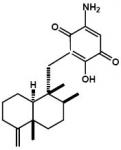
Smenospongine, a Sesquiterpene Aminoquinone from a Marine Sponge, Induces G1 Arrest or Apoptosis in Different Leukemia Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of smenospongine on cell cycle distribution in K562, HL60, and U937 cells
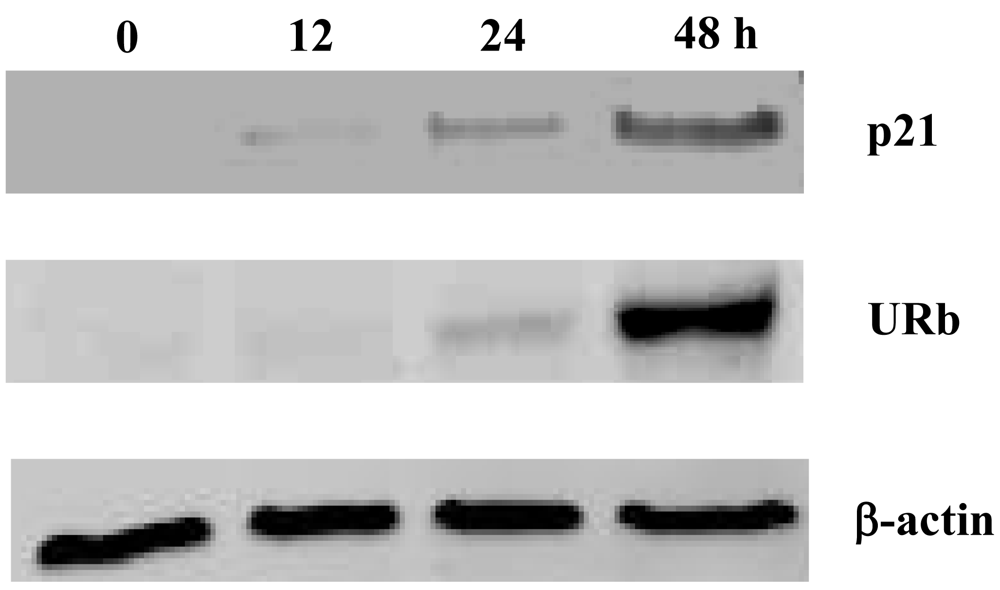
2.2. Effect of smenospongine on expression of p21 and phosphorylation of Rb
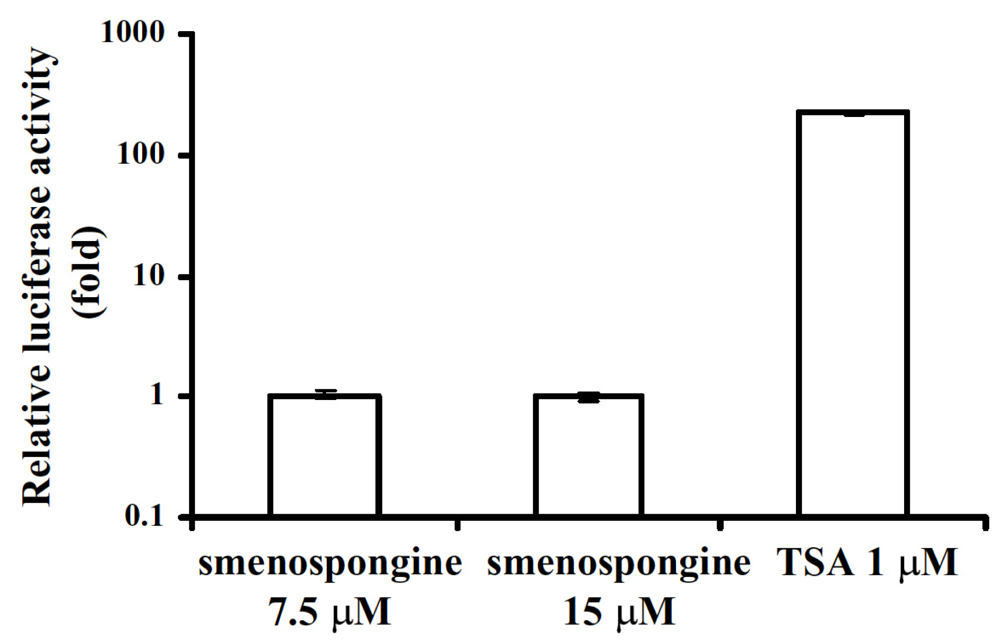
2.3. Effect of smenospongine on activation of p21 promoter
3. Conclusions
4. Experimental Section
4.1. Materials
4.2. Cell culture
4.3. Antibodies
4.4. Isolation and identification of smenospongine
4.5. Flow cytometric analysis of cell cycle
4.6. Western blotting analysis
4.7. Transfection of K562 cells and luciferase assay
Acknowledgements
- Samples Availability: Not available.
