Actinomycetes for Marine Drug Discovery Isolated from Mangrove Soils and Plants in China
Abstract
:1. Introduction
2. Results
2.1 Selective isolation of actinomycetes
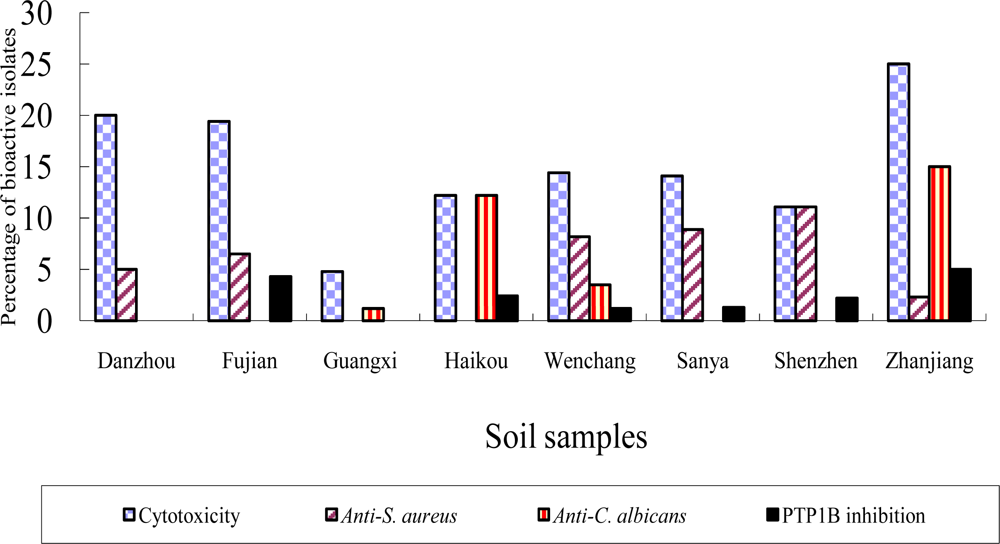
2.2 Bioactivity of strains isolated from the mangrove sites
2.3 Bioactivity of actinomycetes isolated from the 23 mangrove plant species
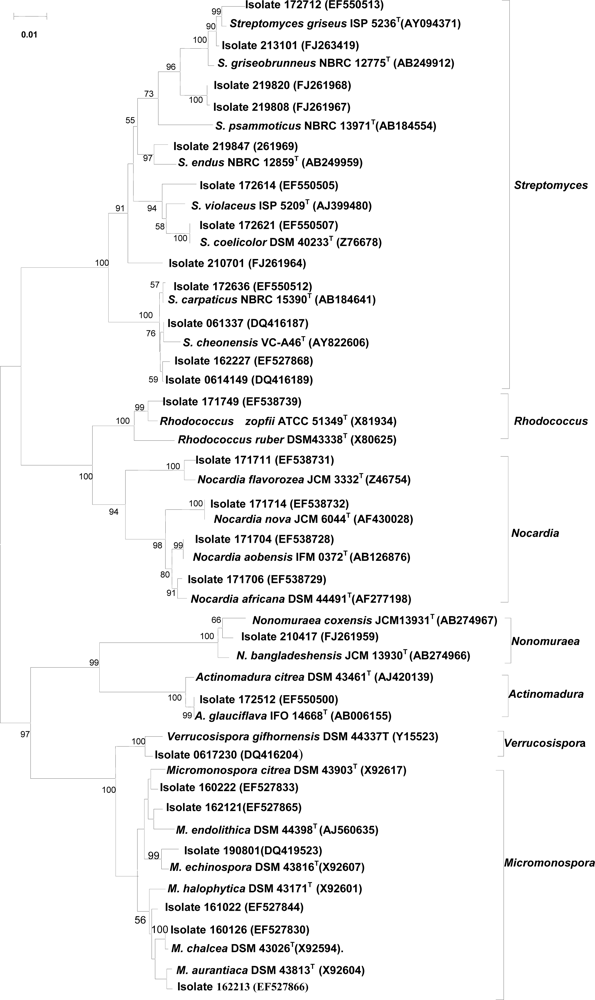
2.4 Taxonomic diversity of bioactive actinomycetes
2.5 16S rRNA gene sequencing of representative isolates showing activity against tumor cells
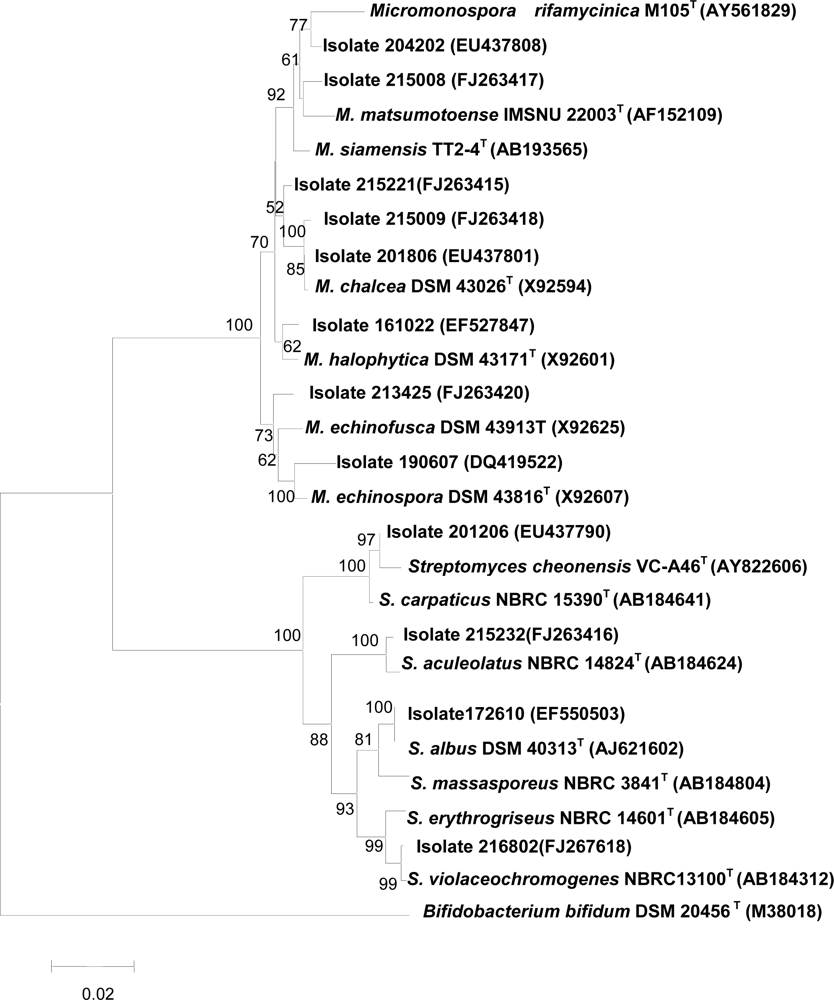
2.6 16S rRNA gene sequences of isolates which inhibited protein tyrosine phosphatase 1B
3. Discussion
4. Experimental Section
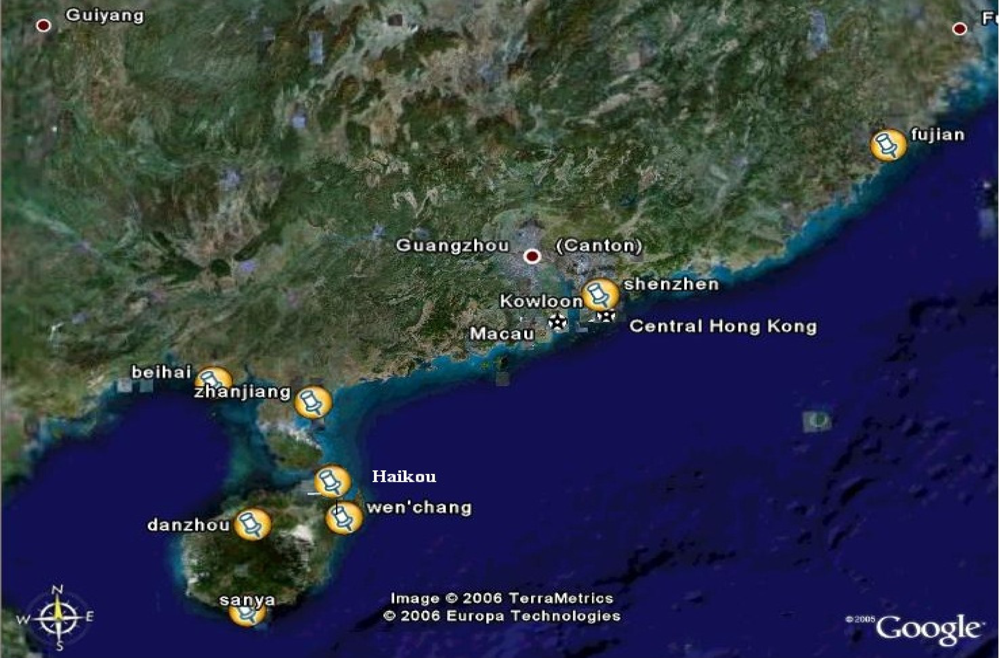
4.1. Collection of samples
4.2. Selective isolation of actinomycetes from environmental samples
4.3. Preparation of crude extracts
4.4. Biological assays
Antimicrobial activity
Cell growth inhibition assay
Biochemical assays
4.5 Identification of isolates
Morphological characterization
Chemotaxonomy
Extraction of DNA from pure cultures and PCR amplification
Cloning, sequencing and phylogenetic analyses
Acknowledgments
References and Notes
- Talbot, GH; Bradley, J; Edwards, JE, Jr; Gilbert, D; Scheld, M; Bartlett, JG. Bad Bugs Need Drugs: An update on the development pipeline from the antimicrobial availability task force of the infectious diseases society of America. Clin. Infect. Dis 2006, 42, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Bentley, SD; Chater, KF; Cerdeno-Tarraga, AM; Challis, GL; Thompson, NR; James, KD; Harris, DE; Quail, MA; Kieser, H; Harper, D; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Omura, S; Ikeda, H; Ishikawa, J; Hanamoto, A; Takahashi, C; Shinose, M; Takahashi, Y; Horikawa, H; Nakazawa, H; Osonoe, T; Kikuchi, H; Shiba, T; Sakaki, Y; Hattori, M; et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. U.S.A 2001, 98, 12215–12220. [Google Scholar] [CrossRef] [PubMed]
- Watve, MG; Tickoo, R; Jog, MM; Bhole, BD. How many antibiotics are produced by the genus Streptomyces. Arch. Microbiol 2001, 176, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Lazzarini, A; Cavaletti, L; Toppo, G; Marinelli, F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie van Leeuwenhoek 2000, 78, 388–405. [Google Scholar]
- Bredholt, H; Fjærvik, E; Johnsen, G; Zotchev, SB. Actinomycetes from sediments in the Trondheim Fjord, Norway: Diversity and biological activity. Mar. Drugs 2008, 6, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Bull, AT; Stach, JEM; Ward, AC; Goodfellow, M. Marine actinobacteria: perspectives, challenges, future directions. Antonie van Leeuwenhoek 2005, 87, 259–276. [Google Scholar] [CrossRef]
- Bull, AT; Stach, JE. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 2007, 15, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Okoro, CK; Brown, R; Jones, AL; Andrews, BA; Asenjo, JA; Goodfellow, M; Bull, AT. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie van Leeuwenhoek.
