Inhibition of Virulence Gene Expression in Staphylococcus aureus by Novel Depsipeptides from a Marine Photobacterium
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of QS Inhibitors from Photobacterium sp.

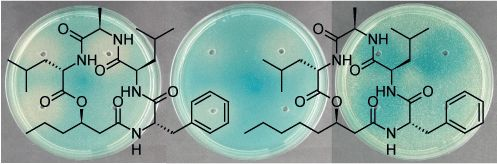
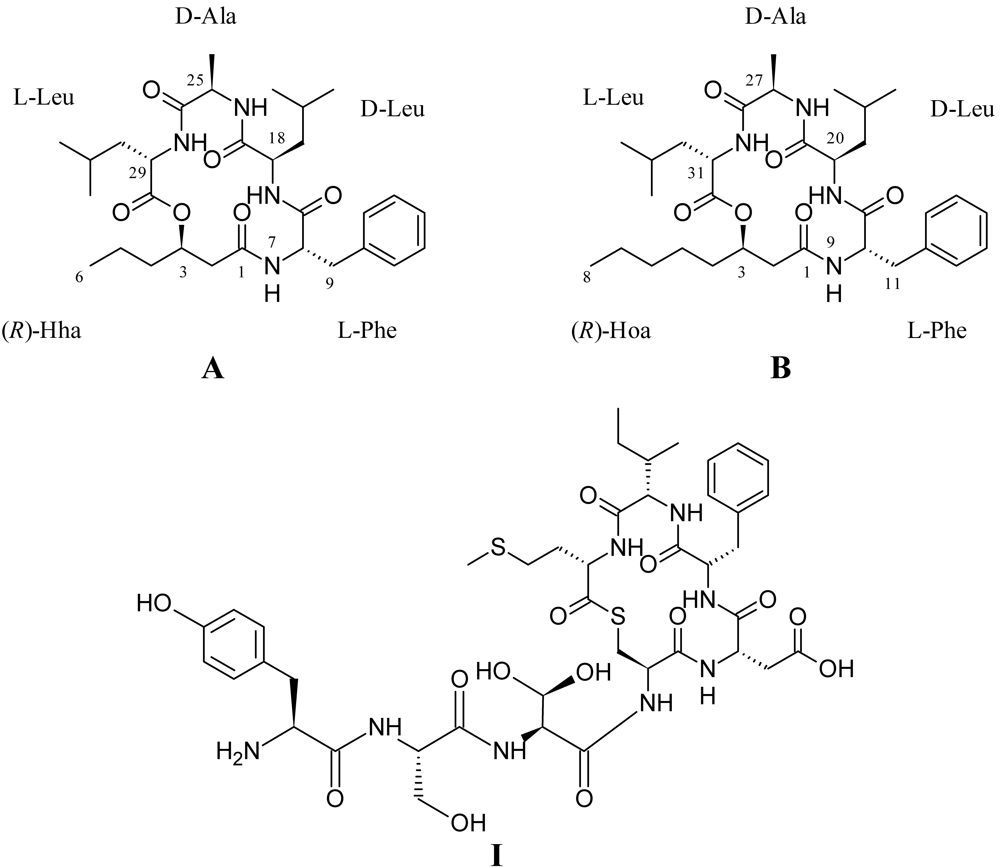
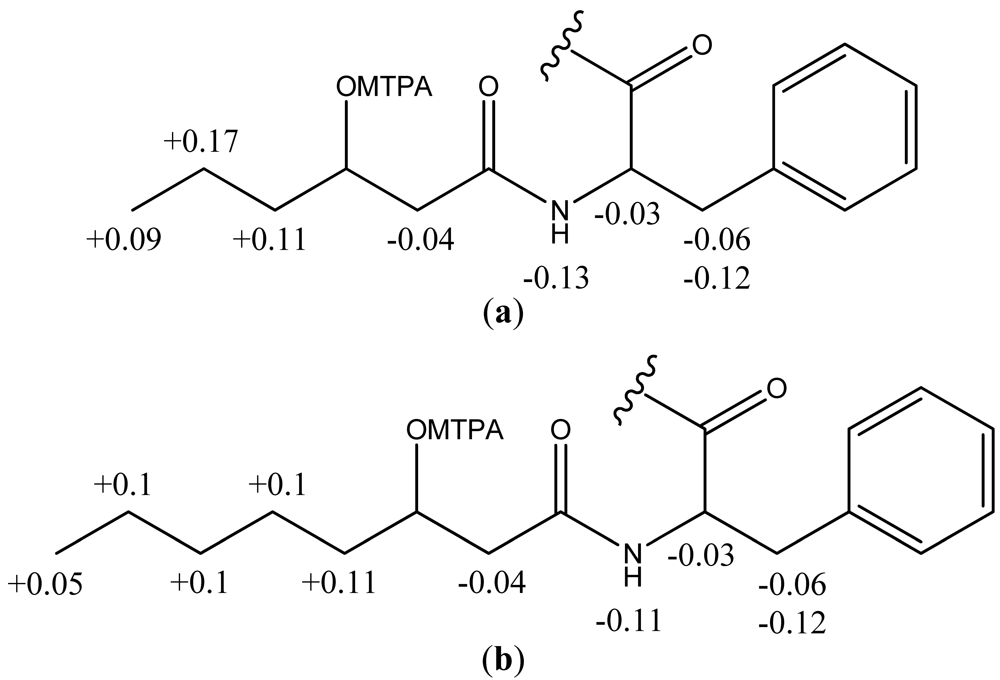
2.2. Structural Elucidation of the Solonamides
2.3. Production of Solonamides by Related Photobacterium Strains
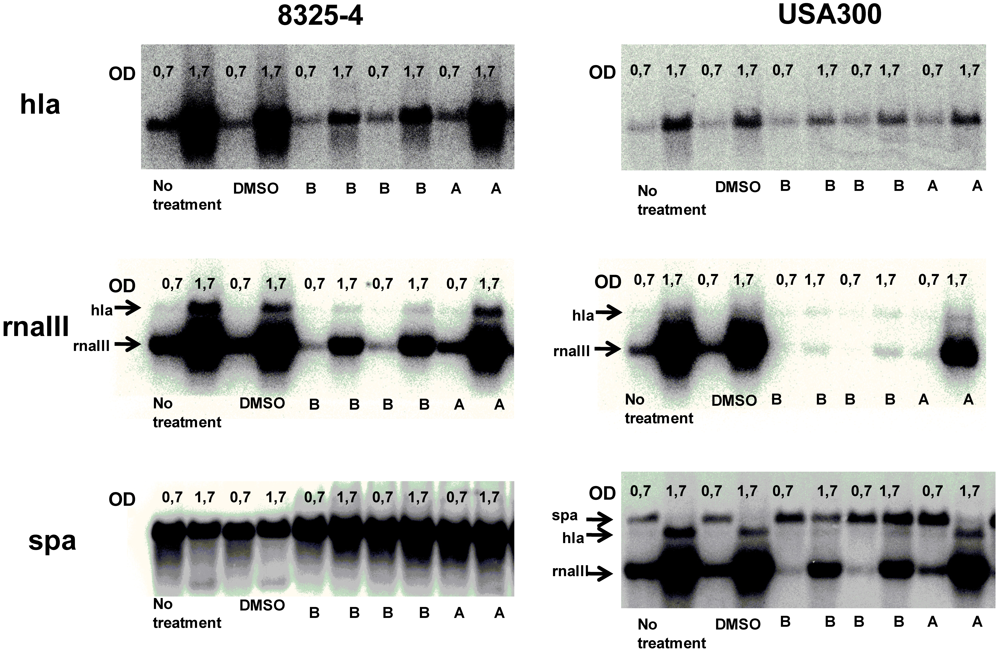
2.4. Solonamides Interfere with agr
3. Experimental Section
3.1. Isolation and Identification of Strain S2753
3.2. Initial Screening for Anti-Virulence Compounds
3.3. Isolation and Structural Elucidation of Solonamide A and B
| A | B | |||||||
|---|---|---|---|---|---|---|---|---|
| Atom | δC (ppm) | δH (ppm)(multiplicity, J (Hz)) | HMBC | Atom | δC (ppm) | δH (ppm)(multiplicity, J (Hz)) | HMBC | |
| Hha/Hoa | 1 | 170.6 | - | 1 | 170.4 | - | - | |
| 2 | 40.7 | 2.67 (1H, dd, 13.5, 3.8) 2.11 (1H, dd, 13.5, 10.1) | 1,3,4 | 2 | 40.5 | 2.69 (1H, dd, 13.5, 3.7) 2.11 (1H, dd, 13.5, 10.3) | 1,3,4 | |
| 3 | 72.1 | 5.14 (1H, m) | 1,5 | 3 | 72.0 | 5.13 (1H, m) | 1,4,5,36 | |
| 4 | 36.2 | 1.62 (1H, m) 1.45 (1H, m) | 2,3,6 | 4 | 33.8 | 1.65 (1H, m) 1.45 (1H, m) | 5,6 | |
| 5 | 17.7 | ~1.22 (1H, m) | 3,4 | 5 | 23.8 | ~1.20 (2H, m) | 6,7 | |
| 6 | 13.8 | 0.85 (3H, t, 7) | 4,5 | 6 | 30.8 | ~1.22 (2H, m) | ND | |
| 7 | 21.8 | 1.25 (2H, m) | ND | |||||
| 8 | 13.6 | 0.84 (3H, m) | 6 | |||||
| L-Phe | 7-NH | - | 8.61 (1H, d, 3.2) | 1 | 9-NH | - | 8.59 (1H, d, 3.0) | 1,10,11,8 |
| 8-CHα | 55.9 | 4.26 (1H, ddd, 10, 6.1, 3.2) | 9,10,14 | 10-CHα | 55.6 | 4.26 (1H, ddd, 10, 6.1, 3.3) | 11,18 | |
| 9-CHβ,1 | 36.4 | 2.94 (1H, dd, 13.3, 6.1) 2.72 (1H, dd, 13.3, 10) | 8,10,11/15,14 | 11-CHβ,1 | 36.1 | 2.95 (1H, dd, 13.3, 6.1) 2.73 (1H, dd, 13.3, 10) | 10,12,13/17,18 | |
| 10 | 135.8 | - | - | 12 | 136.0 | - | - | |
| 11,15 | 129.1 | 7.18 (2H, m) | 9,13 | 13,17 | 128.7 | 7.20 (2H, m) | 11,15 | |
| 12,14 | 128.4 | 7.25 (2H, m) | 10 | 14,16 | 128.0 | 7.26 (2H, m) | 12,14/16 | |
| 13 | 126.5 | 7.18 (1H, m) | 11 | 15 | 126.2 | 7.20 (1H, m) | 13/17 | |
| 16-CO | 174.4 | - | - | 18-CO | 174.2 | - | - | |
| D-Leu | 17-NH | - | 8.63 (1H, d, 5.6) | 16,18,19 | 19-NH | - | 8.64 (1H, d, 5.6) | 18,20,21 |
| 18-CHα | 53.1 | 3.56 (1H, m) | ND | 20-CHα | 52.7 | 3.57 (1H, m) | 21,22,25 | |
| 19-CHβ,1 | 38.9 | 1.36 (1H, m) 1.29 (1H, m) | 18,21,22 | 21-CHβ,1 | 38.6 | 1.31–1.37 (2H, m) | 20,22,24 | |
