Cadmium Modulates Biofilm Formation by Staphylococcus epidermidis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Cadmium
2.3. Bacterial Susceptibility to Cadmium
2.4. Quantification of Biofilm Formation
2.5. Determination of Bacterial Viability in Suspension and in the Biofilm
2.6. Bacterial Viability and Biofilm Thickness Determined by Confocal Laser Scanning Microscopy
2.7. Quantitative RT-PCR
| Gene | GenBank | Primer | Sequence | Product Size (pb) | References |
|---|---|---|---|---|---|
| hsp60 | AF029245 | Forward | 5’ GTTTTAGCACAATCAATGATTCAG 3’ | 491 | [18] |
| Reverse | 5’ GCATCGCCTTCTACTTCATCC 3’ | ||||
| tpi | AF269838 | Forward | 5’ CATCTGATAAACCTTCGACAGCTTT 3’ | 128 | [19] |
| Reverse | 5’ TGCTATCTTCAATCACGGTATGACA 3’ | ||||
| aap | AJ249487 | Forward | 5’ ATACAACTGGTGCAGATGGTTG 3’ | 400 | [20] |
| Reverse | 5’ GTAGCCGTCCAAGTTTTACCAG 3’ | ||||
| agrB | AF012132 | Forward | 5’ TTCGTTTAGGGATGCAGGTA 3’ | 141 | [21] |
| Reverse | 5’ ATGGCACACGTACAGAGGAT 3’ | ||||
| atlE | U71377 | Forward | 5’ TGTCCTGCTTTCACGTATGA 3’ | 139 | [21] |
| Reverse | 5’ AGAAACCTTAACCACGTAAA 3’ | ||||
| embP | AY101364.1 | Forward | 5’AGCGGTACAAATGTCAATATC 3’ | 455 | [22] |
