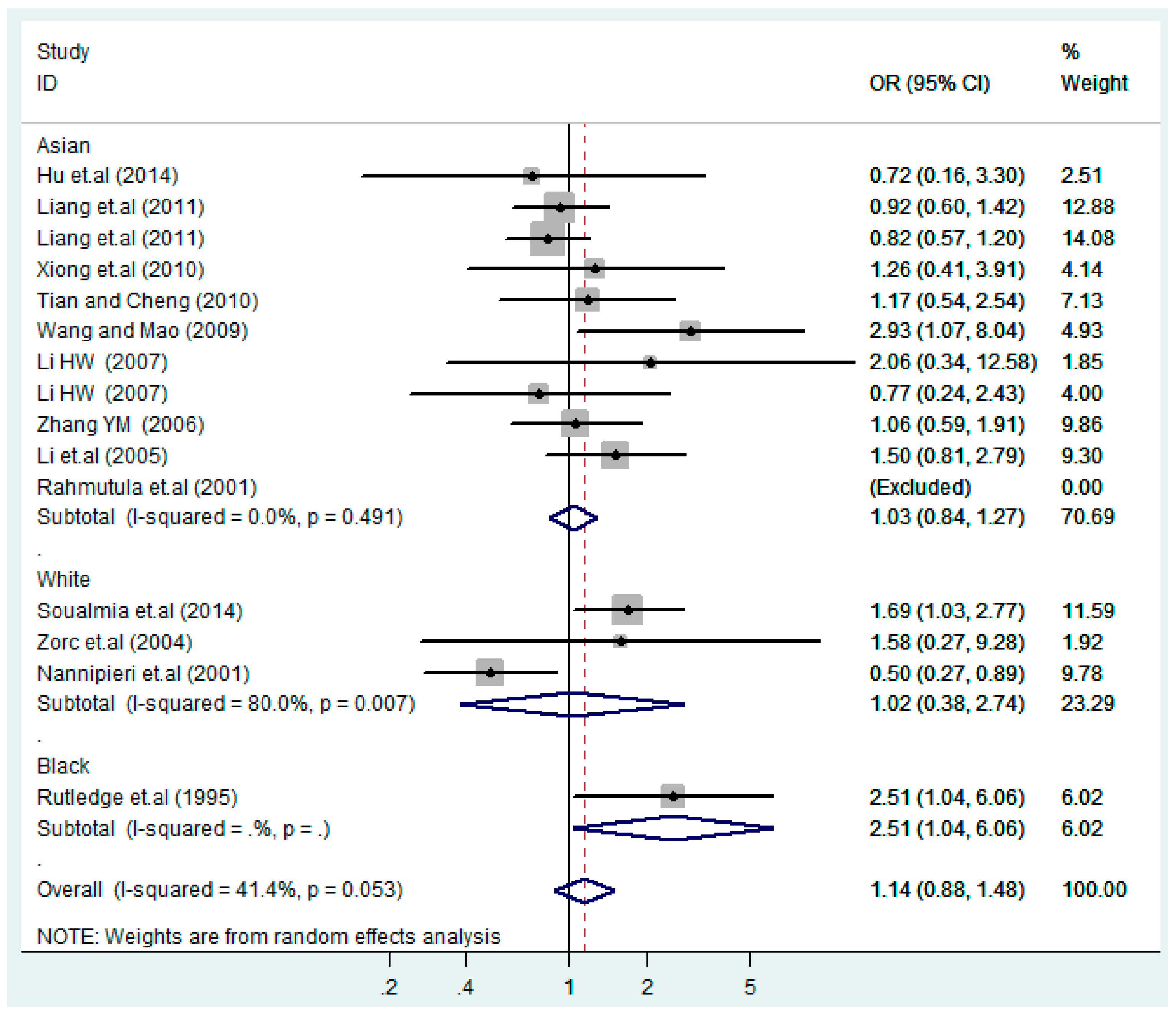
Figure 10.
Ethnic subgroups analysis of T2238C co-dominant model-1 (TC vs. TT).
Figure 10.
Ethnic subgroups analysis of T2238C co-dominant model-1 (TC vs. TT).
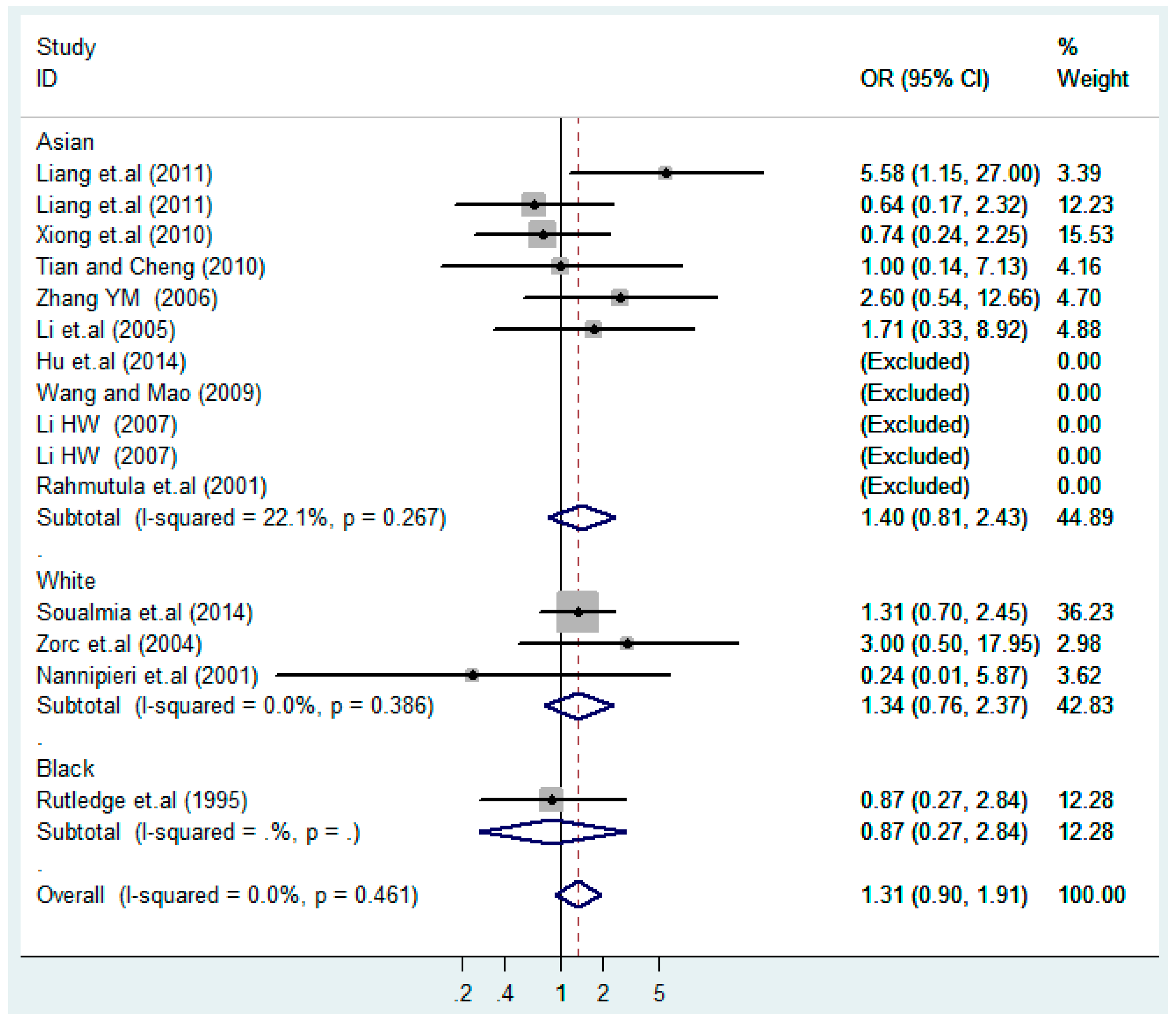
Figure 11.