- Stach, JEM; Maldonado, LA; Ward, AC; Goodfellow, M; Bull, AT. New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ. Microbiol 2003, 5, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Stach, EM; Bull, AT. Estimating and comparing the diversity of marine actinobacteria. Antonie van Leeuwenhoek 2005, 87, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Ara, I; Kudo, T; Matsumoto, A; Takahashi, Y; Omura, S. Nonomuraea maheshkhaliensis sp. nov., a novel actinomycete isolated from mangrove rhizosphere mud. J. Gen. Appl. Microbiol 2007, 53, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Huang, H; Lv, J; Hu, Y; Fang, Z; Zhang, K; Bao, S. Micromonospora rifamycinica sp. nov., a novel actinomycete from mangrove sediment. Int. J. Syst. Evol. Microbiol 2008, 58, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Pathom-aree, W; Nogi, Y; Sutcliffe, IC; Ward, AC; Horikoshi, K; Bull, AT; Goodfellow, M. Williamsia marianensis sp. nov., a novel actinomycete isolated from the Mariana Trench. Int. J. Syst. Evol. Microbiol 2006a, 56, 1123–1126. [Google Scholar] [CrossRef]
- Pathom-aree, W; Nogi, Y; Sutcliffe, IC; Ward, AC; Horikoshi, K; Bull, AT; Goodfellow, M. Dermacoccus abyssi sp. nov., a piezotolerant actinomycete isolated from the Mariana Trench. Int. J. Syst. Evol. Microbiol 2006b, 56, 1233–1237. [Google Scholar] [CrossRef]
- Pathom-aree, W; Nogi, Y; Sutcliffe, IC; Ward, AC; Horikoshi, K; Bull, AT; Goodfellow, M. Dermacoccus barathri sp. nov. and Dermacoccus profundi sp. nov., novel actinomycetes isolated from deep-sea mud of the Mariana Trench. Int. J. Syst. Evol. Microbiol 2006c, 56, 2303–2307. [Google Scholar] [CrossRef]
- Tamura, T; Sakane, T. Asanoa iriomotensis sp. nov., isolated from mangrove soil. Int. J. Syst. Bacteriol 2005, 55, 725–727. [Google Scholar]
- Yi, H; Schumann, P; Sohn, K; Chun, J. Demequina aestuarii gen. nov., sp. nov., a novel actinomycete of the suborder Micrococcineae, and reclassification of Cellulomonas fermentans Bagnara et al. 1985 as Actinotalea fermentans gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol 2007, 57, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Yi, H; Schumann, P; Sohn, K; Chun, J. Serinicoccus marinus gen. nov., sp. nov., a novel actinomycete with L-ornithine and L-serine in the peptidoglycan. Int. J. Syst. Evol. Microbiol 2004, 54, 1585–1589. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, LA; Stach, JEM; Pathom-aree, W; Ward, AC; Bull, AT; Goodfellow, M. Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie van Leeuwenhoek 2005, 87, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y; Li, H; Zeng, Y; Chen, B. Isolation and phylogenetic assignation of actinomycetes in the marine sediments from the Arctic Ocean. Acta Oceanol. Sin 2005, 24, 135–142. [Google Scholar]
