Metabolic Degradation of 1,4-dichloronaphthalene by Pseudomonas sp. HY
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals and Reagents
2.2. Preparation of Bacterial Strain Pseudomonas sp. HY
2.3. Degradation Experiments
2.4. 1,4-DCN Analysis
2.5. Metabolite Analysis
3. Results and Discussion
3.1. Metabolic Degradation of 1,4-DCN
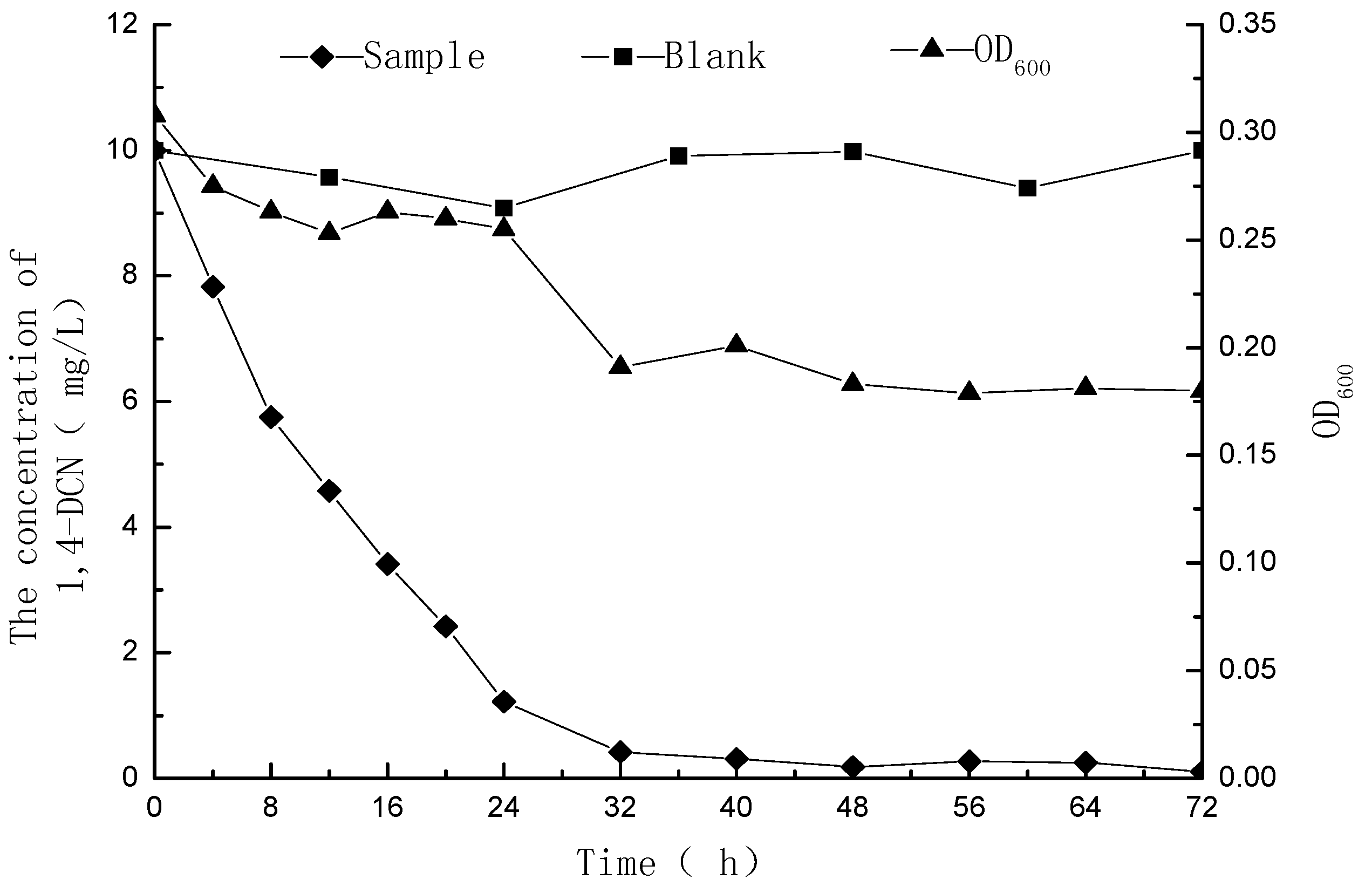

3.2. Metabolites of 1,4-DCN
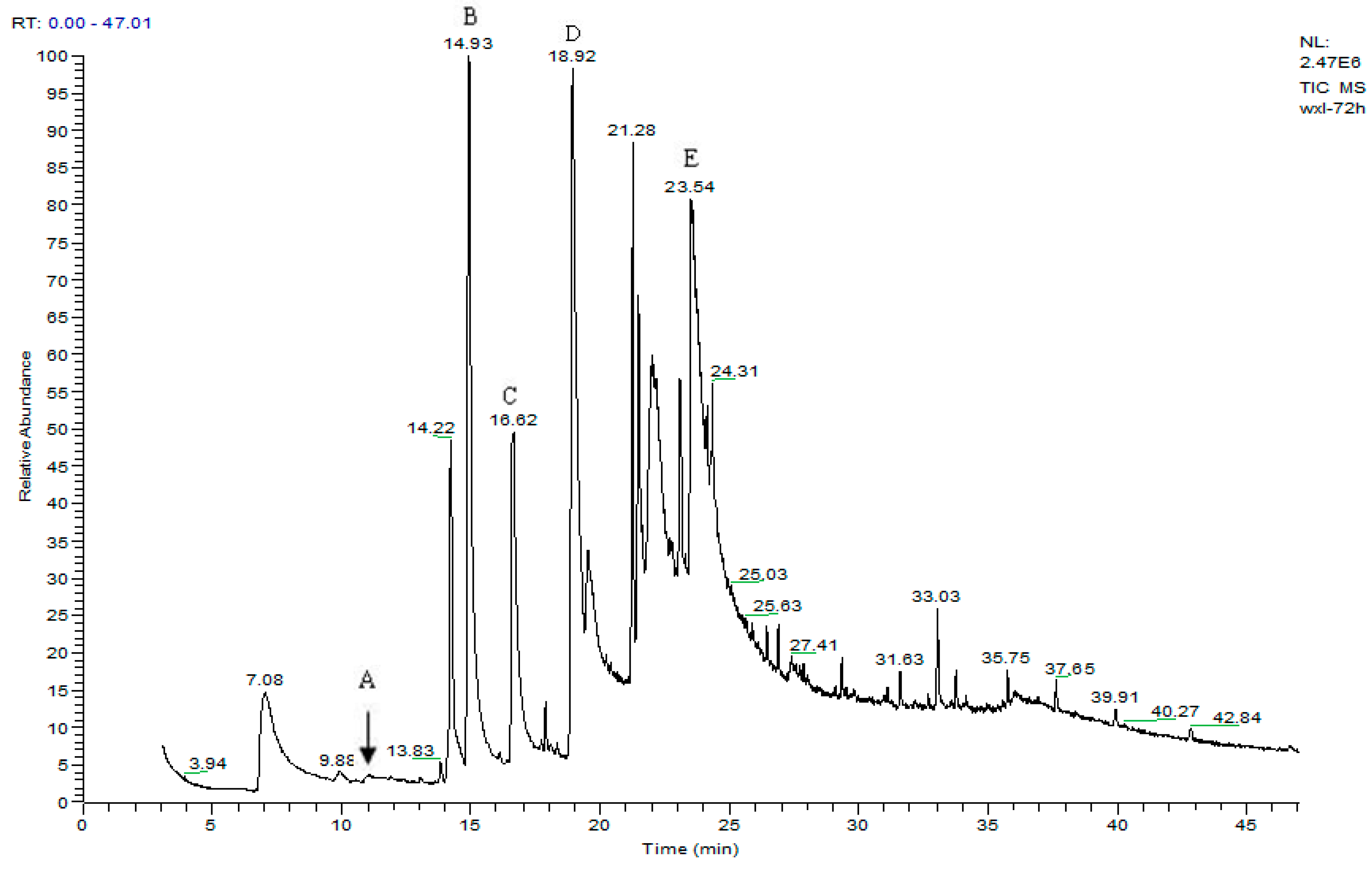
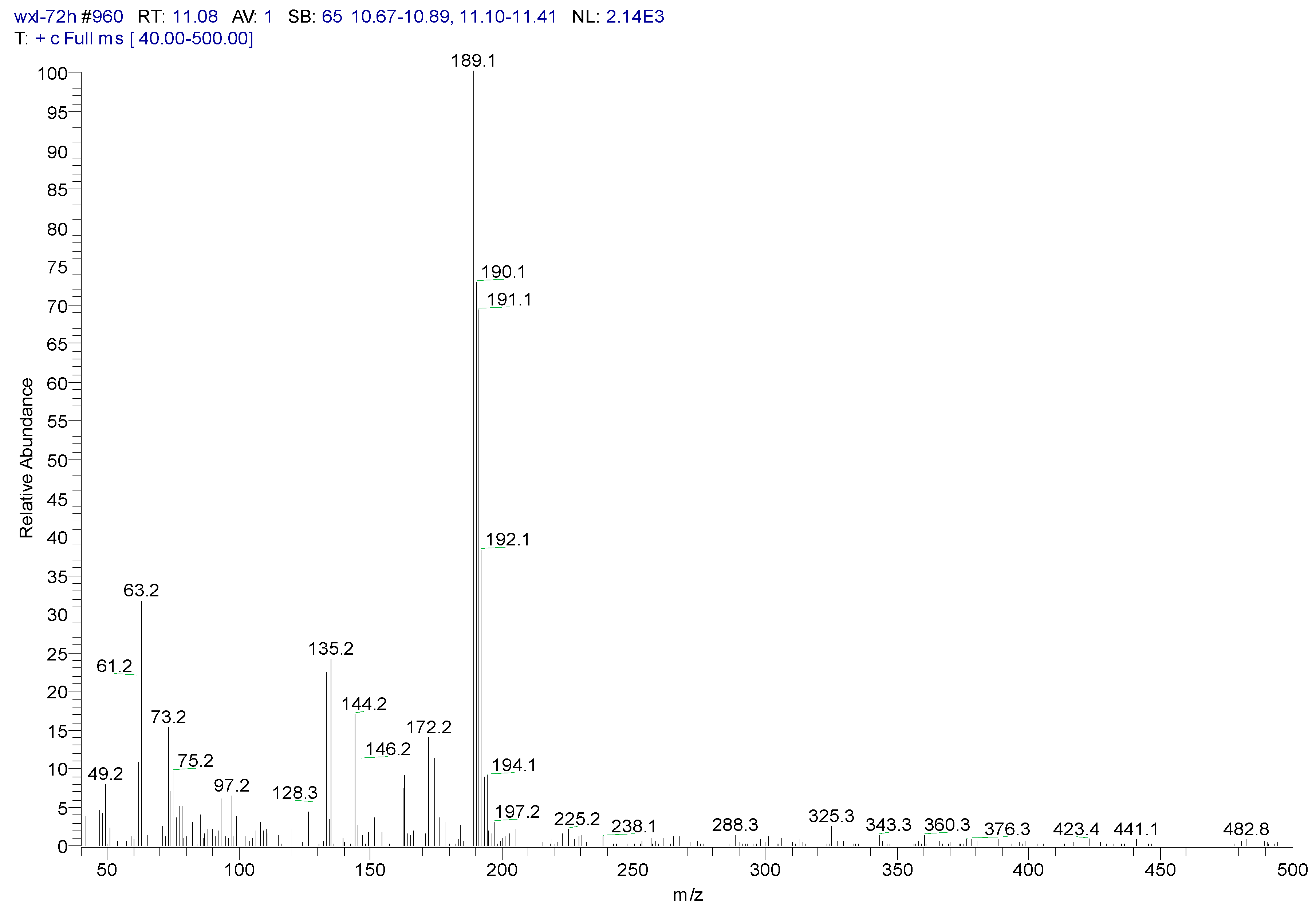
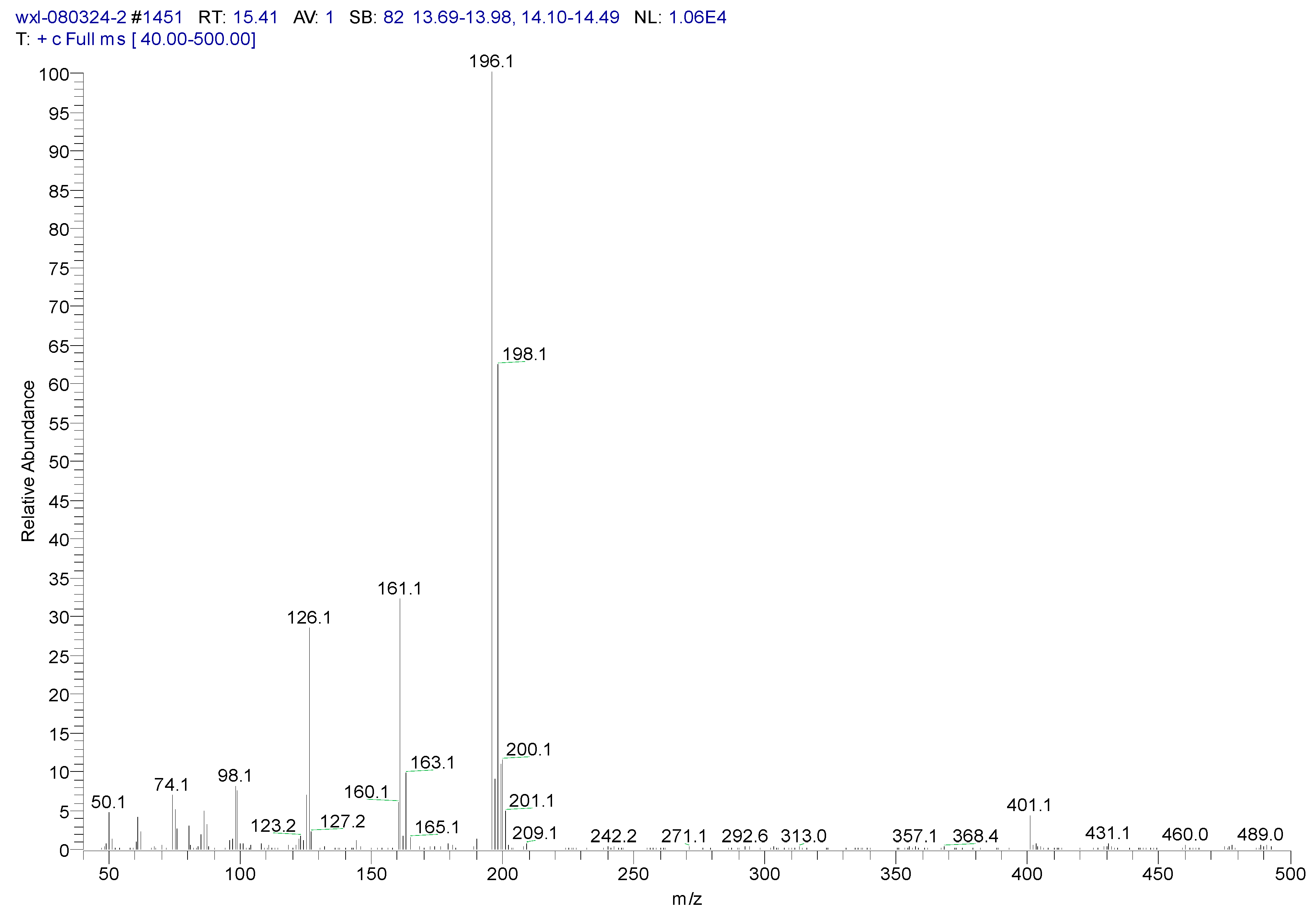

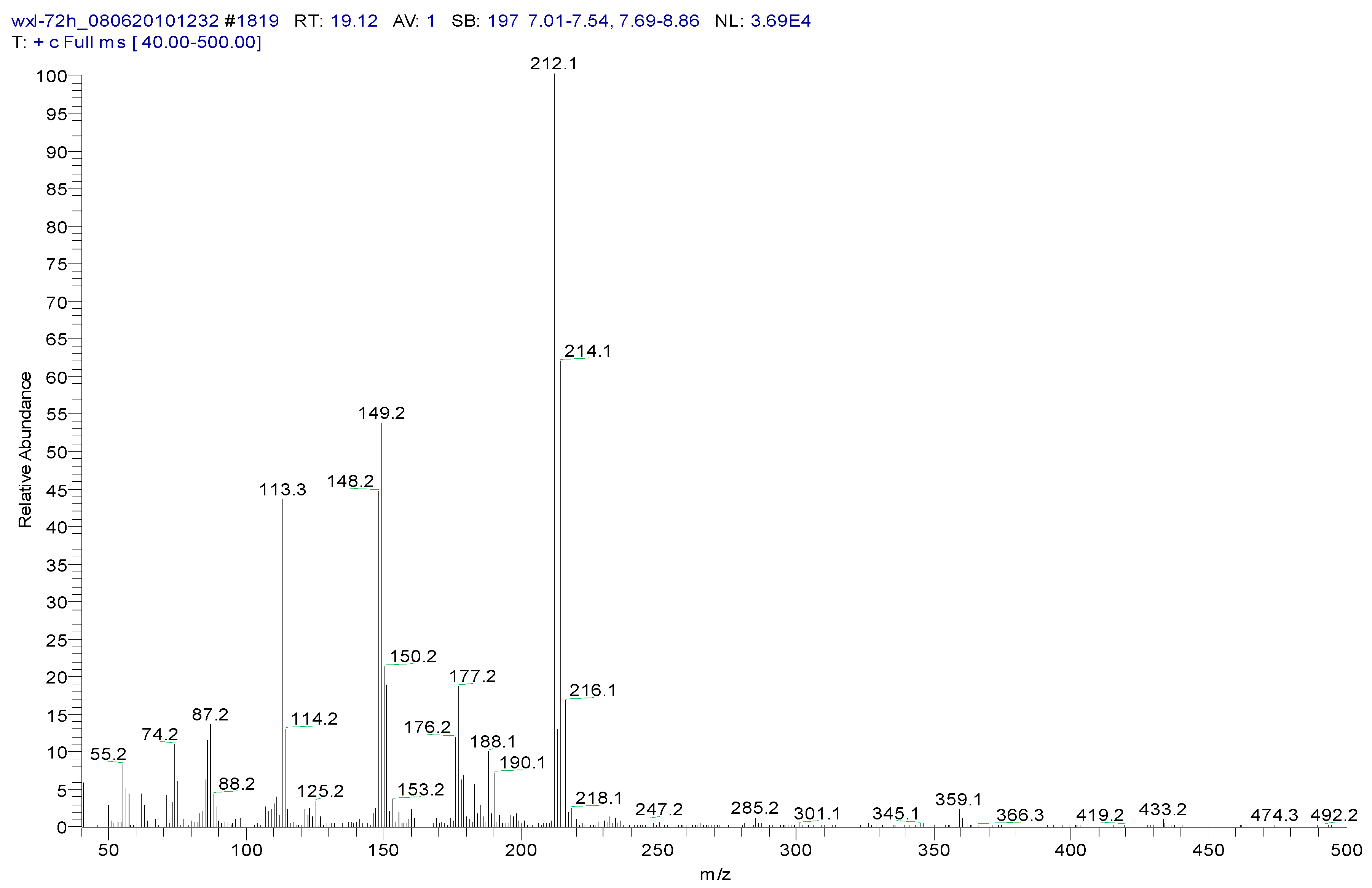