Ethnic subgroups analysis of T2238C co-dominant model-2 (CC vs. TT).
Figure 11.
Ethnic subgroups analysis of T2238C co-dominant model-2 (CC vs. TT).
Figure 12.
Ethnic subgroup analysis of T2238C dominant model (CC + TC) vs. TT.
Figure 12.
Ethnic subgroup analysis of T2238C dominant model (CC + TC) vs. TT.
Figure 13.
Ethnic subgroup analysis of T2238C recessive model CC vs. (TT + TC).
Figure 13.
Ethnic subgroup analysis of T2238C recessive model CC vs. (TT + TC).
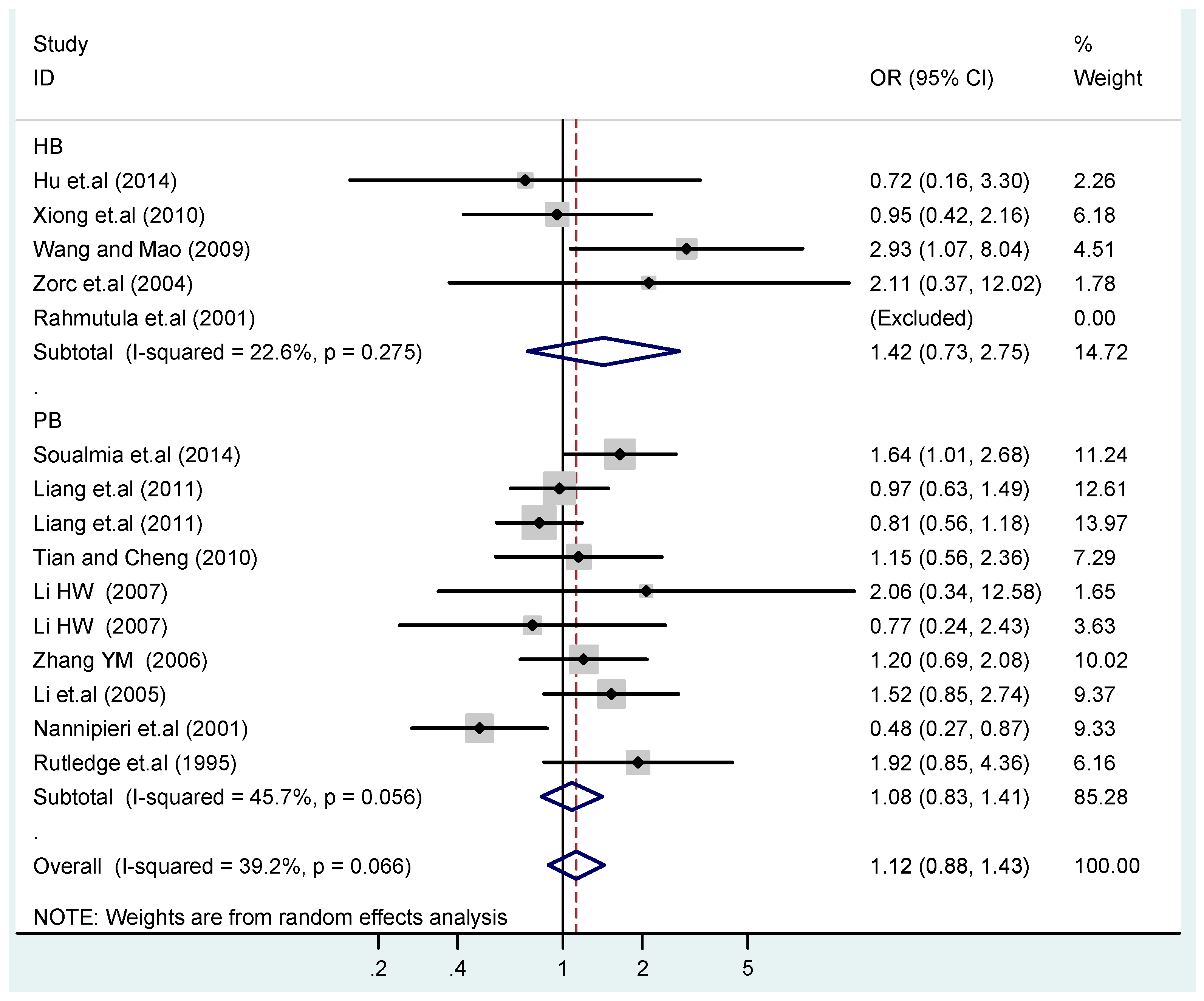
Figure 14.
Subgroups in sources of controls of T2238C additive model (C vs. T).
Figure 14.
Subgroups in sources of controls of T2238C additive model (C vs. T).
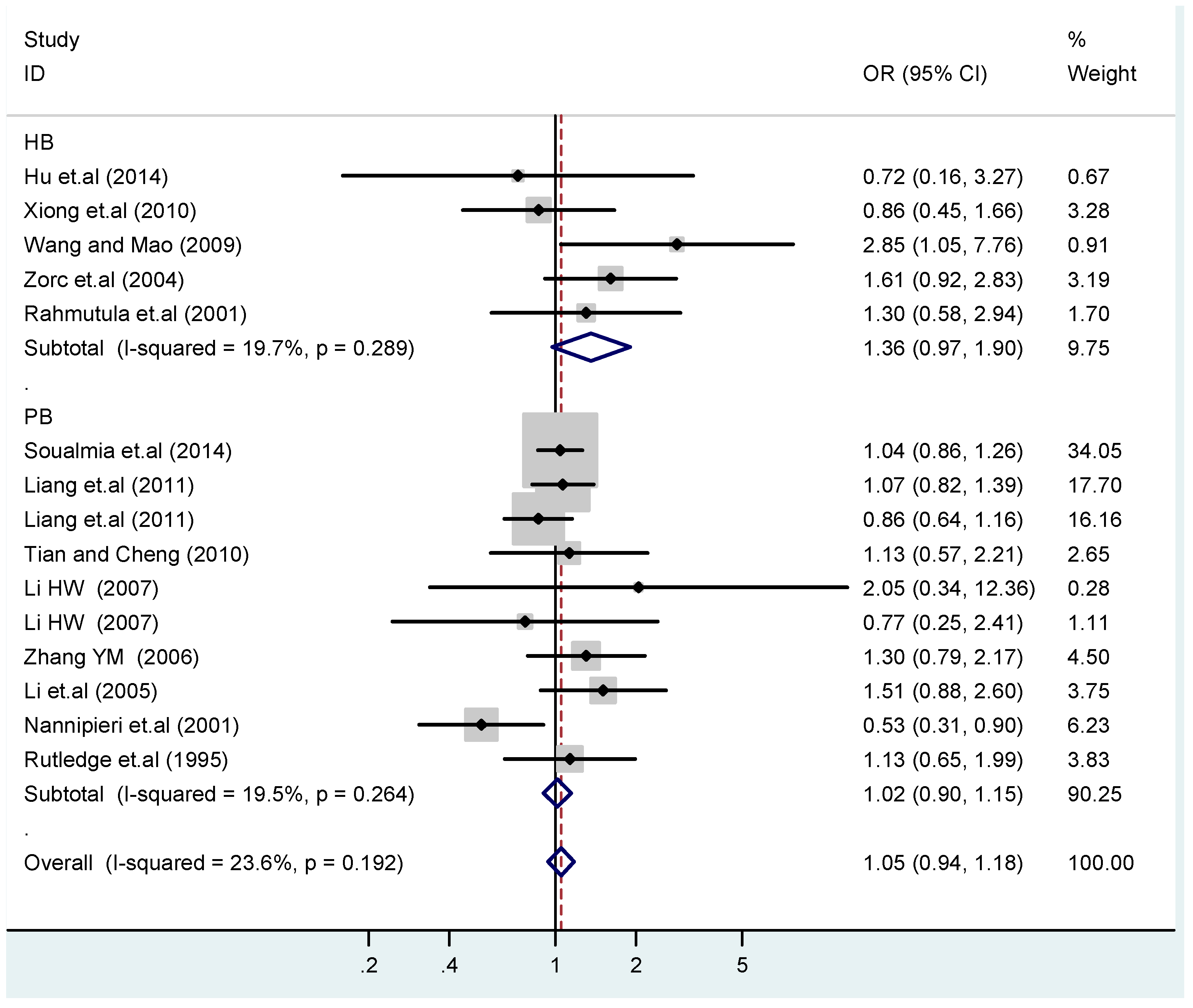
Figure 15.