References
- De Klein, A; Van Kessel, AG; Grosveld, G; Bartram, CR; Hagemeijer, A; Bootsma, D; Spurr, NK; Heisterkamp, N; Groffen, J; Stephenson, JR. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature 1982, 300, 765–767. [Google Scholar]
- Lozzio, CB; Lozzio, BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 1975, 45, 321–334. [Google Scholar]
- Fausel, C. Targeted chronic myeloid leukemia therapy: seeking a cure. J Manag Care Pharm 2007, 13, 8–12. [Google Scholar]
- Dan, S; Naito, M; Tsuruo, T. Selective induction of apoptosis in Philadelphia chromosome-positive chronic myelogenous leukemia cells by an inhibitor of BCR - ABL tyrosine kinase, CGP 57148. Cell Death Differ 1998, 5, 710–715. [Google Scholar]
- Melo, JV; Chuah, C. Resistance to imatinib mesylate in chronic myeloid leukaemia. Cancer Lett 2007, 249, 121–132. [Google Scholar]
- Nardi, V; Azam, M; Daley, GQ. Mechanisms and implications of imatinib resistance mutations in BCR-ABL. Curr Opin Hematol 2004, 11, 35–43. [Google Scholar]
- Okabe, S; Tauchi, T; Nakajima, A; Sashida, G; Gotoh, A; Broxmeyer, HE; Ohyashiki, JH; Ohyashiki, K. Depsipeptide (FK228) preferentially induces apoptosis in BCR/ABL-expressing cell lines and cells from patients with chronic myelogenous leukemia in blast crisis. Stem Cells Dev 2007, 16, 503–514. [Google Scholar]
- Byrd, JC; Marcucci, G; Parthun, MR; Xiao, JJ; Klisovic, RB; Moran, M; Lin, TS; Liu, S; Sklenar, AR; Davis, ME; Lucas, DM; Fischer, B; Shank, R; Tejaswi, SL; Binkley, P; Wright, J; Chan, KK; Grever, MR. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood 2005, 105, 959–967. [Google Scholar]
- Sasakawa, Y; Naoe, Y; Inoue, T; Sasakawa, T; Matsuo, M; Manda, T; Mutoh, S. Effects of FK228, a novel histone deacetylase inhibitor, on human lymphoma U-937 cells in vitro and in vivo. Biochem Pharmacol 2002, 64, 1079–1090. [Google Scholar]
- Aoki, S; Higuchi, K; Isozumi, N; Matsui, K; Miyamoto, Y; Itoh, N; Tanaka, K; Kobayashi, M. Differentiation in chronic myelogenous leukemia cell K562 by spongean sesterterpene. Biochem Biophys Res Commun 2001, 282, 426–431. [Google Scholar]
- Aoki, S; Kong, D; Matsui, K; Kobayashi, M. Erythroid differentiation in K562 chronic myelogenous cells induced by crambescidin 800, a pentacyclic guanidine alkaloid. Anticancer Res 2004, 24, 2325–2330. [Google Scholar]
- Aoki, S; Kong, D; Matsui, K; Kobayashi, M. Smenospongine, a spongean sesquiterpene aminoquinone, induces erythroid differentiation in K562 cells. Anticancer Drugs 2004, 15, 363–369. [Google Scholar]
- Aoki, S; Kong, D; Matsui, K; Rachmat, R; Kobayashi, M. Sesquiterpene aminoquinones, from a marine sponge, induce erythroid differentiation in human chronic myelogenous leukemia, K562 cells. Chem Pharm Bull (Tokyo) 2004, 52, 935–937. [Google Scholar]
- Bi, S; Hughes, T; Bungey, J; Chase, A; De Fabritiis, P; Goldman, JM. p53 in chronic myeloid leukemia cell lines. Leukemia 1992, 6, 839–842. [Google Scholar]
- Sowa, Y; Orita, T; Minamikawa, S; Nakano, K; Mizuno, T; Nomura, H; Sakai, T. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem Biophys Res Commun 1997, 241, 142–150. [Google Scholar]
- Sowa, Y; Orita, T; Minamikawa-Hiranabe, S; Mizuno, T; Nomura, H; Sakai, T. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res 1999, 59, 4266–4270. [Google Scholar]
- Kondracki, ML; GM. Smenospongine: a cytotoxic and antimicrobial aminoquinone isolated from smenospongia SP. Tetrahedron Lett 1987, 28, 5815–5818. [Google Scholar]
- Aoki, S; Kong, D; Suna, H; Sowa, Y; Sakai, T; Setiawan, A; Kobayashi, M. Aaptamine, a spongean alkaloid, activates p21 promoter in a p53-independent manner. Biochem Biophys Res Commun 2006, 342, 101–106. [Google Scholar]

Share and Cite
Kong, D.; Aoki, S.; Sowa, Y.; Sakai, T.; Kobayashi, M. Smenospongine, a Sesquiterpene Aminoquinone from a Marine Sponge, Induces G1 Arrest or Apoptosis in Different Leukemia Cells. Mar. Drugs 2008, 6, 480-488. https://doi.org/10.3390/md6030480
Kong D, Aoki S, Sowa Y, Sakai T, Kobayashi M. Smenospongine, a Sesquiterpene Aminoquinone from a Marine Sponge, Induces G1 Arrest or Apoptosis in Different Leukemia Cells. Marine Drugs. 2008; 6(3):480-488. https://doi.org/10.3390/md6030480
Chicago/Turabian StyleKong, Dexin, Shunji Aoki, Yoshihiro Sowa, Toshiyuki Sakai, and Motomasa Kobayashi. 2008. "Smenospongine, a Sesquiterpene Aminoquinone from a Marine Sponge, Induces G1 Arrest or Apoptosis in Different Leukemia Cells" Marine Drugs 6, no. 3: 480-488. https://doi.org/10.3390/md6030480