| Reverse | 5’AGAAGTGCTCTAGCATCATCC 3’ | ||||
| icaA | U43366 | Forward | 5’ AACAAGTTGAAGGCATCTCC 3’ | 166 | [23] |
| Reverse | 5’ GATGCTTGTTTGATTCCCT 3’ | ||||
| icaB | U43366 | Forward | 5’ AATGGCTTAAAGCACACGAC 3’ | 144 | [21] |
| Reverse | 5’ TTTGTCCTTTCCGTAACAGT 3’ | ||||
| sarA | NC002976.3 | Forward | 5’ TGGTCACTTATGCTGACAGATT 3’ | 313 | [24] |
| Reverse | 5’ TTTGCTTCTGTGATACGGTG 3’ | ||||
| sepA | NC002976.3 | Forward | 5’ CGCACCAGACAACGCTGTA 3’ | 170 | [25] |
| Reverse | 5’ TCAATCGCACATGTAAATAACTTCC 3’ | ||||
| rsbU | NC002976 | Forward | 5’ TCTCTTCATACAGTCCAT 3’ | 172 | [26] |
| Reverse | 5’ ATAGGTTCAGGTATTCCA 3’ |
2.8. Statistical Analysis
3. Results
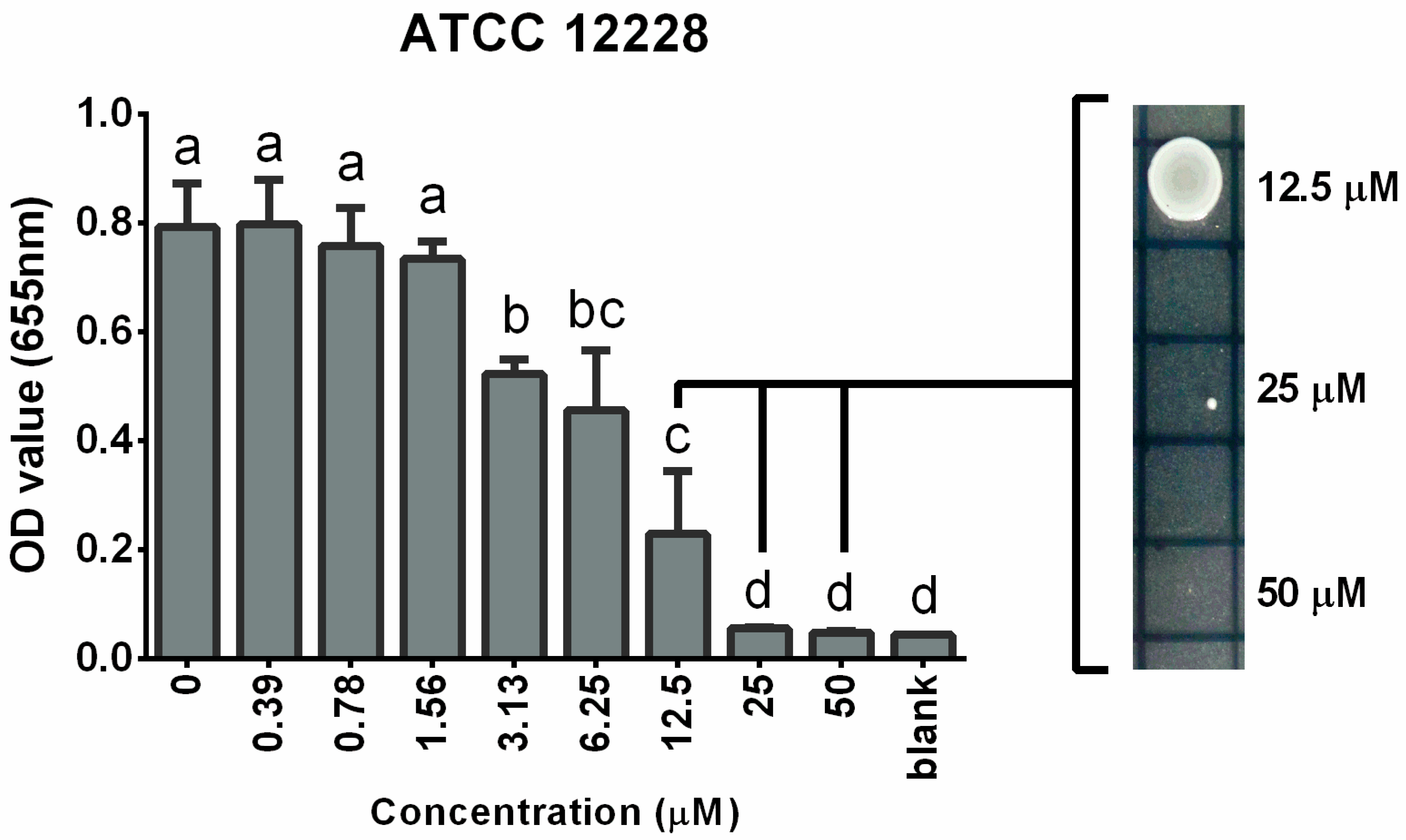
3.1. Bacteriostatic and Bactericidal Effects of Cadmium on S. epidermidis
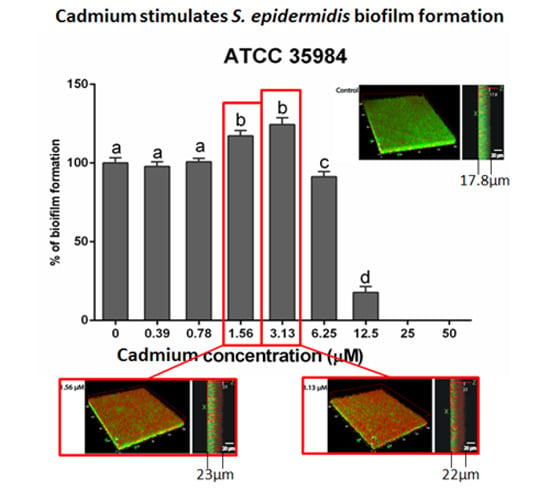
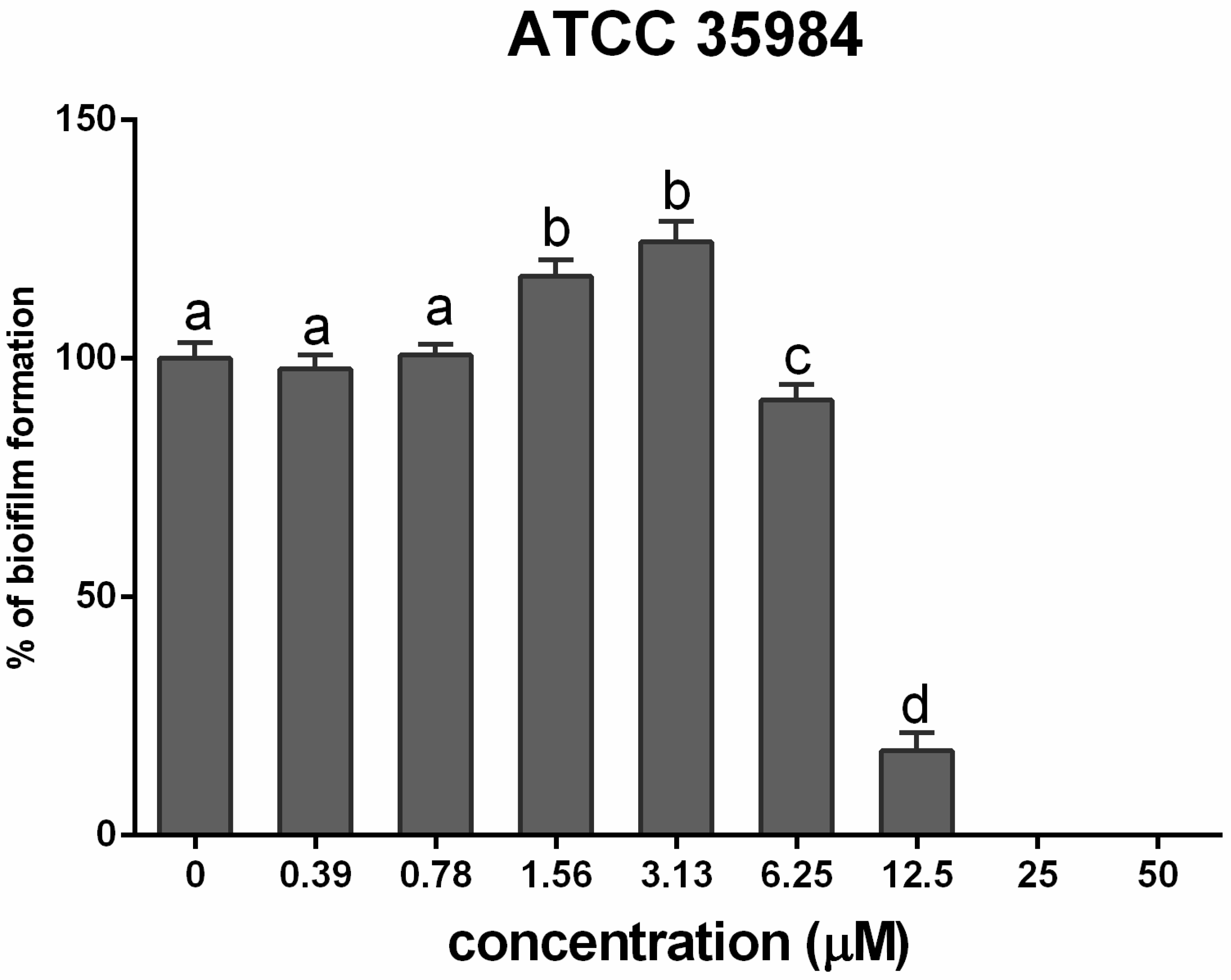
3.2. Cadmium Increases S. epidermidis Biomass
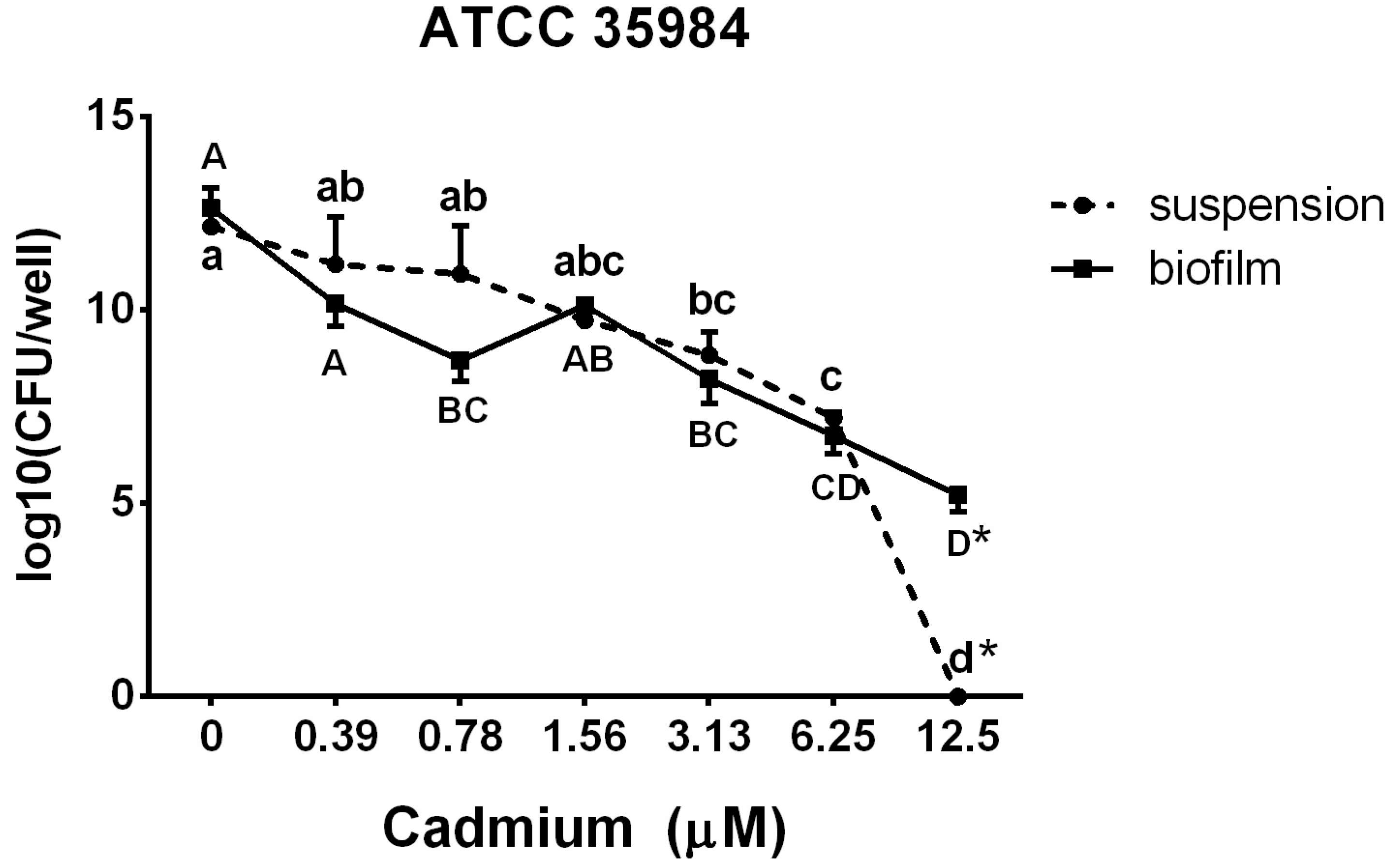
3.3. Viable Counts in Suspension and in the Biofilm
3.4. Bacterial Viability and Biofilm Architecture

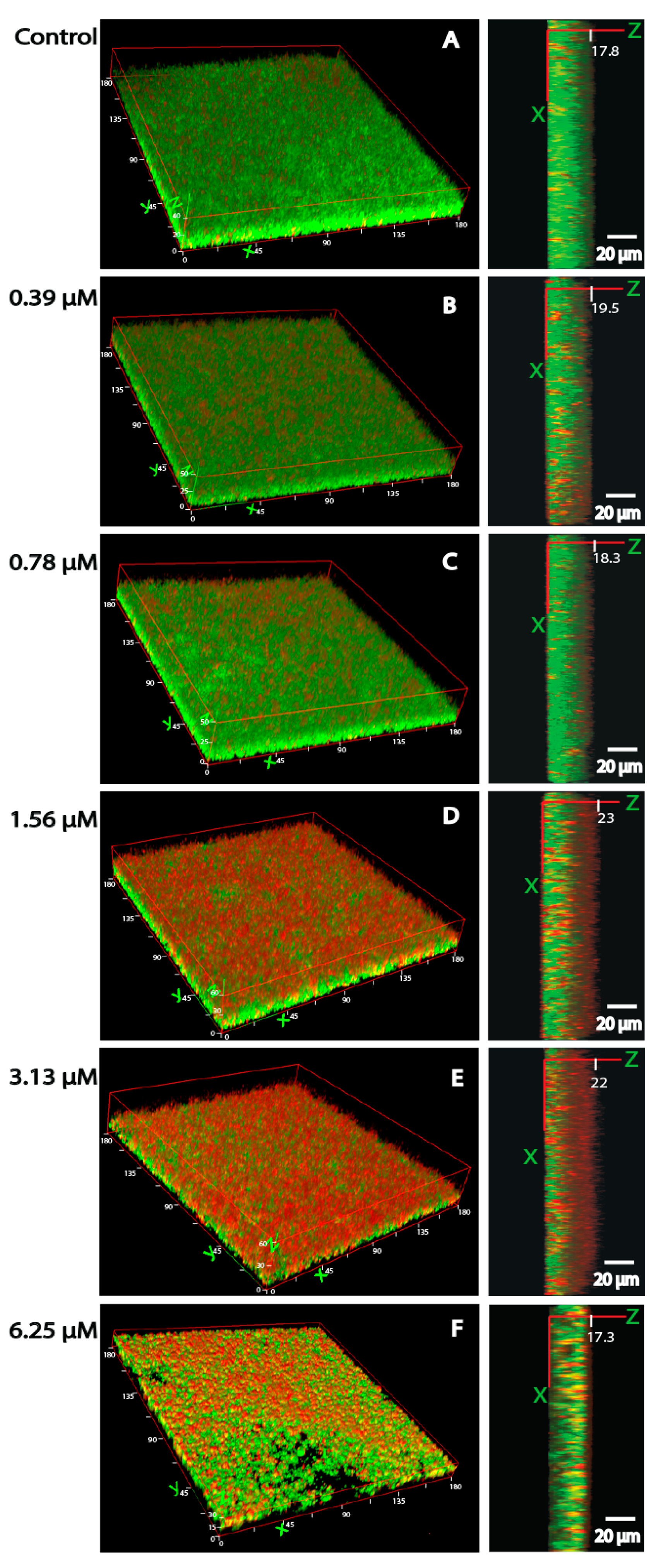
3.5. Gene Expression in Planktonic Cells and Mature Biofilms