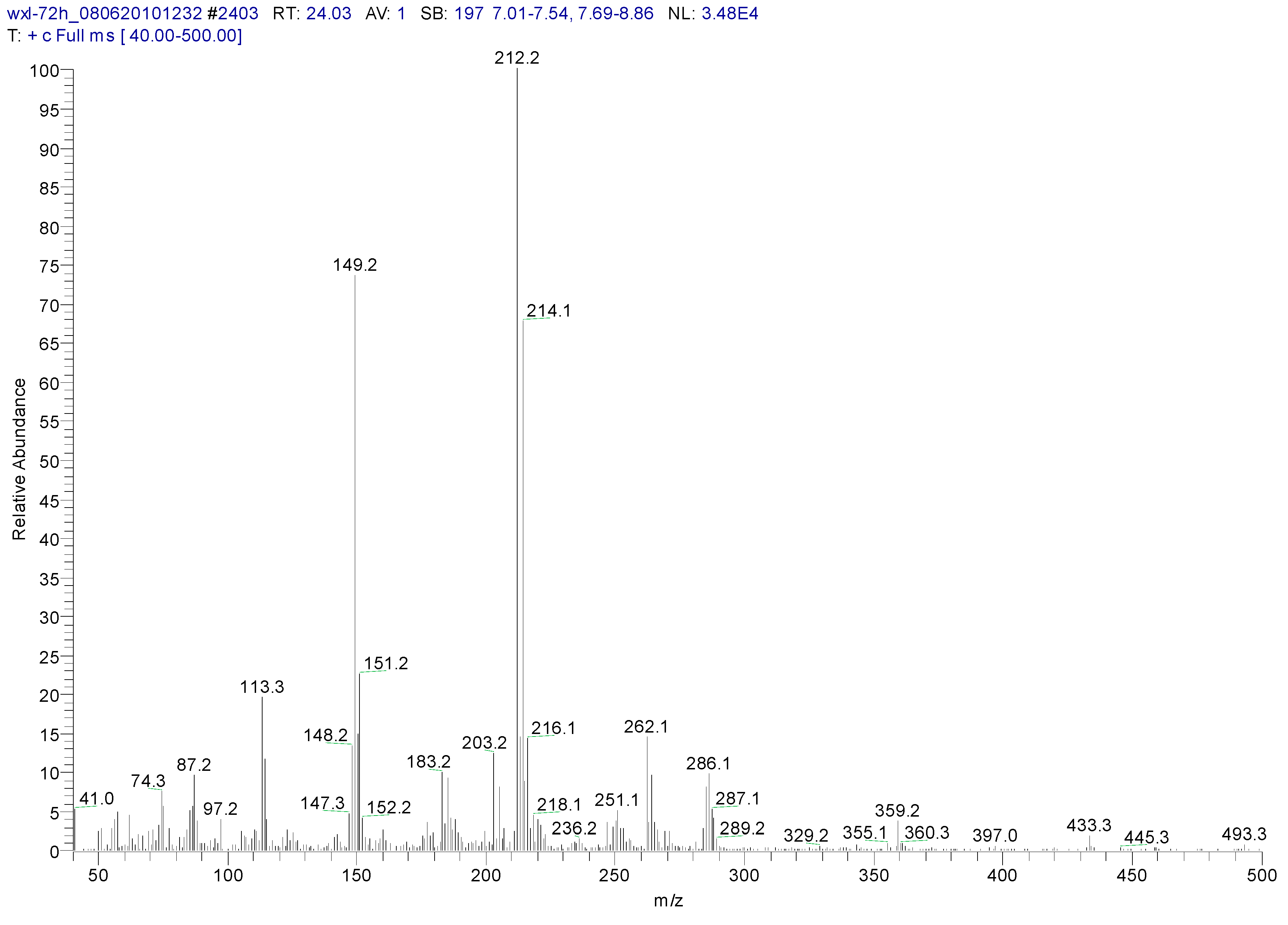

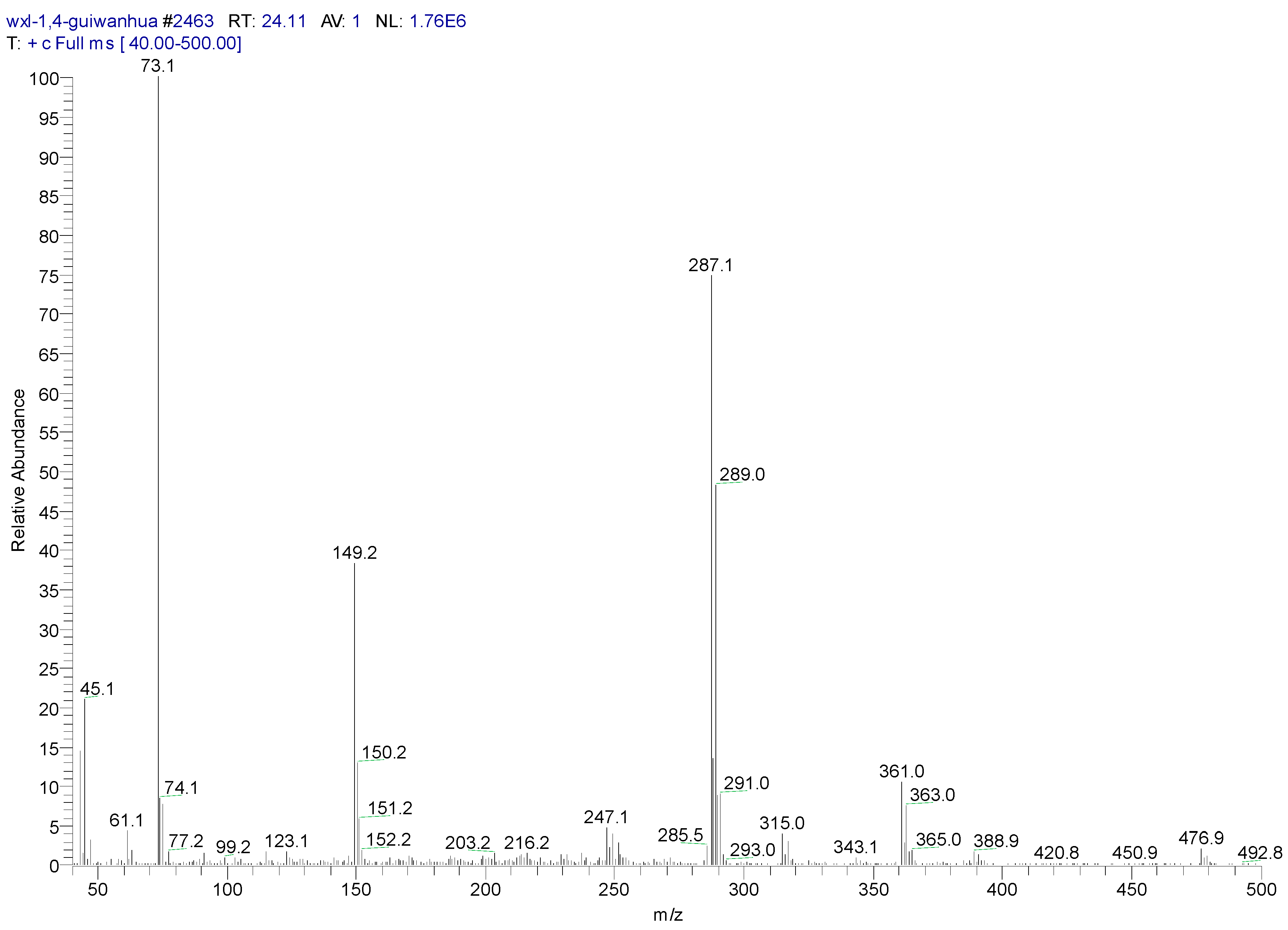
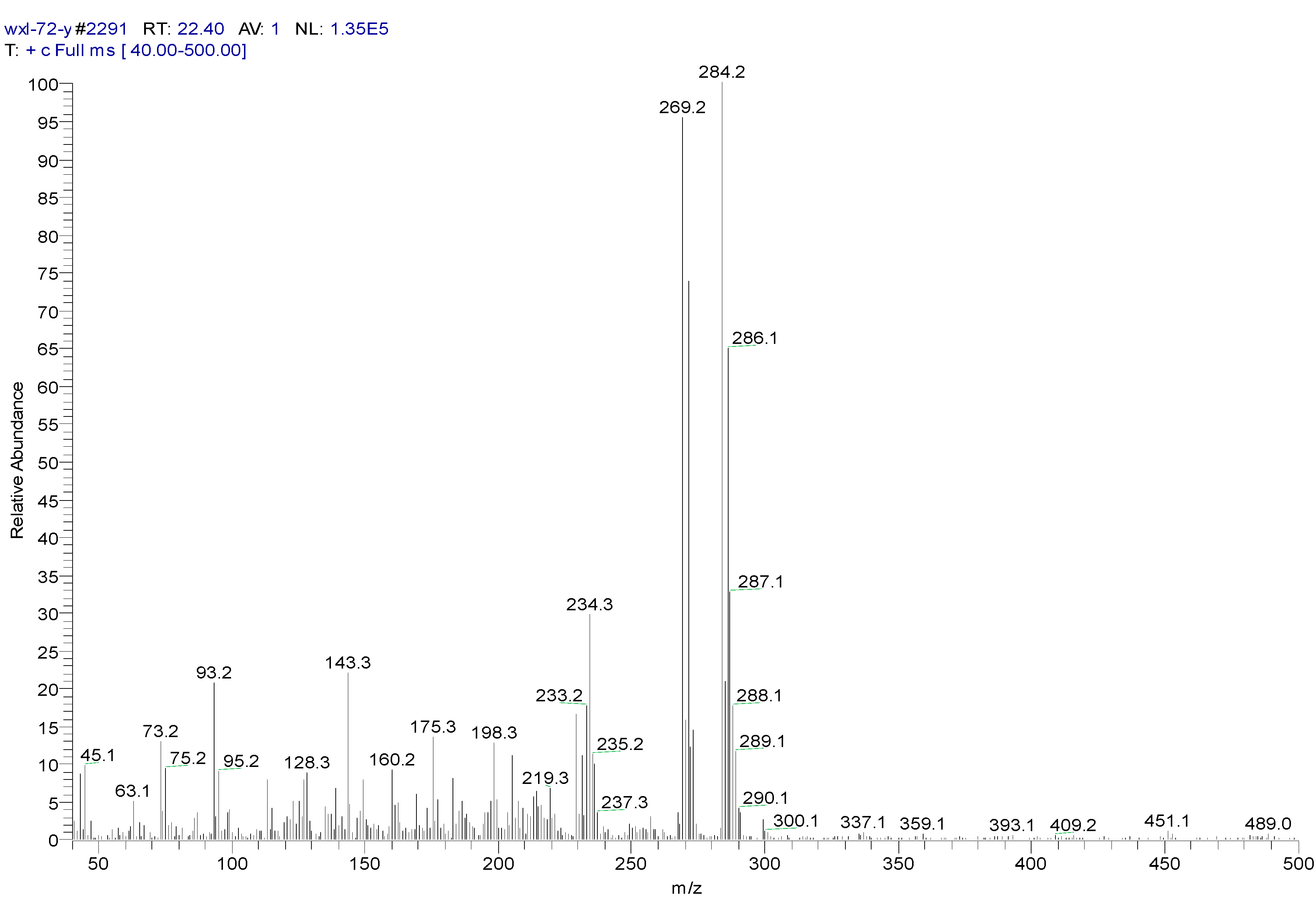

| Peak Time (min) | Peak | Clusters | m/z | Molecular Formula |
|---|---|---|---|---|
| 22.4 | ||||
| a | 284 | C13H14OSiCl2(M) | ||