Subgroups in sources of controls of T2238C co-dominant model-1 (TC vs. TT).
Figure 15.
Subgroups in sources of controls of T2238C co-dominant model-1 (TC vs. TT).
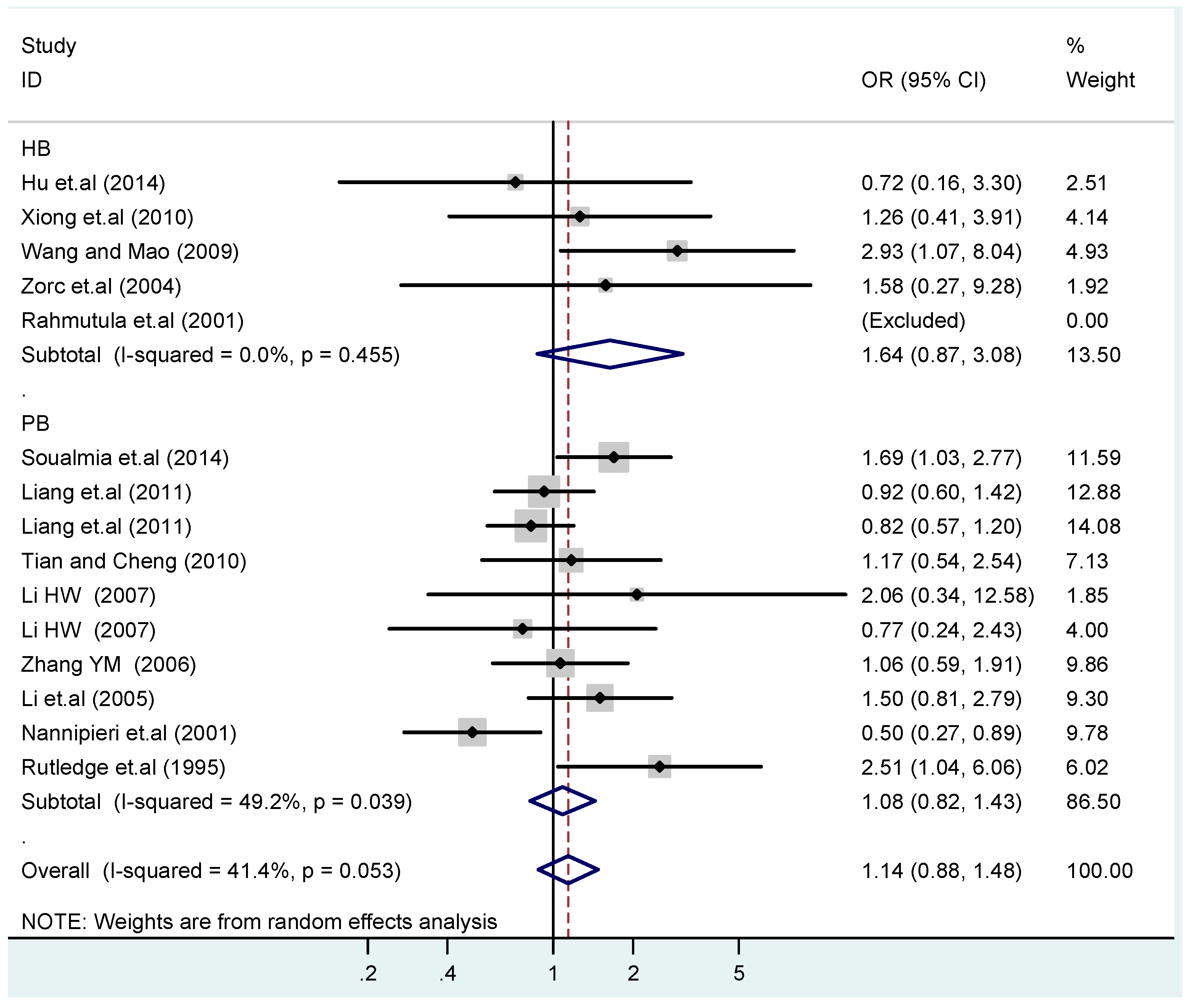
Figure 16.
Subgroups in sources of controls of T2238C co-dominant model-2 (CC vs. TT).
Figure 16.
Subgroups in sources of controls of T2238C co-dominant model-2 (CC vs. TT).
Figure 17.
Subgroups in sources of controls of T2238C dominant model (CC + TC) vs. TT.
Figure 17.
Subgroups in sources of controls of T2238C dominant model (CC + TC) vs. TT.