| 20-CHγ | 23.5 | 0.97 (1H, m) | 21 | 22-CHγ | 23.3 | 0.99 (1H, m) | ND | |
| 21-CHδ,1 | 23.2 | 0.69 (3H, d, 6.6) | 19,20 | 23-CHδ,1 | 22.9 | 0.70 (3H, d, 6.6) | 21,22,24 | |
| 22-CHδ,1 | 20.7 | 0.52 (3H, d, 6.5) | 19,20 | 24-CHδ,1 | 20.4 | 0.53 (3H, d, 6.4) | 21,23 | |
| 23-CO | 171.8 | - | - | 25-CO | 171.6 | - | - | |
| D-Ala | 24-NH | - | 7.53 (1H, d, 8.9) | 27,29 | 26-NH | - | 7.48 (1H, d, 8.9) | 25,27,28 |
| 25-CHα | 48.2 | 4.18 (1H, m) | 26,27 | 27-CHα | 47.8 | 4.18 (1H, m) | 28,29 | |
| 26-CHβ,1 | 16.6 | 1.32 (3H, d, 7.4) | 27 | 28-CHβ,1 | 16.4 | 1.33 (3H, d, 7.3) | 27,29 | |
| 27-CO | 171.3 | - | - | 29-CO | 170.9 | - | - | |
| L-Leu | 28-NH | - | 7.08 (1H, d, 10) | 27,29 | 30-NH | - | 7.05 (1H, d, 10) | 29,31 |
| 29-CHα | 49.0 | 4.46 (1H, dt, 10, 4.4) | 30,31,34 | 31-CHα | 48.6 | 4.47 (1H, dt, 10, 4.3) | 32,33,29/36 | |
| 30-CHβ,1 | 39.3 | 1.63 (1H, m) 1.53 (1H, m) | 29,31,33,27/34 | 32-CHβ,1 | 39.0 | 1.64 (1H, m) 1.52 (1H, m) | 31,33,35,29/36 | |
| 31-CHγ | 23.9 | 1.50 (1H, m) | 29,30,33 | 33-CHγ | 23.8 | 1.50 (1H, m) | 32,34,35 | |
| 32-CHδ,1 | 23.2 | 0.86 (3H, d, 6.5) | 30,31,33 | 34-CHδ,1 | 23.0 | 0.86 (3H, d, 6.6) | 32,33,35 | |
| 33-CHδ,1 | 21.3 | 0.81 (3H, d, 6.5) | 30 | 35-CHδ,1 | 21.1 | 0.82 (3H, d, 6.4) | 32,33 | |
| 34-CO | 171.2 | - | - | 36-CO | 170.8 | - | - | |
3.4. LC-UV/MS Analyses of Related Photobacterium Strains
3.5. Northern Blot Analysis
4. Conclusions
Acknowledgments
- Samples Availability: Available from the authors.
Supplementary Files
References
- Berdy, J. Bioactive microbial metabolites—A personal view. J. Antibiot. 2005, 58, 1–26. [Google Scholar]
- Clardy, J.; Fischbach, M.A.; Walsh, C.T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24, 1541–1550. [Google Scholar]
- Das, S.; Lyla, P.S.; Khan, S.A. Marine microbial diversity and ecology: Importance and future perspectives. Curr. Sci. 2006, 90, 1325–1335. [Google Scholar]
- Gulder, T.A.M.; Moore, B.S. Chasing the treasures of the sea—Bacterial marine natural products. Curr. Opin. Microbiol. 2009, 12, 252–260. [Google Scholar]
- Rahman, H.; Austin, B.; Mitchell, W.J.; Morris, P.C.; Jamieson, D.J.; Adams, D.R.; Spragg, A.M.; Schweizer, M. Novel anti-infective compounds from marine bacteria. Mar. Drugs 2010, 8, 498–518. [Google Scholar]
- de Carvalho, C.C.C.R.; Fernandes, P. Production of metabolites as bacterial responses to the marine environment. Mar. Drugs 2010, 8, 705–727. [Google Scholar]
- Long, R.A.; Azam, F. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 2001, 67, 4975–4983. [Google Scholar]