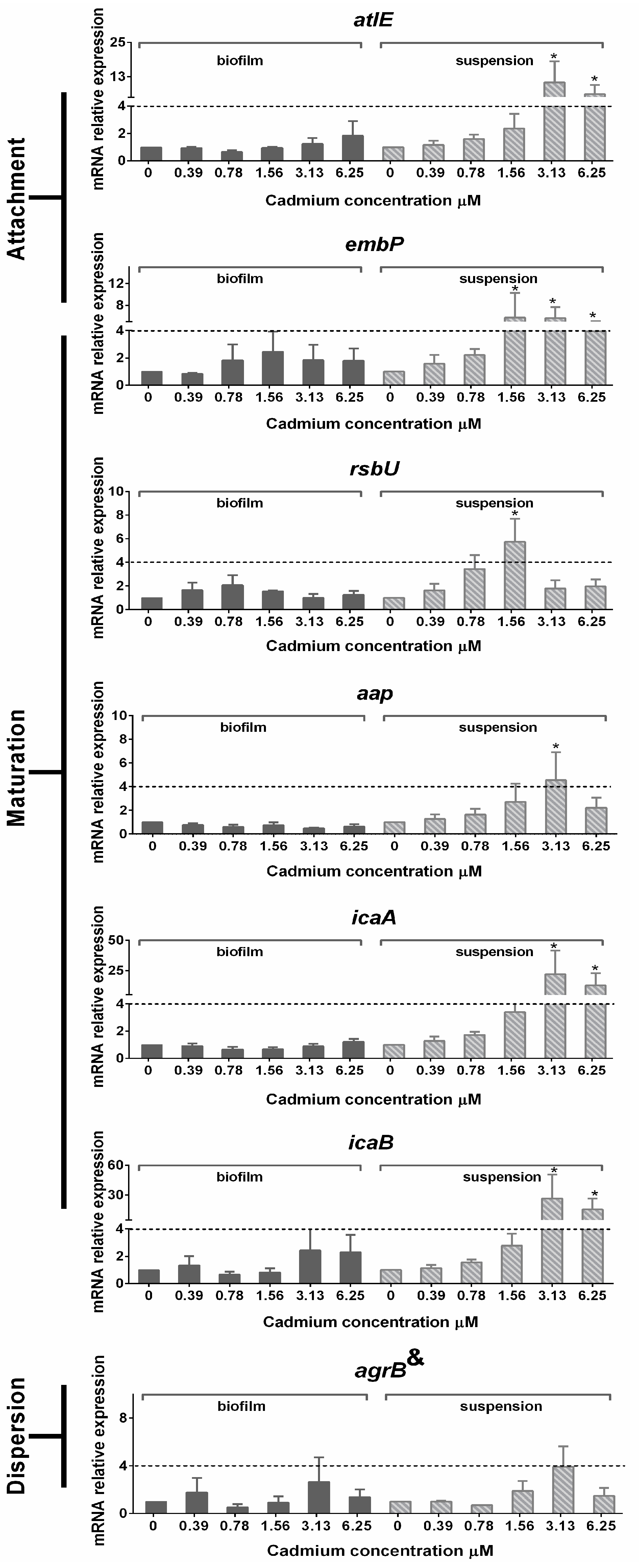
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgements
Author Contributions
Conflicts of Interest
References
- Jarup, L. Hazards of heavy metal contamination. Br. Med. Bull. 2003, 68, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xia, B.; Fan, C.; Zhao, P.; Shen, S. Human health risk from soil heavy metal contamination under different land uses near Dabaoshan mine, Southern China. Sci. Total Environ. 2012, 417–418, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Du, Y. Cadmium and its neurotoxic effects. Oxid. Med. Cell. Longev. 2013, 2013. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, J.; Zhao, B.; Xin, X.; Zhang, C.; Zhang, H. The influence of long-term fertilization on cadmium (Cd) accumulation in soil and its uptake by crops. Environ. Sci. Pollut. Res. Int. 2014, 21, 10377–10385. [Google Scholar] [CrossRef] [PubMed]
- JECFA. Cadmium. Evaluations of the Joint FAO/WHO Expert Committee on Food Additives (JECFA). Available online: http://apps.who.int/food-additives-contaminants-jecfa-database/PrintPreview.aspx?chemID=1376 (accessed on 2 January 2015).