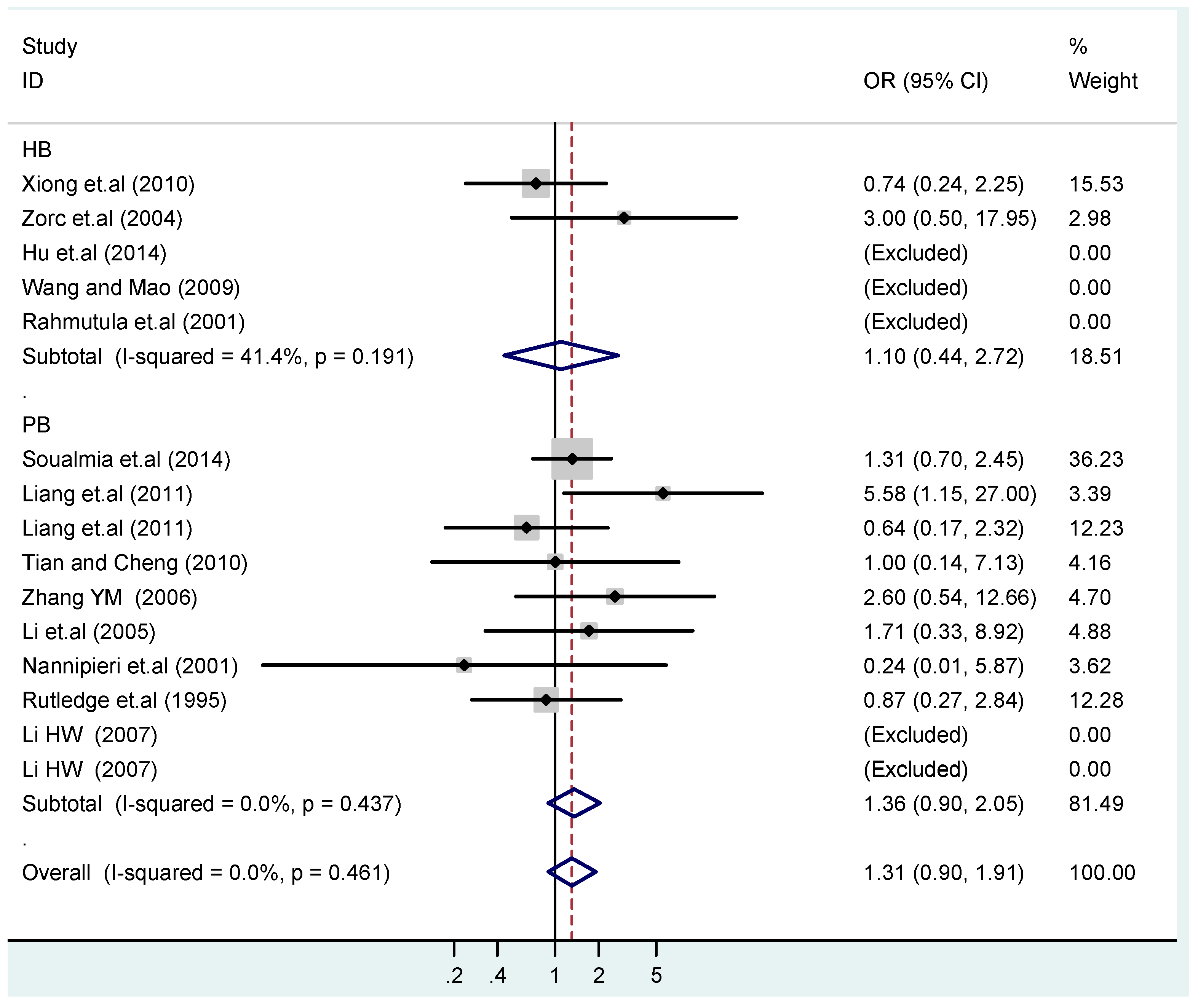
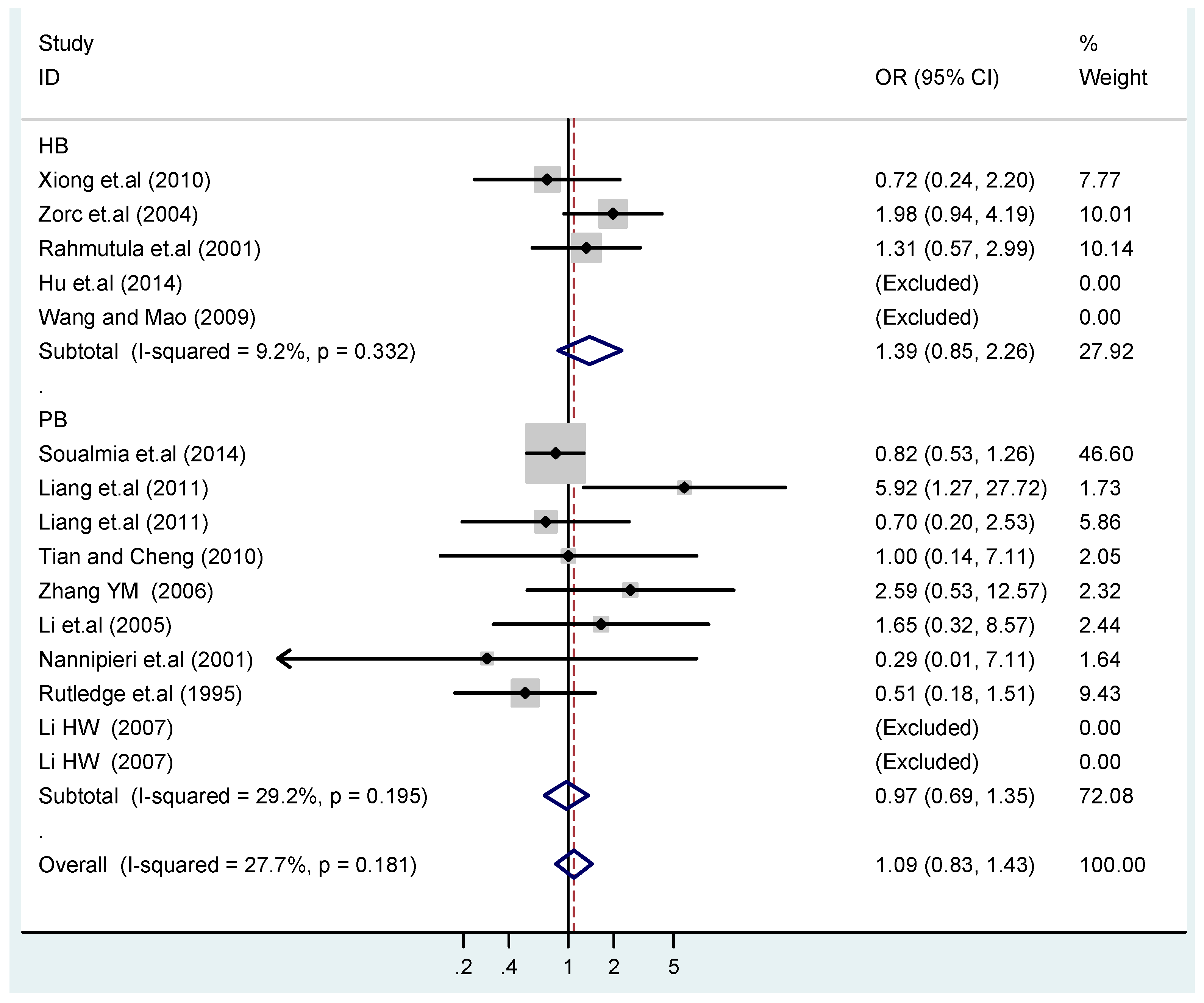
Figure 18.
Subgroups in sources of controls of T2238C recessive model CC vs. (TT + TC).
Figure 18.
Subgroups in sources of controls of T2238C recessive model CC vs. (TT + TC).