- Wright, G.D.; Sutherland, A.D. New strategies for combating multidrug-resistant bacteria. Trends Mol. Med. 2007, 13, 260–267. [Google Scholar]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar]
- Rasmussen, T.B.; Givskov, M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 2006, 296, 149–161. [Google Scholar]
- Sintim, H.O.; Al Smith, J.; Wang, J.X.; Nakayama, S.; Yan, L. Paradigm shift in discovering next-generation anti-infective agents: Targeting quorum sensing, c-di-gmp signaling and biofilm formation in bacteria with small molecules. Future Med. Chem. 2010, 2, 1005–1035. [Google Scholar]
- Bjarnsholt, T.; Givskov, M. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Philos. Trans. R. Soc. Lond. BBiol. Sci. 2007, 362, 1213–1222. [Google Scholar]
- Camara, M.; Williams, P.; Hardman, A. Controlling infection by tuning in and turning down the volume of bacterial small-talk. Lancet Infect. Dis. 2002, 2, 667–676. [Google Scholar]
- Mayville, P.; Ji, G.Y.; Beavis, R.; Yang, H.M.; Goger, M.; Novick, R.P.; Muir, T.W. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 1999, 96, 1218–1223. [Google Scholar]
- Grundmann, H.; Aires-De-Sousa, M.; Boyce, J.; Tiemersma, E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 2006, 368, 874–885. [Google Scholar]
- Hiramatsu, K.; Cui, L.; Kuroda, M.; Ito, T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001, 9, 486–493. [Google Scholar]
- Chan, W.C.; Coyle, B.J.; Williams, P. Virulence regulation and quorum sensing in staphylococcal infections: Competitive agrc antagonists as quorum sensing inhibitors. J. Med. Chem. 2004, 47, 4633–4641. [Google Scholar]
- Novick, R.P.; Morse, S.I. In vivo transmission of drug resistance factors between strains of Staphylococcus aureus. J. Exp. Med. 1967, 125, 45–59. [Google Scholar]
- McDowell, P.; Affas, Z.; Reynolds, C.; Holden, M.T.G.; Wood, S.J.; Saint, S.; Cockayne, A.; Hill, P.J.; Dodd, C.E.R.; Bycroft, B.W.; Chan, W.C.; Williams, P. Structure, activity and evolution of the group i thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol. Microbiol. 2001, 41, 503–512. [Google Scholar]
- Novick, R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar]
- Ji, G.Y.; Beavis, R.C.; Novick, R.P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 1995, 92, 12055–12059. [Google Scholar]
- Ji, G.Y.; Beavis, R.; Novick, R.P. Bacterial interference caused by autoinducing peptide variants. Science 1997, 276, 2027–2030. [Google Scholar]