- Jensen, PR; Gontag, E; Mafnas, C; Mincer, TJ; Fenical, W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ. Microbiol 2005, 7, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Blunt, JW; Copp, BR; Hu, W; Munro, MHG; Northcote, PT; Prinsep, MR. Marine natural products. Nat. Prod. Rep 2007, 24, 31–86. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W; Jensen, PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nature Chem. Biol 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Fiedler, HP; Bruntner, C; Bull, AT; Ward, AC; Goodfellow, M; Potterat, O; Puder, C; Mihm, G. Marine actinomycetes as a source of novel secondary metabolites. Antonie van Leeuwenhoek 2005, 87, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M; Kumar, Y; Labeda, DP; Sembiring, L. The Streptomyces violaceusinger clade: a home for streptomycetes with rugose ornamented spores. Antonie van Leeuwenhoek 2007, 92, 173–199. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y; Goodfellow, M. Five new members of the Streptomyces violaceusniger 16S rRNA gene clade: Streptomyces castelarensis sp. nov., comb. nov., Streptomyces himastatinicus sp. nov., Streptomyces mordarskii sp. nov., Streptomyces rapamycinicus sp. nov. and Streptomyces ruanii sp. nov. Int. J. Syst. Evol. Microbiol 2008, 58, 1369– 1378. [Google Scholar] [CrossRef] [PubMed]
- Ward, AC; Goodfellow, M. Phylogeny and functionality: taxonomy as a roadmap to genes. In “Microbial Diversity and Bioprospecting”; Bull, AT, Ed.; ASM Press: Washington, DC, USA, 2004; pp. 288–313. [Google Scholar]
- Riedlinger, J; Reicke, A; Zahner, H; Krismer, B; Bull, AT; Maldonado, LA; Ward, AC; Goodfellow, M,; Bister, B; Bischoff, D; Süssmuth, RD; Fiedler, H-P. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot 2004, 57, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Williams, PG; Buchanan, GO; Feling, RH; Kauffman, CA; Jensen, PR; Fenical, W. New cytotoxic salinosporamides from marine actinomycete Salinispora tropica. J. Org. Chem 2005, 70, 6196–6203. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K; Keller, S; Wolter, FE; Roglin, L; Beil, W; Seitz, O; Nicholson, G; Bruntner, C; Riedlinger, J; Fiedler, H-P; Süssmuth, RD. Proximicins A, B, and C- antitumor furan analogues of netropsin from the marine actinomycete Verrocosispora induce upregulation of p53 and the cyclin kinase inhibitor p21. Angew. Chem. Int. Ed 2008, 47, 3258–3261. [Google Scholar] [CrossRef]
- Williams, PG. Panning for chemical gold: marine bacteria as a source of new therapeutics. Trends Biotechnol 2009, 27, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, YH; Zhang, HJ; Wu, F; Chen, YH; Ma, XQ; Du, JQ; Zhou, ZL; Li, JY; Nan, FJ; Li, J. Isoquinoline-1,3,4-trione and its derivatives attenuate □-amyloid-induced apoptosis of neuronal cells. FEBS J 2006, 273, 4842–4852. [Google Scholar] [CrossRef] [PubMed]