| b | 269 | 284-15 | C12H11OSiCl2(M-CH3) | |
| c | 233 | 269-36 | C12H10OSiCl (M-CH3-HCl) | |
| d | 219 | 234-15 | C11H8OSiCl(M-CH3-Cl- CH3) | |
| e | 198 | 234-36 | C12H10OSi(M-CH3-Cl-HCl) | |
| f | 175 | 219-44 | C9H4SiCl(M-(CH3)2-Cl- CH3CHO) | |
| g | 73 | -O-Si-(CH3)3 | ||
| 26.44 | ||||
| a | 372 | C16H22O2Si2Cl2(M) | ||
| b | 357 | 372-15 | C15H19O2Si2Cl2(M-CH3) | |
| c | 284 | 372-16-72 | C13H13OSiCl2 (M-O-Si-(CH3)3) |
3.3. Metabolic Pathway of 1,4-DCN
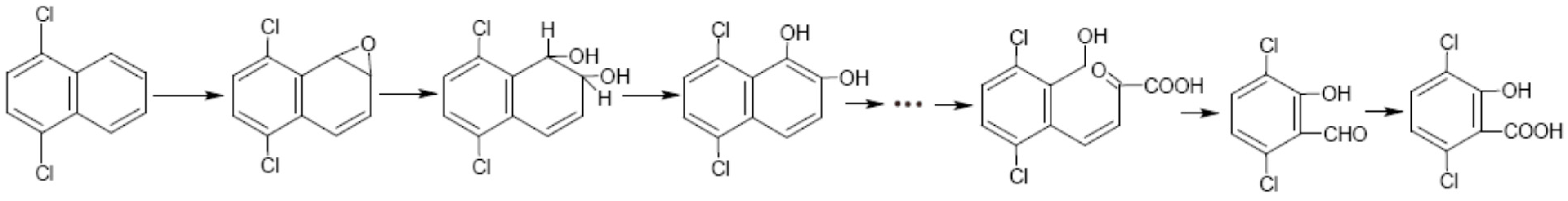
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jerzy, F. Polychlorinated naphthalenes: An environmental update. Environ. Pollut. 1998, 101, 77–90. [Google Scholar]
- Meijer, S.N.; Harner, T.; Helm, P.A.; Halshall, C.J.; Johnston, A.E.; Jones, K.C. Polychlorinated naphthalenes in U.K. soils: Time trends, markers of source, and equilibrium status. Environ. Sci. Technol. 2001, 35, 4205–4213. [Google Scholar] [CrossRef] [PubMed]
- Howe, P.D.; Melber, C. Chlorinated Naphthalenes; World Health Organization: Geneva, Switzerland, 2011; Available online: http://www.who.int/ipcs/publications/cicad/en/cicad34.pdf (accessed on 21 August 2015).