- Lyon, G.J.; Wright, J.S.; Muir, T.W.; Novick, R.P. Key Determinants of Receptor Activation in the Agr Autoinducing Peptides of Staphylococcus aureus. Biochemistry 2002, 41, 10095–10104. [Google Scholar]
- Muir, T.W. Turning virulence on and off in staphylococci. J. Pept. Sci. 2003, 9, 612–619. [Google Scholar]
- Kiran, M.D.; Adikesavan, N.V.; Cirioni, O.; Giacometti, A.; Silvestri, C.; Scalise, G.; Ghiselli, R.; Saba, V.; Orlando, F.; Shoham, M.; Balaban, N. Discovery of a quorum-sensing inhibitor of drug-resistant staphylococcal infections by structure-based virtual screening. Mol. Pharmacol. 2008, 73, 1578–1586. [Google Scholar]
- Nakayama, J.; Uemura, Y.; Nishiguchi, K.; Yoshimura, N.; Igarashi, Y.; Sonomoto, K. Ambuic acid inhibits the biosynthesis of cyclic peptide quormones in gram-positive bacteria. Antimicrob. Agents Chemother. 2009, 53, 580–586. [Google Scholar]
- Qazi, S.; Middleton, B.; Muharram, S.H.; Cockayne, A.; Hill, P.; O’Shea, P.; Chhabra, S.R.; Camara, M.; Williams, P. N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 2006, 74, 910–919. [Google Scholar]
- Nielsen, A.; Nielsen, K.F.; Frees, D.; Larsen, T.O.; Ingmer, H. Method for screening compounds that influence virulence gene expression in Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 509–512. [Google Scholar]
- Gram, L.; Melchiorsen, J.; Bruhn, J.B. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar. Biotechnol. 2010, 12, 439–451. [Google Scholar]
- Thompson, F.L.; Iida, T.; Swings, J. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 2004, 68, 403–435. [Google Scholar]
- Faruque, S.M.; Albert, M.J.; Mekalanos, J.J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 1998, 62, 1301–1314. [Google Scholar]
- Linkous, D.A.; Oliver, J.D. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 1999, 174, 207–214. [Google Scholar]
- Su, Y.C.; Liu, C.C. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar]
- Fidopiastis, P.M.; von Boletzky, S.; Ruby, E.G. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 1998, 180, 59–64. [Google Scholar]
- Haygood, M.G.; Distel, D.L. Bioluminescent symbionts of flashlight fishes and deep-sea anglerfishes form unique lineages related to the genus Vibrio. Nature 1993, 363, 154–156. [Google Scholar]
- Ruby, E.G. Lessons from a cooperative, bacterial-animal association: The Vibrio fischeri Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 1996, 50, 591–624. [Google Scholar]