- Johnson, TO; Ermolieff, J; Jirousek, MR. Protein tyrosine phosphatase 1B inhibitors for diabetes. Nat. Rev., Drug. Discov 2002, 1, 696–709. [Google Scholar] [CrossRef]
- Taylor, SD; Hill, B. Recent advances in protein tyrosine phosphatase 1B inhibitors. Expert Opin. Investig. Drugs 2004, 13, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W; Hong, D; Zhou, YY; Zhang, YN; Shen, Q; Li, JY; Hu, LH; Li, J. Ursolic acid and its derivative inhibit protein tyrosine phosphatase 1B enhancing insulin receptor phosphorylation and stimulating glucose uptake. Biochim. Biophys. Acta 2006, 1760, 1505–1512. [Google Scholar] [CrossRef] [PubMed]
- Costanza, R; d’Arge, R; Groot, R; Farberk, S; Grasso, M; Hannon, B; Limburg, K; Naeem, S; O’Neill, RV; Paruelo, J; Raskin, RG; Sutton, P; van den Belt, M. The value of the world’s ecosystem services and natural capital. Nature 1997, 387, 253–260. [Google Scholar] [CrossRef]
- Wang, B; Liang, S; Zhang, W; Zan, Q. Mangrove flora of the world. Acta Botanica Sinica 2003, 45, 644– 653. [Google Scholar]
- Hong, K; Yan, B. Uncultured microorganisms in Hainan mangrove soil: diversity and functional genes. “Microbes and the Environment: Perspective and Challenges”, Liu, SJ, Drake, HL, Eds.; Science Press: Beijing, China, 2008; 52–58. [Google Scholar]
- Hyde, KD; Lee, SY. Ecology of mangrove fungi and their role in nutrient cycling: what gaps occur in our knowledge? Hydrobiologia 1995, 295, 107–118. [Google Scholar] [CrossRef]
- Yan, B; Hong, K; Yu, Z. Archaeal communities in mangrove soil characterized by 16S rRNA gene colones. J. Microbiol 2006, 44, 566–571. [Google Scholar] [PubMed]
- Eccleston, GP; Brooks, PR; Kurtböke, DI. The occurrence of bioactive micromonosporae in aquatic habitats of the Sunshine Coast in Australia. Mar. Drugs 2008, 6, 243–261. [Google Scholar] [CrossRef] [PubMed]
- Han, L; Huang, XS; Sattler, I; Fu, HZ; Grabley, S; Lin, WH. Two new constituents from mangrove Bruguiera gymnorrhiza. J. Asian Nat. Prod. Res 2007, 9, 327–31. [Google Scholar] [CrossRef] [PubMed]
- Huo, C; Liang, H; Tu, G; Zhao, Y; Lin, W. A new 5, 11-epoxymegastigmane glucoside from Acanthus ilicifolius. Nat. Prod. Res 2008, 22, 896–900. [Google Scholar] [CrossRef] [PubMed]
- Wu, J; Xiao, Q; Huang, J; Xiao, Z; Qi, S; Li, Q; Zhang, S. Xyloccensin O and P, unique 8,9,30-phragmalin ortho esters from Xylocarpus granatum. Org. Lett 2004, 6, 1841–1844. [Google Scholar] [CrossRef] [PubMed]
- Gao, H; Hong, K; Zhang, X; Liu, HW; Wang, NL; Zhang, L; Yao, XS. Polyhydroxylated sterols and new sterol fatty esters from the mangrove fungus Aspergillus awamori exhibiting potent cytotoxic activity. Helv. Chim. Acta 2007, 90, 1165–1178. [Google Scholar] [CrossRef]
- Krohn, K; Steingröver, K; Zsila, F. Five unique compounds: Xyloketals from the mangrove fungus Xylaria sp. from the South China Sea coast. J Org Chem 2001, 66, 6252–6256. [Google Scholar] [CrossRef] [PubMed]