- Pan, J.; Yang, Y.-L.; Xu, Q.; Chen, D.-Z.; Xi, D.-L. PCBs, PCNs and PBDEs in sediment and mussels from Qingdao coastal sea in the frame of current circulation and influence of sewage sludge. Chemosphere 2007, 66, 1971–1982. [Google Scholar] [CrossRef] [PubMed]
- Eljarrat, E.; Barceó, D. Priority lists for persistent organic pollutants and emerging contaminants based on their relative toxic potency in environmental samples. Trends Anal. Chem. 2003, 22, 655–665. [Google Scholar] [CrossRef]
- Li, N.; Ma, M.; Wang, Z.; Satyanarayanan, S.K. In vitro assay for human thyroid hormone receptor β agonist and antagonist effects of individual polychlorinated naphthalenes and Halowax mixtures. Chin. Sci. Bull. 2011, 56, 508–513. [Google Scholar] [CrossRef]
- Morris, C.M.; Barnsley, E.A. The cometabolism of 1- and 2-chloronaphalene by pseudomnads. Can. J. Microb. 1982, 28, 73–79. [Google Scholar] [CrossRef]
- Mori, T.; Kitano, S.; Kondo, R. Metabolism of chloronaphthalenes and polycyclic aromatic hydrocarbons by the white-rot fungus Phlebia lindtneri. Appl. Microbiol. Biotechnol. 2003, 61, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Durham, D.R.; Stewart, D.B. Recruitment of naphthalene dissimilatory enzymes for the oxidation of 1,4-dichloronaphthalen to 3,6-dichloro-salicylate, a precursor for the herbicide dicamba. J. Bacteriol. 1987, 169, 2889–2892. [Google Scholar] [PubMed]
- Mori, T.; Nakamura, K.; Kondo, R. Fungal hydroxylation of polychlorinated naphthalenes with chlorine migration by wood rotting fungi. Chemosphere 2009, 77, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lei, Z.; He, W.; Song, Y. The metabolism experiment of 1-chloronaphthalene and 2-chloronaphthalene. In Proceedings of the 2012 International Conference on Biomedical Engineering and Biotechnology, Macau, Macau, 28–30 May 2012; Volume 3, pp. 1770–1777.
- Tang, H.L.; Chen, H. Nitrification at full-scale municipal wastewater treatment plants: Evaluation of inhibition and bioaugmentation of nitrifiers. Bioresour. Technol. 2015, 190, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Hao, O.J. Cometabolic metabolism of chlorophenols by Acinetobacter species. Water Res. 1999, 33, 562–574. [Google Scholar] [CrossRef]
- Li, C.; Lan, Y.; Zhang, J.; Chen, Z.; Tang, D.; Xu, H. Biodegradation of methidathion by Serratia sp. in pure cultures using an orthogonal experiment design, and its application in detoxification of the insecticide on crops. Ann. Microbiol. 2013, 63, 451–459. [Google Scholar] [CrossRef]
- Peng, X.; Zhang, Z.; Luo, W.; Jia, X. Biodegradation of tetrabromobisphenol A by a novel Comamonas sp. strain, isolated from anaerobic sludge. Bioresour. Technol. 2013, 128, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.-G.; Han, B.; Duan, S.; Zhao, Z.; Wang, Y. Degradation of the endocrine-disrupting dimethyl phthalate carboxylic ester by Sphingomonas yanoikuyae DOS01 isolated from the South China Sea and the biochemical pathway. Int. Biodeterior. Biodegrad. 2009, 63, 450–455. [Google Scholar] [CrossRef]
- Kim, S.; Picardal, F.W. A novel bacterium that utilizes monochlorobiphenyls and 4-chlorobenzoate as growth substrates. FEMS Microbiol. Lett. 2000, 185, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Trìska, J.; Kuncovà, G.; Mackovà, M.; Nováková, H.; Paasivirta, J.; Lahtiperä, M.; Vrchotová, N. Isolation and identification of intermediates from metabolism of low chlorinated biphenyl (Delor103). Chemosphere 2004, 54, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Bosch, R.; García-Valdés, E.; Moore, E.R.B. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-metabolism lower pathway from Pseudomonas stutzeri AN10. Gene 2000, 245, 65–74. [Google Scholar] [CrossRef]
- Commandeur, L.C.M.; May, R.J.; Mokross, H.; Bedard, D.L.; Reineke, W.; Govers, H.A.J.; Parsons, J.R. Aerobic degradation of polychlorinated biphenyls by Alcaligenes sp. JB1: Metabolites and enzymes. Metabolism 1996, 7, 435–443. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Wu, X.; Song, Y.; Ren, W.; Tang, H.L. Metabolic Degradation of 1,4-dichloronaphthalene by Pseudomonas sp. HY. Int. J. Environ. Res. Public Health 2015, 12, 10300-10313. https://doi.org/10.3390/ijerph120910300
Yu J, Wu X, Song Y, Ren W, Tang HL. Metabolic Degradation of 1,4-dichloronaphthalene by Pseudomonas sp. HY. International Journal of Environmental Research and Public Health. 2015; 12(9):10300-10313. https://doi.org/10.3390/ijerph120910300
Chicago/Turabian StyleYu, Jian, Xiaoli Wu, Youqun Song, Wenhui Ren, and Hao L. Tang. 2015. "Metabolic Degradation of 1,4-dichloronaphthalene by Pseudomonas sp. HY" International Journal of Environmental Research and Public Health 12, no. 9: 10300-10313. https://doi.org/10.3390/ijerph120910300