- Ast, J.C.; Dunlap, P.V. Phylogenetic resolution and habitat specificity of members of the Photobacterium phosphoreum species group. Environ. Microbiol. 2005, 7, 1641–1654. [Google Scholar]
- Boisvert, H.; Chatelai, R.; Bassot, J.M. Studies on a Photobacterium isolated from light organ of fish Leiognathidae. Ann. Inst. Pasteur 1967, 112, 521–525. [Google Scholar]
- Magarinos, B.; Romalde, J.L.; Lopez-Romalde, S.; Morinigo, M.A.; Toranzo, A.E. Pathobiological characterisation of Photobacterium damselae subsp piscicida isolated from cultured sole (Solea senegalensis). Bull. Eur. Assoc. Fish Pathol. 2003, 23, 183–190. [Google Scholar]
- Thompson, F.L.; Thompson, C.C.; Naser, S.; Hoste, B.; Vandemeulebroecke, K.; Munn, C.; Bourne, D.; Swings, J. Photobacterium rosenbergii sp. nov. and Enterovibrio coralii sp. nov., vibrios associated with coral bleaching. Int. J. Syst. Evol. Microbiol. 2005, 55, 913–917. [Google Scholar]
- Richards, G.P.; Watson, M.A.; Crane, E.J., III; Burt, I.G.; Bushek, D. Shewanella and Photobacterium spp. in oysters and seawater from delaware bay. Appl. Environ. Microbiol. 2008, 74, 3323–3327. [Google Scholar]
- Mansson, M.; Gram, L.; Larsen, T.O. Production of bioactive secondary metabolites by marine Vibrionaceae. Mar. Drugs 2011, 9, 1440–1468. [Google Scholar]
- Long, R.A.; Rowley, D.C.; Zamora, E.; Liu, J.Y.; Bartlett, D.H.; Azam, F. Antagonistic interactions among marine bacteria impede the proliferation of Vibrio cholerae. Appl. Environ. Microbiol. 2005, 71, 8531–8536. [Google Scholar]
- Oclarit, J.M.; Okada, H.; Ohta, S.; Kaminura, K.; Yamaoka, Y.; Iizuka, T.; Miyashiro, S.; Ikegami, S. Anti-bacillus substance in the marine sponge, hyatella species, produced by an associated Vibrio species bacterium. Microbios 1994, 78, 7–16. [Google Scholar]
- Wietz, M.; Mansson, M.; Gotfredsen, C.H.; Larsen, T.O.; Gram, L. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar. Drugs 2010, 8, 2946–2960. [Google Scholar]
- Needham, J.; Kelly, M.T.; Ishige, M.; Andersen, R.J. Andrimid and moiramides A–C, metabolites produced in culture by a marine isolate of the bacterium Pseudomonas fluorescens—Structure elucidation and biosynthesis. J. Org. Chem. 1994, 59, 2058–2063. [Google Scholar]
- Pohlmann, J.; Lampe, T.; Shimada, M.; Nell, P.G.; Pernerstorfer, J.; Svenstrup, N.; Brunner, N.A.; Schiffer, G.; Freiberg, C. Pyrrolidinedione derivatives as antibacterial agents with a novel mode of action. Bioorg. Med. Chem. Lett. 2005, 15, 1189–1192. [Google Scholar]