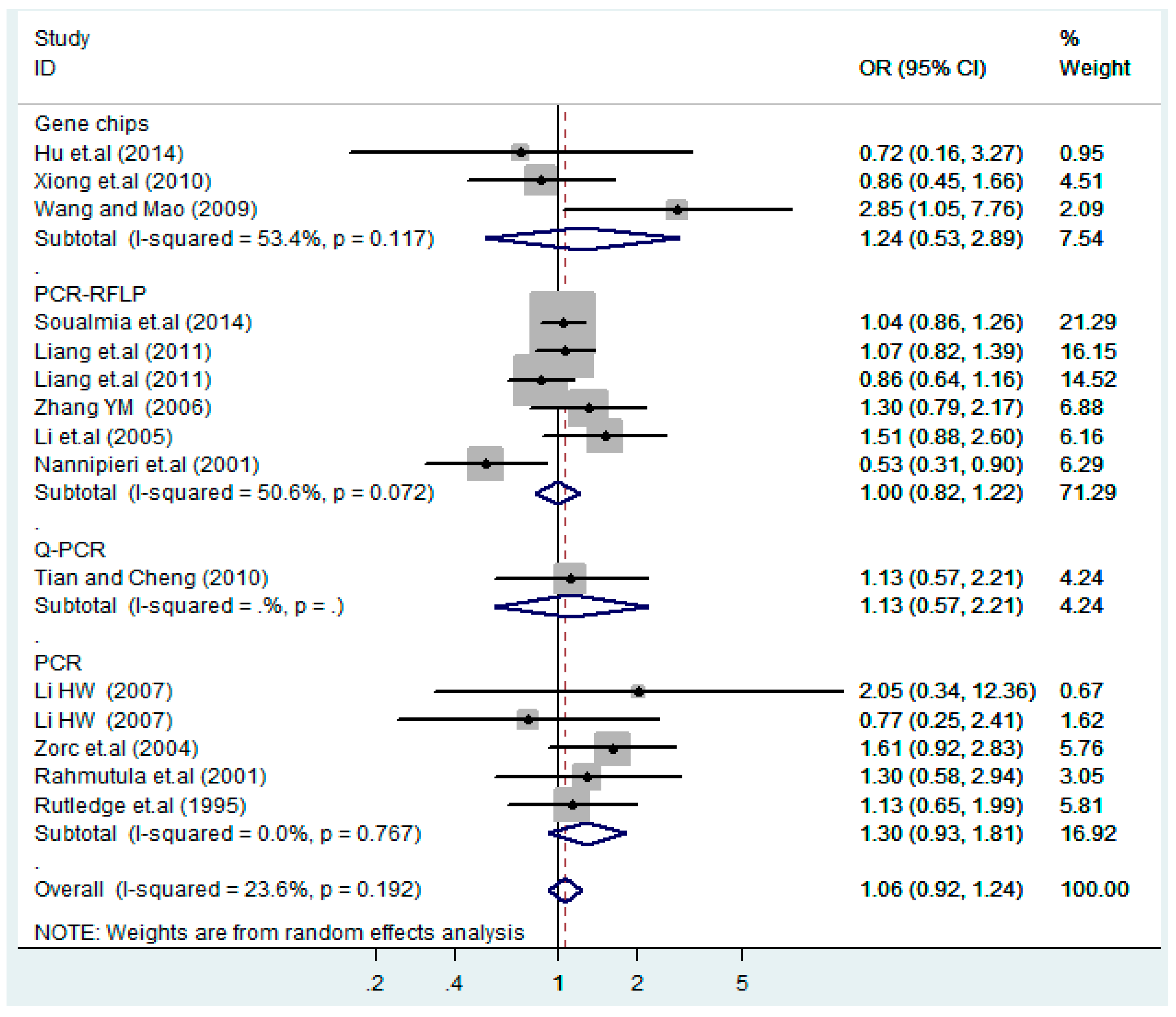
Figure 19.
Subgroups of genotyping methods of T2238C additive model (C vs. T).
Figure 19.
Subgroups of genotyping methods of T2238C additive model (C vs. T).
Figure 20.
Subgroups of genotyping methods of T2238C co-dominant model-1 (TC vs. TT).
Figure 20.
Subgroups of genotyping methods of T2238C co-dominant model-1 (TC vs. TT).
Figure 21.
Subgroups of genotyping methods of T2238C co-dominant model-2 (CC vs. TT).
Figure 21.
Subgroups of genotyping methods of T2238C co-dominant model-2 (CC vs. TT).
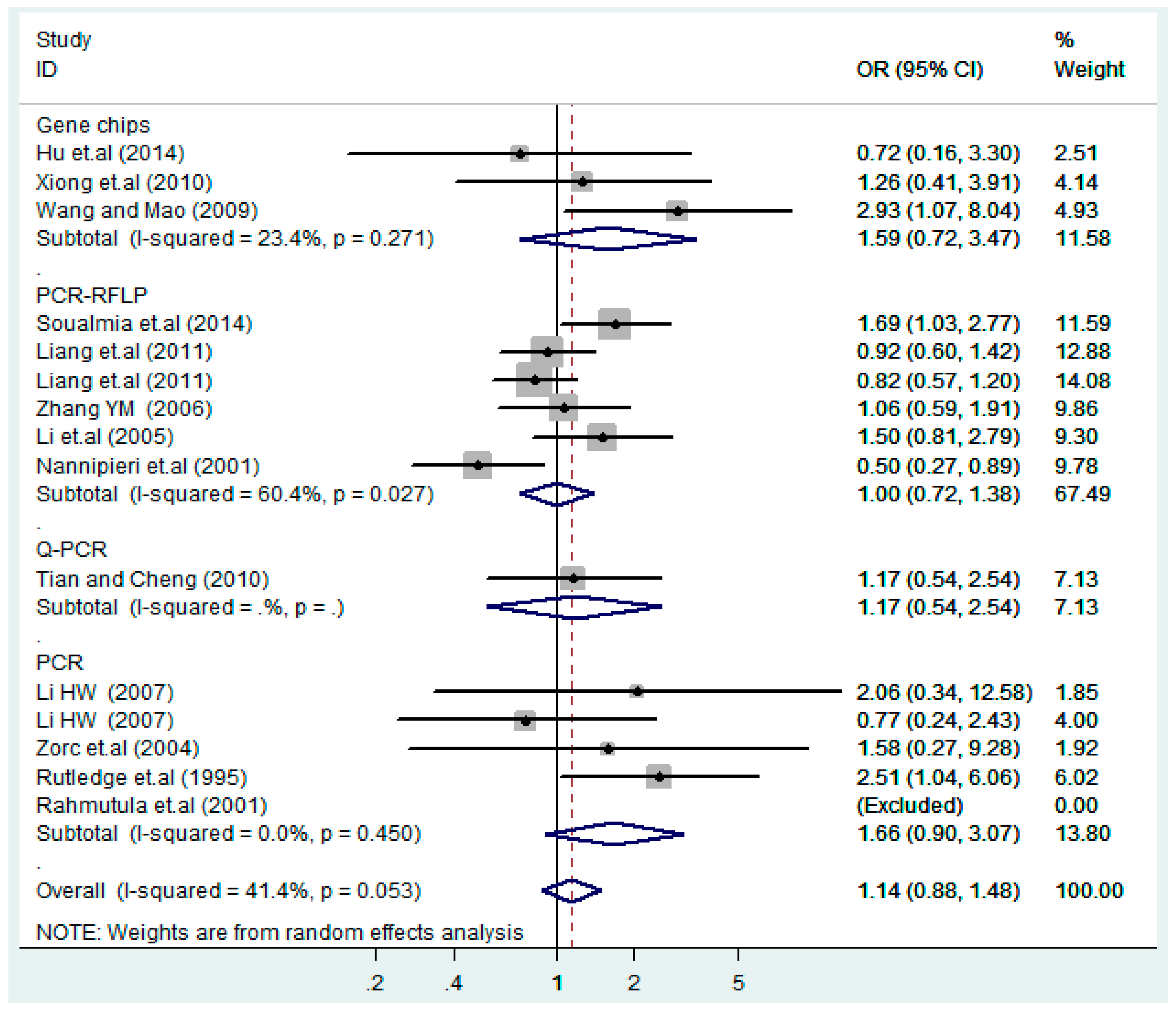
Figure 22.