- Perrin, C.; Briandet, R.; Jubelin, G.; Lejeune, P.; Mandrand-Berthelot, M.A.; Rodrigue, A.; Dorel, C. Nickel promotes biofilm formation by Escherichia coli K-12 strains that produce curli. Appl. Environ. Microbiol. 2009, 75, 1723–1733. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Nelis, H.J. In vitro and in vivo model systems to study microbial biofilm formation. J. Microbiol. Methods 2010, 83, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.D.; Olson, M.E. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Rabah, A.B.; Oyeleke, S.B.; Manga, S.B.; Hassan, L.G.; Ijah, U.J.J. Microbiological and physico-chemical assessment of soil contaminated with abattoir effluents in sokoto metroplis, Nigeria. Sci. World J. 2010, 5, 1–4. [Google Scholar]
- Ayed, L.; Chaieb, K.; Cheref, K.; Bakhrouf, A. Biodegradation and decolorization of triphenylmethane dyes by Staphylococcus epidermidis. Desalination 2010, 260, 137–146. [Google Scholar] [CrossRef]
- Cargill, J.S.; Upton, M. Low concentrations of vancomycin stimulate biofilm formation in some clinical isolates of Staphylococcus epidermidis. J. Clin. Pathol. 2009, 62, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Schue, M.; Fekete, A.; Ortet, P.; Brutesco, C.; Heulin, T.; Schmitt-Kopplin, P.; Achouak, W.; Santaella, C. Modulation of metabolism and switching to biofilm prevail over exopolysaccharide production in the response of Rhizobium alamii to cadmium. PLoS One 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; Deboy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef] [PubMed]
- Mertens, A.; Ghebermedhin, B. Genetic determinants and biofilm formation of clinical Staphylococcus epidermidis isolates from blood cultures and indwelling devises. Eur. J. Microbiol. Immunol. 2013, 3, 111–119. [Google Scholar] [CrossRef]
- Melchior, M.B.; Vaarkamp, H.; Fink-Gremmels, J. Biofilms: A role in recurrent mastitis infections? Vet. J. 2006, 171, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Santos, R.R.; Fink-Gremmels, J. Staphylococcus epidermidis biofilm quantification: Effect of different solvents and dyes. J. Microbiol. Methods 2014, 101, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Cabal, B.; Cafini, F.; Esteban-Tejeda, L.; Alou, L.; Bartolome, J.F.; Sevillano, D.; Lopez-Piriz, R.; Torrecillas, R.; Moya, J.S. Inhibitory effect on in vitro Streptococcus oralis biofilm of a soda-lime glass containing silver nanoparticles coating on titanium alloy. PLoS One 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.M.; Noble, L.; Kreiswirth, B.N.; Eisner, W.; McClements, W.; Jansen, K.U.; Anderson, A.S. Evaluation of a multilocus sequence typing system for Staphylococcus epidermidis. J. Med. Microbiol. 2003, 52, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Vandecasteele, S.J.; Peetermans, W.E.; R Merckx, R.; Rijnders, B.J.; Van Eldere, J. Reliability of the ica, aap and atlE genes in the discrimination between invasive, colonizing and contaminant Staphylococcus epidermidis isolates in the diagnosis of catheter-related infections. Clin. Microbiol. Infect. 2003, 9, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Vandecasteele, S.J.; Peetermans, W.E.; Merckx, R.; Van Eldere, J. Quantification of expression of Staphylococcus epidermidis housekeeping genes with taqman quantitative PCR during in vitro growth and under different conditions. J. Bacteriol. 2001, 183, 7094–7101. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.D.; Colton, E.; Ebert, M.; Anderson, J.M. Gene expression during S. epidermidis biofilm formation on biomaterials. J. Biomed. Mater. Res. A 2012, 100, 2863–2869. [Google Scholar] [CrossRef] [PubMed]
- Mekni, M.A.; Bouchami, O.; Achour, W.; Ben Hassen, A. Strong biofilm production but not adhesion virulence factors can discriminate between invasive and commensal Staphylococcus epidermidis strains. APMIS 2012, 120, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Tormo, M.A.; Marti, M.; Valle, J.; Manna, A.C.; Cheung, A.L.; Lasa, I.; Penades, J.R. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 2005, 187, 2348–2356. [Google Scholar] [CrossRef] [PubMed]