- Lin, YC; Wu, XY; Deng, ZJ; Wang, J; Zhou, SN; Vrijmoed, LLP; Jones, EBG. The metabolites of the mangrove fungus Verruculina enalia No. 2606 from a salt lake in the Bahamas. Phytochem 2002, 59, 469–471. [Google Scholar] [CrossRef]
- Lin, Z; Zhu, T; Fang, Y; Gu, Q. 1H and 13C NMR assignments of two new indolic enamide diastereomers from a mangrove endophytic fungus Aspergillus sp. Magn. Reson. Chem 2008, 46, 1212–1216. [Google Scholar] [CrossRef] [PubMed]
- Tang, JS; Gao, H; Hong, K; Yu, Y; Jiang, MM; Lin, HP; Ye, WC; Yao, XS. Complete assignments of 1H and 13C NMR spectral data of nine surfactin isomers. Magn. Reson. Chem 2007, 45, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Xie, XC; Mei, WL; Zhao, YX; Hong, K; Dai, HF. A new degraded sesquiterpene from marine actinomycete Streptomyces sp. 0616208. Chinese Chem. Lett 2006, 17, 1463–1465. [Google Scholar]
- Lozupone, CA; Knight, R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z; Shi, Y; Zhang, Y; Zhou, Z; Li, W; Huang, Y; Rodrigues, C; Goodfellow, M. Classification of Streptomyces grieseus (Krainsky, 1914) Waksman and Henrici 1948 and related species and the transfer of “Microstreptospora cinerea” to the genus Streptomyces as Streptomyces yanii sp. nov. Int. J. Syst. Evol. Microbiol 2005, 55, 1605–1610. [Google Scholar] [CrossRef] [PubMed]
- Antony-Babu, S; Stach, JEM; Goodfellow, M. Genetic and phenotypic evidence for Streptomyces griseus ecovars isolated from a beach and dune sand system. Antonie van Leeuwenhoek 2008, 94, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Pisano, AM; Sommer, JM; Lopez, MM. Application of pretreatment for the isolation of bioactive actinomycetes from marine sediments. Appl. Microbiol. Biotechnol 1986, 25, 285–288. [Google Scholar]
- Hayakawa, M; Iino, H; Takeuchi, S; Yamazaki, T. Application of a method incorporating treatment with chloramine-T for the selective isolation of Stretpsporangiaceae from soil. J. Ferment. Bioengin 1997, 84, 599–602. [Google Scholar] [CrossRef]
- Takahashi, Y; Matsumoto, A; Seino, A; Iwai, Y; Omura, S. Rare actinomycetes isolated from desert soils. Actinomycetologica 1996, 10, 91–97. [Google Scholar] [CrossRef]
- Shirling, EB; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Ivantiskaya, LP; Singal, SM; Bibikova, MV; Vostrov, SN. Direct isolation of Micromonospora on selective media with gentamicin. Antibiotiki 1978, 23, 690–692. [Google Scholar] [PubMed]
- Williams, ST; Lanning, S; Wellington, EMH. Ecology of actinomycetes. In “The Biology of the Actinomycetes”; Goodfellow, M, Mordarski, M, Williams, ST, Eds.; Academic Press: London, UK, 1984; pp. 481–528. [Google Scholar]
- Küster, E; Williams, ST. Media for the isolation of streptomycetes: starch casein medium. Nature 1964, 202, 928–929. [Google Scholar] [CrossRef] [PubMed]