Subgroups of genotyping methods of T2238C dominant model (CC + TC) vs. TT.
Figure 22.
Subgroups of genotyping methods of T2238C dominant model (CC + TC) vs. TT.
Figure 23.
Subgroups of genotyping of T2238C recessive model CC vs. (TT + TC).
Figure 23.
Subgroups of genotyping of T2238C recessive model CC vs. (TT + TC).
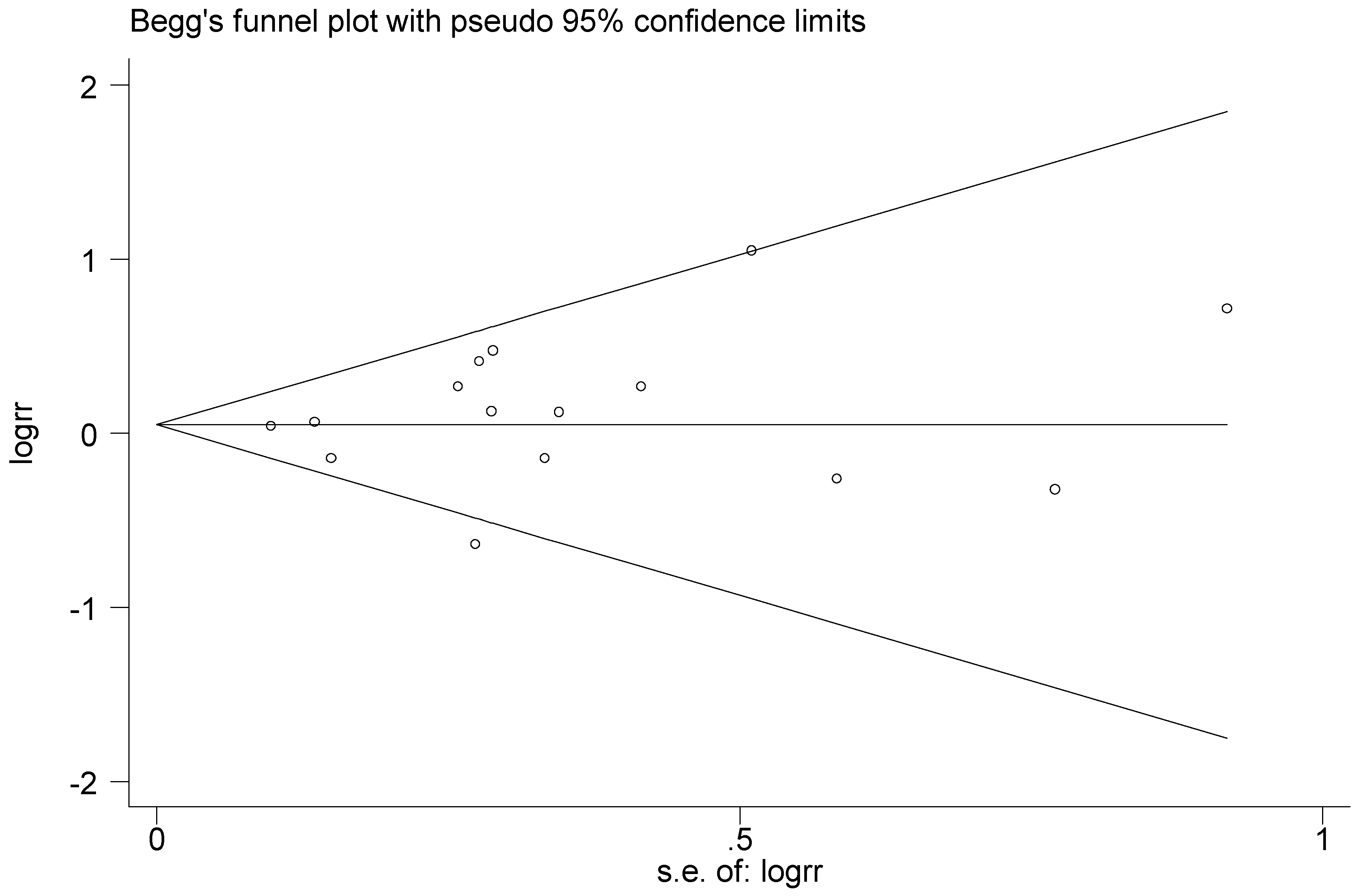
Figure 24.
Begg’s funnel plot of T2238C polymorphism for the publication bias test (C vs. T).
Figure 24.
Begg’s funnel plot of T2238C polymorphism for the publication bias test (C vs. T).
Figure 25.
Forest plot of sensitivity analysis of overall population of T2238C polymorphism and EH (C vs. T).
Figure 25.
Forest plot of sensitivity analysis of overall population of T2238C polymorphism and EH (C vs. T).
Table 1.
Characteristics of studies included in the meta-analysis.
Table 1.
Characteristics of studies included in the meta-analysis.