- Mansson, M.; Phipps, R.K.; Gram, L.; Munro, M.H.G.; Larsen, T.O.; Nielsen, K.F. Explorative Solid-Phase Extraction (E-SPE) for accelerated microbial natural product discovery, dereplication, and purification. J. Nat. Prod. 2010, 73, 1126–1132. [Google Scholar]
- Nyberg, N.T.; Duus, J.O.; Sorensen, O.W. Heteronuclear two-bond correlation: Suppressing heteronuclear three-bond or higher nmr correlations while enhancing two-bond correlations even for vanishing (2)J(CH). J. Am. Chem. Soc. 2005, 127, 6154–6155. [Google Scholar]
- Fenical, W. Chemical studies of marine-bacteria—Developing a new resource. Chem. Rev. 1993, 93, 1673–1683. [Google Scholar]
- Oku, N.; Kawabata, K.; Adachi, K.; Katsuta, A.; Shizuri, Y. Unnarmicins A and C, new antibacterial depsipeptides produced by marine bacterium Photobacterium sp. MBIC06485. J. Antibiot. 2008, 61, 11–17. [Google Scholar]
- de Nys, R.; Kumar, N.; Sharara, K.A.; Srinivasan, S.; Ball, G.; Kjelleberg, S. A new metabolite from the marine bacterium Vibrio angustum S14. J. Nat. Prod. 2001, 64, 531–532. [Google Scholar]
- Matsuura, S.; Odaka, M.; Sugimoto, T.; Goto, T. Structure of pteridines from Photobacterium phosphorium. Chem. Lett. 1973, 2, 343–346. [Google Scholar]
- Suzuki, A.; Goto, M. Photolumazines, new naturally occurring inhibitors of riboflavin synthetase. Biochim. Biophys. Acta 1973, 313, 229–234. [Google Scholar]
- Diep, B.A.; Gill, S.R.; Chang, R.F.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Carleton, H.A.; Mongodin, E.F.; Sensabaugh, G.F.; Perdreau-Remington, F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 2006, 367, 731–739. [Google Scholar]
- Li, M.; Diep, B.A.; Villaruz, A.E.; Braughton, K.R.; Jiang, X.F.; Deleo, F.R.; Chambers, H.F.; Lu, Y.; Otto, M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2009, 106, 5883–5888. [Google Scholar]
- Cao, J.G.; Meighen, E.A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 1989, 264, 21670–21676. [Google Scholar]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar]
- Higgins, D.A.; Pomianek, M.E.; Kraml, C.M.; Taylor, R.K.; Semmelhack, M.F.; Bassler, B.L. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 2007, 450, 883–886. [Google Scholar]
- Kuo, A.; Blough, N.V.; Dunlap, P.V. Multiple N-Acyl-L-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J. Bacteriol. 1994, 176, 7558–7565. [Google Scholar]