- Tan, GYA; Robinson, S; Lacey, E; Goodfellow, M. Exploration of Amycolatopsis diversity in soil using genus-specific primers and novel selective media. Syst Appl Microbiol 2006, 29, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, M; Ohara, Y. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol 1987, 65, 501–509. [Google Scholar] [CrossRef]
- Duangmal, K; Ward, AC; Goodfellow, M. Selective isolation of members of the Streptomyces violaceoruber clade from soil. FEMS Microbiol. Lett 2005, 245, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Williams, ST; Davies, FL. Use of antibiotics for selective isolation and enumeration of actinomycetes in soil. J. Gen Microbiol 1965, 38, 251–261. [Google Scholar] [PubMed]
- Garcia, GD; Romero, MF; Perez, BJ; Garcia, DT. Thiodepsipeptide isolated from a marine actinomycete WO9527730, 1999; Patent Number: US5681813.
- Yu, JS; Hong, K; Lin, HP; Yuan, GJ. Optimization study on fermentative medium for precursor of daptomycin A21978C produced by Streptomyces roseosporus NRRL 11397. J. Anhui Agric. Sci 2008, 36, 7974–7976. [Google Scholar]
- Chen, H; Hong, K; Zhuang, L; Zhong, QP. Growth characteristics and fermentation condition optimization of mangrove actinomycete strain 0616167. Microbiology 2006, 33, 16–20. [Google Scholar]
- Hong, K; Xiao, C. A rapid method for detection of biological activity of anti- yeast-like pathogen. China Patent 2006, ZL03128096. [Google Scholar]
- Shi, L; Yu, HP; Zhou, YY; Du, JQ; Shen, Q; Li, JY; Li, J. Discovery of a novel competitive inhibitor of PTP1B by high-throughput screening. Acta Pharmacol. Sin 2008, 29, 278–84. [Google Scholar] [CrossRef] [PubMed]
- Du, JQ; Wu, J; Zhang, HJ; Zhang, YH; Qiu, BY; Wu, F; Chen, YH; Li, JY; Nan, FJ; Ding, JP; Li, J. Isoquinoline-1,3,4-trione derivatives inactivate caspase-3 by generation of reactive oxygen species. J. Biol. Chem 2008, 283, 30205–30215. [Google Scholar] [CrossRef] [PubMed]
- Gautschi, O; Heighway, J; Mack, PC; Purnell, PR; Lara, PN, Jr; Gandara, DR. Aurora kinases as anticancer drug targets. Clin. Cancer. Res 2008, 14, 1639–1648. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T; Takizawa, M; Tanida, S. A rapid analysis for chemical grouping of aerobic actinoycetes. J. Gen. Appl. Microbiol 1983, 329, 1319–1322. [Google Scholar]
- Lane, DJ. 16S/23S rRNA sequencing. In “Nucleic Acid Techniques in Bacterial Systematics”; Stackebrandt, E, Goodfellow, M, Eds.; Wiley: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Thompson, JD; Gibson, TJ; Plewniak, F; Jeanmougin, F; Higgins, DG. The CLUSTAL _X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997, 25, 4876–4876. [Google Scholar] [CrossRef] [PubMed]
- Hall, UA. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser 1999, 41, 95–98. [Google Scholar]