| Author | Year | Locus | Source | Country | Ethnicity | Number | Genotyping Methods | Quality Score |
|---|
| T2238C | Case | Control | Case | Control |
|---|
| Hu et al. [17] | 2014 | | H-B | H-B | China | Asian (Han) | 100 | 97 | Gene chips | 6 |
| Soualmia et al. [14] | 2014 | | H-B | P-B | Tunisia | White (Tunisian) | 384 | 453 | PCR-RFLP | 7 |
| Liang et al. [20] | 2011 | | P-B | P-B | China | Asian (Han) | 205 | 260 | PCR-RFLP | 6 |
| Liang et al. [20] | 2011 | | P-B | P-B | China | Asian (Kazakh) | 218 | 232 | PCR-RFLP | 6 |
| Xiong et al. [21] | 2010 | | H-B | H-B | China | Asian (Han) | 81 | 120 | Gene chips | 5 |
| Tian and Cheng [22] | 2010 | | H-B | P-B | China | Asian (Han) | 976 | 976 | Q-PCR | 6 |
| Wang and Mao [23] | 2009 | | H-B | H-B | China | Asian (Han) | 238 | 184 | Gene chips | 4 |
| Li [24] | 2007 | | P-B | P-B | China | Asian (Yi) | 99 | 134 | PCR | 5 |
| Li [24] | 2007 | | P-B | P-B | China | Asian (Hani) | 172 | 133 | PCR | 5 |
| Zhang YM [25] | 2006 | | P-B | P-B | China | Asian (Kazakh) | 314 | 229 | PCR-RFLP | 6 |
| Li et.al. [26] | 2005 | | P-B | P-B | China | Asian (Kazakh) | 313 | 205 | PCR-RFLP | 6 |
| Zorc et.al. [27] | 2004 | | H-B | H-B | Slovenia | Caucasian | 58 | 57 | PCR | 6 |
| Nannipieri et.al. [28] | 2001 | | P-B | P-B | Europeans | White | 121 | 105 | PCR-RFLP | 6 |
| Rahmutula et.al. [29] | 2001 | | H-B | H-B | Japan | Asian | 233 | 213 | PCR | 3 |
| Rutledge et.al. [30] | 1995 | | H-B | P-B | American | Black | 60 | 44 | PCR | 6 |
| | | G1837A | case | control | | | case | control | | |
| Li [24] | 2007 | | P-B | P-B | China | Asian (Yi) | 99 | 134 | PCR | 5 |
| Li [24] | 2007 | | P-B | P-B | China | Asian (Hani) | 172 | 133 | PCR | 5 |
| Zhang et.al. [31] | 2005 | | P-B | P-B | China | Asian (Kazakh) | 287 | 190 | PCR-RFLP | 5 |
| Rahmutula et.al. [29] | 2001 | | H-B | H-B | Japan | Asian | 233 | 213 | PCR | 3 |
| Bernard et.al. [32] | 1999 | | H-B | P-B | China | Asian | 108 | 109 | PCR | 6 |
| Rutledge et.al. [30] | 1995 | | H-B | P-B | American | Black | 60 | 44 | PCR | 6 |
| | | T1766C | case | control | | | case | control | | |
| He [33] | 2007 | | P-B | P-B | China | Asian (Kazakh) | 199 | 198 | PCR-RFLP | 5 |
| He et.al. [34] | 2007 | | P-B | P-B | China | Asian (Kazakh) | 246 | 244 | PCR-RFLP | 5 |
| Benedicta et.al. [35] | 2002 | | H-B | P-B | African | Black | 289 | 278 | PCR-RFLP | 6 |
| Kato et.al. [36] | 2000 | | H-B | H-B | Japan | Asian | 255 | 225 | PCR | 5 |
Table 2.
The allele gene and genotype frequency of ANP polymorphisms in the meta-analysis.
Table 2.
The allele gene and genotype frequency of ANP polymorphisms in the meta-analysis.
| Author | Year | Locus | Allele Number | Gene Number | HWE |
|---|
| Case | Control | Case | Control |
|---|
| T2238C | T | C | T | C | TT | TC | CC | TT | TC | CC |
|---|
| Hu et.al. [17] | 2014 | | 197 | 3 | 190 | 4 | 97 | 3 | 0 | 93 | 4 | 0 | YES |
| Soualmia et.al. [14] | 2014 | | 372 | 396 | 448 | 458 | 27 | 318 | 39 | 50 | 348 | 55 | NO |
| Liang et.al. [20] | 2011 | | 246 | 164 | 320 | 200 | 50 | 146 | 9 | 62 | 196 | 2 | YES |
| Liang et.al. [20] | 2011 | | 322 | 114 | 329 | 135 | 108 | 106 | 4 | 103 | 123 | 6 | YES |
| Xiong et.al. [21] | 2010 | | 146 | 16 | 213 | 27 | 70 | 6 | 5 | 103 | 7 | 10 | YES |
| Tian and Cheng [22] | 2010 | | 1934 | 18 | 1936 | 16 | 960 | 14 | 2 | 962 | 12 | 2 | YES |
| Wang and Mao [23] | 2009 | | 458 | 18 | 363 | 5 | 220 | 18 | 0 | 179 | 5 | 0 | YES |
| Li [24] | 2007 | | 195 | 3 | 266 | 2 | 96 | 3 | 0 | 132 | 2 | 0 | YES |
| Li [24] | 2007 | | 338 | 6 | 260 | 6 | 166 | 6 | 0 | 127 | 6 | 0 | YES |
| Zhang YM [25] | 2006 | | 584 | 44 | 433 | 25 | 277 | 30 | 7 | 206 | 21 | 2 | YES |
| Li et.al. [26] | 2005 | | 581 | 45 | 390 | 20 | 273 | 35 | 5 | 187 | 16 | 2 | YES |
| Zorc et.al. [27] | 2004 | | 30 | 86 | 41 | 73 | 2 | 26 | 30 | 4 | 33 | 20 | YES |
| Nannipieri et.al. [28] | 2001 | | 216 | 26 | 171 | 39 | 95 | 26 | 0 | 67 | 37 | 1 | YES |
| Rahmutula et.al. [29] | 2001 | | 11 | 455 | 13 | 413 | 0 | 11 | 222 | 0 | 13 | 200 | YES |
| Rutledge et.al. [30] | 1995 | | 70 | 50 | 54 | 34 | 17 | 36 | 7 | 19 | 16 | 9 | YES |
| | | G1837A | G | A | G | A | GG | GA | AA | GG | GA | AA | |
| Li [24] | 2007 | | 178 | 20 | 245 | 23 | 79 | 20 | 0 | 113 | 19 | 2 | YES |
| Li [24] | 2007 | | 296 | 48 | 233 | 33 | 127 | 42 | 3 | 101 | 31 | 1 | YES |
| Zhang et.al. [31] | 2005 | | 514 | 60 | 346 | 34 | 228 | 58 | 1 | 158 | 30 | 2 | YES |
| Rahmutula et.al. [29] | 2001 | | 42 | 424 | 47 | 379 | 3 | 36 | 194 | 1 | 45 | 167 | YES |
| Bernard et.al. [32] | 1999 | | 191 | 25 | 195 | 23 | 86 | 19 | 3 | 87 | 21 | 1 | YES |
| Rutledge et.al. [30] | 1995 | | 90 | 30 | 85 | 3 | 30 | 30 | 0 | 41 | 3 | 0 | YES |
| | | T1766C | T | C | T | C | TT | TC | CC | TT | TC | CC | |
| He [33] | 2007 | | 304 | 94 | 291 | 105 | 108 | 88 | 3 | 95 | 101 | 2 | YES |
| He et.al. [34] | 2007 | | 290 | 202 | 267 | 221 | 49 | 192 | 5 | 29 | 209 | 6 | YES |
| Benedicta et.al. [35] | 2002 | | 333 | 245 | 311 | 245 | 87 | 159 | 43 | 85 | 141 | 52 | YES |
| Kato et.al. [36] | 2000 | | 506 | 4 | 440 | 10 | 251 | 4 | 0 | 215 | 10 | 0 | YES |
Table 3.
Meta-analysis of T2238C polymorphism and EH.
Table 3.