- Milton, D.L.; Hardman, A.; Camara, M.; Chhabra, S.R.; Bycroft, B.W.; Stewart, G.S.A.B.; Williams, P. Quorum sensing in Vibrio anguillarum: Characterization of the VanI/VanR locus and identification of the autoinducer N-(3-Oxodecanoyl)-L-homoserine lactone. J. Bacteriol. 1997, 179, 3004–3012. [Google Scholar]
- Schaefer, A.L.; Greenberg, E.P.; Oliver, C.M.; Oda, Y.; Huang, J.J.; Bittan-Banin, G.; Peres, C.M.; Schmidt, S.; Juhaszova, K.; Sufrin, J.R.; Harwood, C.S. A new class of homoserine lactone quorum-sensing signals. Nature 2008, 454, 595–599. [Google Scholar]
- Dobretsov, S.; Teplitski, M.; Paul, V. Mini-review: Quorum sensing in the marine environment and its relationship to biofouling. Biofouling 2009, 25, 413–427. [Google Scholar]
- Harraghy, N.; Kerdudou, S.; Herrmann, M. Quorum-sensing systems in staphylococci as therapeutic targets. Anal. Bioanal. Chem. 2007, 387, 437–444. [Google Scholar]
- Otto, M. Quorum-sensing control in staphylococci—A target for antimicrobial drug therapy. FEMS Microbiol. Lett. 2004, 241, 135–141. [Google Scholar]
- Boles, B.R.; Horswill, A.R. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008, 4, 1–13. [Google Scholar]
- Defoirdt, T.; Boon, N.; Bossier, P. Can bacteria evolve resistance to quorum sensing disruption. PLoS Pathog. 2010, 6, 1–6. [Google Scholar]
- Köhler, T.; Perron, G.G.; Buckling, A.; van Delden, C. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 2010, 6, 1–6. [Google Scholar]
- Rumbaugh, K.P.; Diggle, S.P.; Watters, C.M.; Ross-Gillespie, A.; Griffin, A.S.; West, S.A. Quorum sensing and social evolution of bacterial virulence. Curr. Biol. 2009, 19, 341–345. [Google Scholar]
- Ostling, J.; Goodman, A.; Kjelleberg, S. Behavior of incp-1 plasmids and a minimu transposon in a marine Vibrio sp.—Isolation of starvation inducible lac operon fusions. FEMS Microbiol. Ecol. 1991, 86, 83–94. [Google Scholar]
- Wietz, M.; Mansson, M.; Gram, L. Chitin stimulates production of the antibiotic andrimid in a Vibrio coralliilyticus strain. Environ. Microbiol. Rep. 2011, 3, 559–564. [Google Scholar]
- Weyland, H.; Ruger, H.-J.; Schwarz, H. Zur Isolierung und identifizierung mariner bakterien. Ein beitrag zur standardisierung und entwicklung adaequater methoden. Veroff. Inst. Meeresforsch. Bremerh. 1970, 12, 269–296. [Google Scholar]
- Hjelm, M.; Bergh, O.; Riaza, A.; Nielsen, J.; Melchiorsen, J.; Jensen, S.; Duncan, H.; Ahrens, P.; Birkbeck, H.; Gram, L. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 2004, 27, 360–371. [Google Scholar]