- Saitou, N; Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K; Dudley, J; Nei, M; Kumar, S. MEGA4, Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. PHYLIP (phylogenetic inference package) version 3.2. Cladistics 1989, 5, 164–166. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
| Number of samples / Number of isolated actinomycetes
| ||||
|---|---|---|---|---|
| Sites | Composite soil | Rizhosphere soil | Plant tissues | Total |
| Danzhou | 5/40 | ND | ND | 5/40 |
| Fujian | 3/93 | ND | ND | 3/93 |
| Guangxi | 17/62 | 5/27 | 4/4 | 26/93 |
| Haikou | 3/25 | 5/16 | 2/11 | 10/52 |
| Sanya | 3/78 | ND | ND | 3/78 |
| Shenzhen | 4/57 | 5/44 | 8/48 | 17/149 |
| Wenchang | 26/205 | 31/917 | 62/256 | 119/1378 |
| Zhanjiang | 5/43 | ND | 23/115 | 28/158 |
| Total | 66/603 | 46/1004 | 99/434 | 211/2041 |
| Sampling sites | Sample type | Anti-C. albicans | Anti-S. aureus | Anti-tumor cell | PTP1B inhibition |
|---|---|---|---|---|---|
| Wenchang | Plant tissues | 9.8 | 16.0 | 22.2 | 1.6 |
| Rhizosphere soil | 3.5 | 8.2 | 14.4 | 1.2 | |
| Zhangjiang | Plant tissues | 20.0 | 37.4 | 4.3 | / |
| Rhizosphere soil | 15.0 | 2.3 | 25.0 | 5.0 |
| Number of Bioactive actinomycetes
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Rhizosphere soil | Plant tissues | |||||||
| Mangrove plant species | Anti-C. albicans | Anti-S. aureus | Anti- tumor cell | PTP1B inhibition | Anti-C. albicans | Anti-S. aureus | Anti- tumor cell | PTP1B inhibition |
| Acanthus ilicifolius | 1 | 5 | 13 | 2 | 6 | 3 | 5 | 3 |
| Acrostichum aureum | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 |
| Acrostichum speciosum | 0 | 2 | 7 | 0 | 0 | 2 | 2 | 0 |
| Aegiceras corniculatum | 3 | 15 | 5 | 0 | 5 | 11 | 10 | 0 |
| Avicennia marina | 1 | 0 | 0 | 0 | 4 | 7 | 1 | 0 |
| Barringtonia racemosa | 0 | 0 | 1 | 0 | ND | ND | ND | ND |
| Bruguiera gymnorrhiza | 2 | 4 | 5 | 0 | 12 | 15 | 6 | 1 |
| Bruguiera sexangula | 1 | 2 | 12 | 2 | 0 | 0 | 1 | 0 |
| Cerbera manghas | 5 | 0 | 4 | 3 | 1 | 4 | 4 | 0 |
| Ceriops tagal | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 |
| Excoecaria agallocha | 0 | 0 | 3 | 0 | 0 | 3 | 2 | 0 |
| Heritiera littoralis | 13 | 26 | 22 | 2 | 3 | 3 | 2 | 0 |
| Hibiscus tilisaceus | 6 | 16 | 33 | 4 | 7 | 4 | 7 | 0 |
| Kandelia candel | 0 | 0 | 1 | 0 | 2 | 4 | 1 | 0 |
| Lumnitzera racemosa | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 1 |
| Pongamia pinnata | 0 | 7 | 6 | 0 | 0 | 2 | 2 | 0 |
| Rhizophora apiculata | 0 | 2 | 11 | 3 | 0 | 1 | 2 | 0 |
| Rhizophora stylosa | ND | ND | ND | ND | 7 | 14 | 3 | 0 |
| Sonneratia alba | 7 | 12 | 10 | 0 | 0 | 2 | 3 | 0 |
| Sonneratia caseolaris | 1 | 4 | 6 | 1 | 0 | 3 | 4 | 0 |
| Sonneratia hainanensis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sonneratia paracaseolaris | 2 | 4 | 5 | 0 | 0 | 4 | 1 | 0 |
| Xylocarpus granatum | 0 | 0 | 0 | 0 | 4 | 2 | 3 | 0 |
| Total | 42 | 99 | 144 | 17 | 55 | 88 | 62 | 5 |
| Genera | Families | Suborders | Number of the identified representative strains