Meta-analysis of T2238C polymorphism and EH.
| Stratification Factors | No. | Additive Model (C vs. T) | p | Co-Dominant Model-1 (TC vs. TT) | p | Co-Dominant Model-2 (CC vs. TT) | p | Dominant Model (CC + TC) vs. TT | p | Recessive Model CC vs. (TT + TC) | p |
|---|
| OR(95%CI) a | OR(95%CI) a | OR(95%CI) a | OR(95%CI) a | OR(95%CI) a |
|---|
| Overall | 15 | 1.1(0.94–1.2) | 0.38 | 1.1(0.88–1.5) | 0.32 | 1.3(0.90–1.9) | 0.16 | 1.1(0.88–1.4) | 0.35 | 1.1(0.83–1.4) | 0.55 |
|---|
| Ethnicity | | | | | | | | | | | |
|---|
| Asian | 11 | 1.1(0.92–1.3) | 0.38 | 1.0(0.84–1.3) | 0.75 | 1.4(0.81–2.4) | 0.23 | 1.1(0.86–1.3) | 0.62 | 1.3(0.81–2.2) | 0.26 |
| White | 3 | 0.96(0.58–1.6) | 0.89 | 1.0(0.38–2.7) | 0.96 | 1.3 (0.76–2.4) | 0.32 | 1.1(0.39–2.9) | 0.91 | 1.1(0.51–2.4) | 0.80 |
| Black | 1 | 1.1(0.65–2.0) | 0.66 | 2.5(1.0–6.1) | 0.040 | 0.87(0.27–2.8) | 0.82 | 1.9(0.85–4.4) | 0.12 | 0.51(0.18–1.5) | 0.23 |
| Source of controls | | | | | | | | | | |
| HB | 5 | 1.4(0.97–1.9) | 0.073 | 1.6(0.87–3.1) | 0.13 | 1.1(0.45–2.7) | 0.83 | 1.4(0.73–2.8) | 0.30 | 1.4(0.85–2.3) | 0.19 |
| PB | 10 | 1.0(0.90–1.1) | 0.77 | 1.1(0.82–1.4) | 0.58 | 1.4(0.90–2.1) | 0.15 | 1.1(0.83–1.4) | 0.57 | 0.97(0.70–1.4) | 0.86 |
| Genotyping methods | | | | | | | | | | |
| Gene chips | 3 | 1.2(0.53–2.9) | 0.62 | 1.6(0.72–3.5) | 0.25 | 0.74(0.24–2.2) | 0.59 | 1.3(0.58–3.1) | 0.50 | 0.72(0.24–2.2) | 0.57 |
| PCR-RFLP | 6 | 1.0(0.82–1.2) | 1.0 | 1.0(0.72–1.4) | 0.99 | 1.5(0.93–2.3) | 0.096 | 1.0(0.73–1.4) | 0.91 | 1.3(0.64–2.6) | 0.48 |
| Q-PCR | 1 | 1.1(0.57–2.2) | 0.73 | 1.2(0.54–2.5) | 0.69 | 1.0(0.14–7.1) | 1.0 | 1.1(0.56–2.4) | 0.71 | 1.0(0.14–7.1) | 1.0 |
| PCR | 5 | 1.3(0.93–1.8) | 0.13 | 1.7(0.90–3.1) | 0.10 | 1.3(0.49–3.4) | 0.61 | 1.5(0.85–2.8) | 0.15 | 1.2(0.58–2.5) | 0.62 |
Table 4.
Meta-analysis of G1837A polymorphism and EH.
Table 4.
Meta-analysis of G1837A polymorphism and EH.
| Gene Type | Genetic Model | OR a | 95%CI | p | I2 | p* | Model |
|---|
| Overall | | | | | | | |
| A vs. G | additive model | 1.3 | 0.96–1.9 | 0.090 | 54.7% | 0.051 | RE |
| GA vs. GG | co-dominant model-1 | 1.5 | 0.83–2.6 | 0.19 | 70.1% | 0.005 | RE |
| AA vs. GG | co-dominant model-2 | 0.87 | 0.34–2.3 | 0.78 | 0.0% | 0.48 | FE |
| (AA + GA) vs. GG | dominant model | 1.5 | 0.86–2.5 | 0.17 | 67.5% | 0.009 | RE |
| AA vs. (GG + GA) | recessive model | 1.3 | 0.85–2.0 | 0.22 | 0.0% | 0.53 | FE |
| Work of Rutledge removed | | | | | | | |
| A vs. G | additive model | 1.2 | 0.95–1.5 | 0.14 | 0.0% | 1.0 | FE |
| GA vs. GG | co-dominant model-1 | 1.2 | 0.88–1.6 | 0.29 | 0.0% | 0.56 | FE |
| AA vs. GG | co-dominant model-2 | 0.87 | 0.34–2.3 | 0.78 | 0.0% | 0.48 | FE |
| (AA + GA) vs. GG | dominant model | 1.2 | 0.88–1.5 | 0.28 | 0.0% | 0.82 | FE |
| AA vs. (GG + GA) | recessive model | 1.3 | 0.85–2.0 | 0.22 | 0.0% | 0.53 | FE |
Table 5.
Meta-analysis of T1766C polymorphism and EH.
Table 5.
Meta-analysis of T1766C polymorphism and EH.
| Gene Type | Genetic Model | OR a | 95%CI | p | I2 | p* | Model |
|---|
| Overall | |
| C vs. T | additive model | 0.87 | 0.75–1.0 | 0.063 | 0.0% | 0.42 | FE |
| TC vs. TT | co-dominant model-1 | 0.73 | 0.49–1.1 | 0.12 | 58.0% | 0.068 | RE |
| CC vs. TT | co-dominant model-2 | 0.78 | 0.50–1.2 | 0.29 | 0.0% | 0.66 | FE |
| (CC + TC) vs. TT | dominant model | 0.73 | 0.51–1.0 | 0.084 | 51.1% | 0.11 | RE |
| CC vs. (TT + TC) | recessive model | 0.79 | 0.53–1.2 | 0.26 | 0.0% | 0.77 | FE |
| Work of Benedicta removed | |
| C vs. T | additive model | 0.82 | 0.68–1.0 | 0.052 | 8.4% | 0.34 | FE |
| TC vs. TT | co-dominant model-1 | 0.64 | 0.47–0.86 | 0.003 | 12.7% | 0.32 | FE |
| CC vs. TT | co-dominant model-2 | 0.69 | 0.25–1.9 | 0.48 | 0.0% | 0.38 | FE |
| (CC + TC) vs. TT | dominant model | 0.64 | 0.48–0.87 | 0.004 | 18.1% | 0.30 | FE |
| CC vs. (TT + TC) | recessive model | 0.99 | 0.37–2.7 | 0.99 | 0.0% | 0.59 | FE |
Table 6.
Sensitivity analysis of G1837A polymorphism and EH (GA vs. GG).
Table 6.
Sensitivity analysis of G1837A polymorphism and EH (GA vs. GG).
| Study Omitted | Estimate | 95%Confidence Interval |
|---|
| Li (2007) [24] | 1.4 | 1.0–1.9 |
| Li (2007) [24] | 1.5 | 1.1–2.1 |
| Zhang et.al. (2005) [31] | 1.4 | 1.0–2.0 |
| Rahmutula et.al. (2001) [29] | 1.4 | 1.1–1.9 |
| Bernard et.al. (1999) [32] | 1.5 | 1.1–2.0 |
| Rutledge et.al. (1995) [30] | 1.2 | 0.88–1.6 |
| Combined | 1.4 | 1.1–1.8 |
Table 7.
Sensitivity analysis of T1766C polymorphism and EH (TC vs. TT).
Table 7.
Sensitivity analysis of T1766C polymorphism and EH (TC vs. TT).
| Study Omitted | Estimate | 95%Confidence Interval |
|---|
| He (2007) [33] | 0.80 | 0.60–1.1 |
| He et.al.(2007) [34] | 0.88 | 0.67–1.1 |
| Benedicta et.al.(2002) [35] | 0.64 | 0.47–0.86 |
| Kato et.al.(2000) [36] | 0.82 | 0.65–1.0 |
| Combined | 0.79 | 0.62–0.99 |