- Frebourg, N.B.; Lefebvre, S.; Baert, S.; Lemeland, J.F. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 2000, 38, 877–880. [Google Scholar] [PubMed]
- Christner, M.; Heinze, C.; Busch, M.; Franke, G.; Hentschke, M.; Bayard Duhring, S.; Buttner, H.; Kotasinska, M.; Wischnewski, V.; Kroll, G.; et al. sarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Mol. Microbiol. 2012, 86, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, J.K.; Bartscht, K.; Sabottke, A.; Rohde, H.; Feucht, H.H.; Mack, D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: Differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 2001, 183, 2624–2633. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Campoccia, D.; Gamberini, S.; Donati, M.E.; Pirini, V.; Visai, L.; Speziale, P.; Montanaro, L. Antibiotic resistance in exopolysaccharide-forming Staphylococcus epidermidis clinical isolates from orthopaedic implant infections. Biomaterials 2005, 26, 6530–6535. [Google Scholar] [CrossRef] [PubMed]
- Shaivastave, A.; Singh, V.; Jadon, S.; Bhadauria, S. Heavy metal tolerance of three different bacteria isolated from industrial effluent. Int. J. Pharm. Bio-Sci. 2013, 2, 137–147. [Google Scholar]
- AbubAkr, S.; Crupper, S.S. Prevalence of cadmium resistance in Stahphylococcus sciuri isolated from the Gray Treefrog, Hyla chrysoscelis (Anura: Hylidae). Phyllomedusa 2010, 9, 141–146. [Google Scholar] [CrossRef]
- Teitzel, G.M.; Parsek, M.R. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2003, 69, 2313–2320. [Google Scholar] [CrossRef] [PubMed]
- Delumeau, O.; Dutta, S.; Brigulla, M.; Kuhnke, G.; Hardwick, S.W.; Volker, U.; Yudkin, M.D.; Lewis, R.J. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J. Biol. Chem. 2004, 279, 40927–40937. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.J.; Henderson, B.; Sharp, L.J.; Nair, S.P. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 2002, 70, 6805–6810. [Google Scholar] [CrossRef] [PubMed]
- Gerke, C.; Kraft, A.; Sussmuth, R.; Schweitzer, O.; Gotz, F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 1998, 273, 18586–18593. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Xu, T.; Zhu, T.; Lou, Q.; Wang, X.; Wu, Y.; Huang, R.; Liu, J.; Liu, H.; Yu, F.; et al. Monoclonal antibodies against accumulation-associated protein affect EPS biosynthesis and enhance bacterial accumulation of Staphylococcus epidermidis. PLoS One 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Yang, L.; Parsons, C.; Findlay, V.J.; Molin, S.; Qin, Z. Staphylococcus epidermidis recovered from indwelling catheters exhibit enhanced biofilm dispersal and “self-renewal” through downregulation of agr. BMC Microbiol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Dieppois, G.; Ducret, V.; Caille, O.; Perron, K. The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS One 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Pintens, V.; Massonet, C.; Merckx, R.; Vandecasteele, S.; Peetermans, W.E.; Knobloch, J.K.; Van Eldere, J. The role of sigmaB in persistence of Staphylococcus epidermidis foreign body infection. Microbiology 2008, 154, 2827–2836. [Google Scholar] [CrossRef] [PubMed]
- Waite, R.D.; Papakonstantinopoulou, A.; Littler, E.; Curtis, M.A. Transcriptome analysis of Pseudomonas aeruginosa growth: Comparison of gene expression in planktonic cultures and developing and mature biofilms. J. Bacteriol. 2005, 187, 6571–6576. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Santos, R.R.; Fink-Gremmels, J. Cadmium Modulates Biofilm Formation by Staphylococcus epidermidis. Int. J. Environ. Res. Public Health 2015, 12, 2878-2894. https://doi.org/10.3390/ijerph120302878
Wu X, Santos RR, Fink-Gremmels J. Cadmium Modulates Biofilm Formation by Staphylococcus epidermidis. International Journal of Environmental Research and Public Health. 2015; 12(3):2878-2894. https://doi.org/10.3390/ijerph120302878
Chicago/Turabian StyleWu, Xueqing, Regiane R. Santos, and Johanna Fink-Gremmels. 2015. "Cadmium Modulates Biofilm Formation by Staphylococcus epidermidis" International Journal of Environmental Research and Public Health 12, no. 3: 2878-2894. https://doi.org/10.3390/ijerph120302878