| ||||
|---|---|---|---|---|---|---|---|
| Anti-C. albicans | Anti-S. aureus | Anti-tumor cell | Caspase 3 inhibition | PTP1B- inhibition | |||
| Actinomadura | Thermomonosporaceae | Streptosporangineae | 3 | ||||
| Microbispora | 1 | ||||||
| Nonomuraea | Streptosporangiaceae | 1 | |||||
| Actinoplanes | Micromonosporaceae | Micromonosporineae | 1 | ||||
| Micromonospora | 7 | 10 | 25 | 1 | 8 | ||
| Verrucosispora | 1 | ||||||
| Arthrobacter | Micrococcaceae | Micrococcineae | 1 | ||||
| Isoptericola | 1 | ||||||
| Micrococcus | 1 | ||||||
| Microbacterium | Microbacteriaceae | 1 | |||||
| Nocardia | Nocardiaceae | Corynebacterineae | 5 | ||||
| Rhodococcus | 1 | 1 | |||||
| Strepomyces | Streptomycetaceae | Streptomycineae | 1 | 7 | 45 | 1 | 4 |
| Genus and Species | Samples |
|---|---|
| Acanthus ilicifolius | Rhizosphere soil, roots, leaves and flowers |
| Acrostichum aureum | }Rhizosphere soil, roots and leaves |
| Acrostichum speciosum | |
| Aegiceras corniculatum | |
| Avicennia marina | Rhizosphere soil, leaves, and fruit |
| Barringtonia racemosa | Rhizosphere soil |
| Bruguiera gymnorrhiza | }Rhizosphere soil, roots and leaves |
| Bruguiera sexangula | |
| Cerbera manghas | }Rhizosphere soil, roots, leaves, and fruit |
| Ceriops tagal | |
| Excoecaria agallocha | Rhizosphere soil, roots and leaves |
| Heritiera littoralis | Rhizosphere soil, leaves, and fruit |
| Hibiscus tilisaceus | Rhizosphere soil, roots and leaves |
| Kandelia candel | Rhizosphere soil and leaves |
| Lumnitzera racemosa | Rhizosphere soil, roots and leaves |
| Pongamia pinnata | Rhizosphere soil and leaves |
| Rhizophora apiculata | Rhizosphere soil and leaves, |
| Rhizophora stylosa | Roots and leaves |
| Sonneratia alba | }Rhizosphere soil, leaves, and fruit |
| Sonneratia caseolaris | |
| Sonneratia hainanensis | Rhizosphere soil and leaves |
| Sonneratia paracaseolaris | Roots and leaves |
| Xylocarpus granatum | Roots, leaves, and fruit |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hong, K.; Gao, A.-H.; Xie, Q.-Y.; Gao, H.G.; Zhuang, L.; Lin, H.-P.; Yu, H.-P.; Li, J.; Yao, X.-S.; Goodfellow, M.; et al. Actinomycetes for Marine Drug Discovery Isolated from Mangrove Soils and Plants in China. Mar. Drugs 2009, 7, 24-44. https://doi.org/10.3390/md7010024
Hong K, Gao A-H, Xie Q-Y, Gao HG, Zhuang L, Lin H-P, Yu H-P, Li J, Yao X-S, Goodfellow M, et al. Actinomycetes for Marine Drug Discovery Isolated from Mangrove Soils and Plants in China. Marine Drugs. 2009; 7(1):24-44. https://doi.org/10.3390/md7010024
Chicago/Turabian StyleHong, Kui, An-Hui Gao, Qing-Yi Xie, Hao Gao Gao, Ling Zhuang, Hai-Peng Lin, Hai-Ping Yu, Jia Li, Xin-Sheng Yao, Michael Goodfellow, and et al. 2009. "Actinomycetes for Marine Drug Discovery Isolated from Mangrove Soils and Plants in China" Marine Drugs 7, no. 1: 24-44. https://doi.org/10.3390/md7010024