- Fujii, K.; Ikai, Y.; Oka, H.; Suzuki, M.; Harada, K. A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: Combination of marfey’s method with mass spectrometry and its practical application. Anal. Chem. 1997, 69, 5146–5151. [Google Scholar]
- Bonnard, I.; Manzanares, I.; Rinehart, K.L. Stereochemistry of kahalalide F. J. Nat. Prod. 2003, 66, 1466–1470. [Google Scholar]
- Dale, J.A.; Dull, D.L.; Mosher, H.S. Alpha-methoxy-alpha-trifluoromethylphenylacetic acid, a versatile reagent for determination of enantiomeric composition of alcohols and amines. J. Org. Chem. 1969, 34, 2543–2549. [Google Scholar]
- Sullivan, G.R.; Dale, J.A.; Mosher, H.S. Correlation of configuration and f-19 chemical-shifts of alpha-methoxy-alpha-trifluoromethylphenylacetate derivatives. J. Org. Chem. 1973, 38, 2143–2147. [Google Scholar]
- Jelsbak, L.; Ingmer, H.; Valihrach, L.; Cohn, M.T.; Christiansen, M.H.G.; Kallipolitis, B.H.; Frees, D. The chaperone clpx stimulates expression of Staphylococcus aureus protein a by rot dependent and independent pathways. PLoS One 2010, 5, e12752:1–e12752:11. [Google Scholar]
- Tenover, F.C.; Goering, R.V. Methicillin-resistant Staphylococcus aureus strain USA300: Origin and epidemiology. J. Antimicrob. Chemother. 2009, 64, 441–446. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mansson, M.; Nielsen, A.; Kjærulff, L.; Gotfredsen, C.H.; Wietz, M.; Ingmer, H.; Gram, L.; Larsen, T.O. Inhibition of Virulence Gene Expression in Staphylococcus aureus by Novel Depsipeptides from a Marine Photobacterium. Mar. Drugs 2011, 9, 2537-2552. https://doi.org/10.3390/md9122537
Mansson M, Nielsen A, Kjærulff L, Gotfredsen CH, Wietz M, Ingmer H, Gram L, Larsen TO. Inhibition of Virulence Gene Expression in Staphylococcus aureus by Novel Depsipeptides from a Marine Photobacterium. Marine Drugs. 2011; 9(12):2537-2552. https://doi.org/10.3390/md9122537
Chicago/Turabian StyleMansson, Maria, Anita Nielsen, Louise Kjærulff, Charlotte H. Gotfredsen, Matthias Wietz, Hanne Ingmer, Lone Gram, and Thomas O. Larsen. 2011. "Inhibition of Virulence Gene Expression in Staphylococcus aureus by Novel Depsipeptides from a Marine Photobacterium" Marine Drugs 9, no. 12: 2537-2552. https://doi.org/10.3390/md9122537
APA StyleMansson, M., Nielsen, A., Kjærulff, L., Gotfredsen, C. H., Wietz, M., Ingmer, H., Gram, L., & Larsen, T. O. (2011). Inhibition of Virulence Gene Expression in Staphylococcus aureus by Novel Depsipeptides from a Marine Photobacterium. Marine Drugs, 9(12), 2537-2552. https://doi.org/10.3390/md9122537