Structure and Effects of Cyanobacterial Lipopolysaccharides
Abstract
:1. Introduction
2. Gram-Negative LPSs
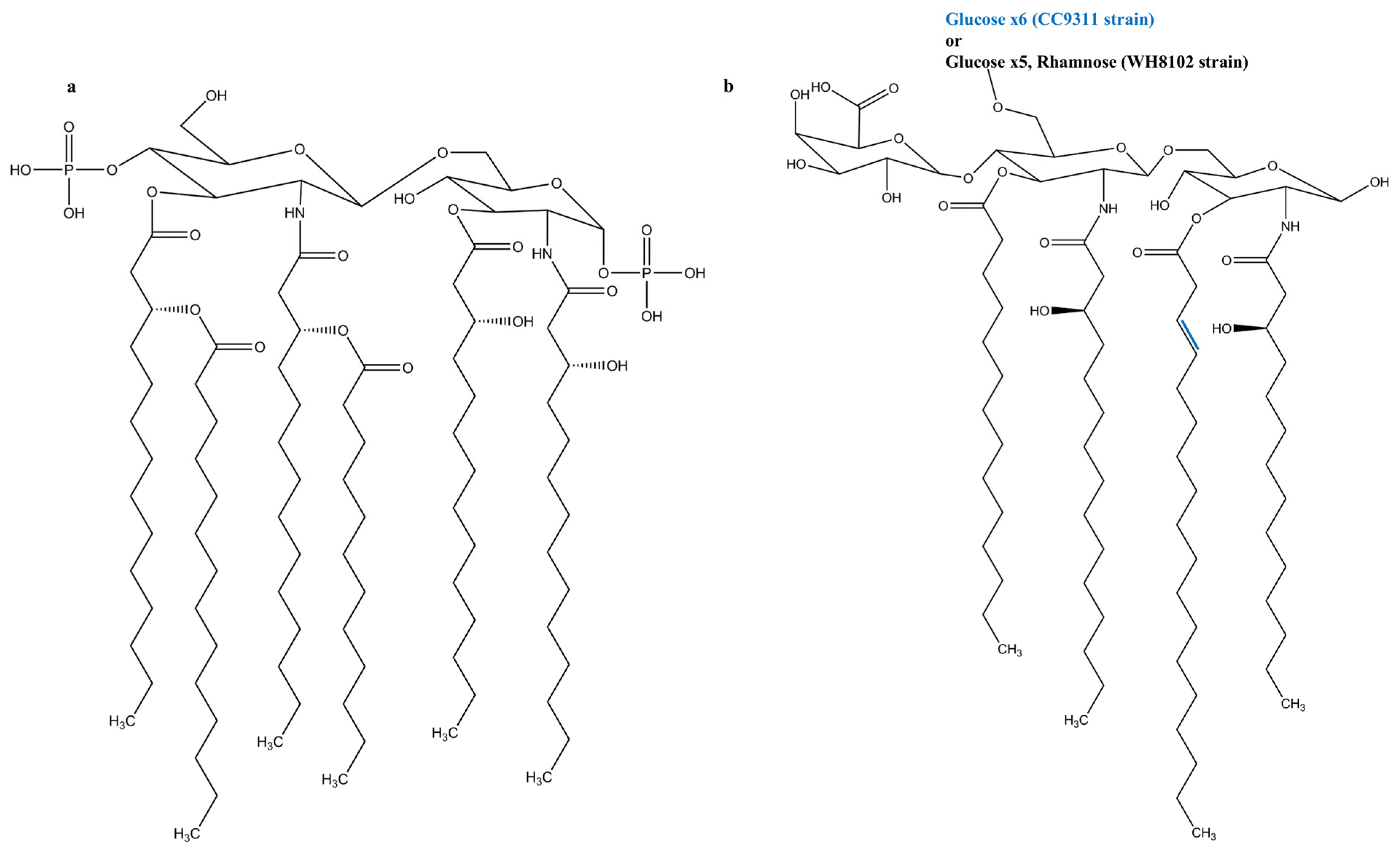
3. Cyanobacterial LPSs
| Cyanobacterial Species | Carbohydrates (%) | Phosphorus (%) | KDO (%) | Proteins (%) | Fatty Acids (%) | References |
|---|---|---|---|---|---|---|
| Schizothrix calcicola | 63 | <0.1 | Absent | 7.8 | 8 | [26] |
| Phormidium spp. | 60 | <1 | 0.5 | 7.20 | NA | [30] |
| Agmenellum quadruplicatum | 59.5 | 2.9 | 0.13 | 0.13 | 15.1 | [25] |
| Anabaena variabilis | 80.3 | 0.03 | Absent | 8.4 | 10.7 | [27] |
| Spirulina platensis | 31.6 | 0.6 | NA | 0.6 | 14.3 | [28] |
| Anacystis nidulans | 60 | 0.03 | 1.5 | NA | 12.4 | [23,24] |
| Microcystis aeruginosa | 36.0 | 0.7 | Absent | 0.4 | 18.2 | [20,21] |
| Anabaena flos-aquae | 65 | Absent | 12.5 | NA | NA | [27] |
4. Role of TLRs in Sepsis
5. LPSs and TLR4 Signaling
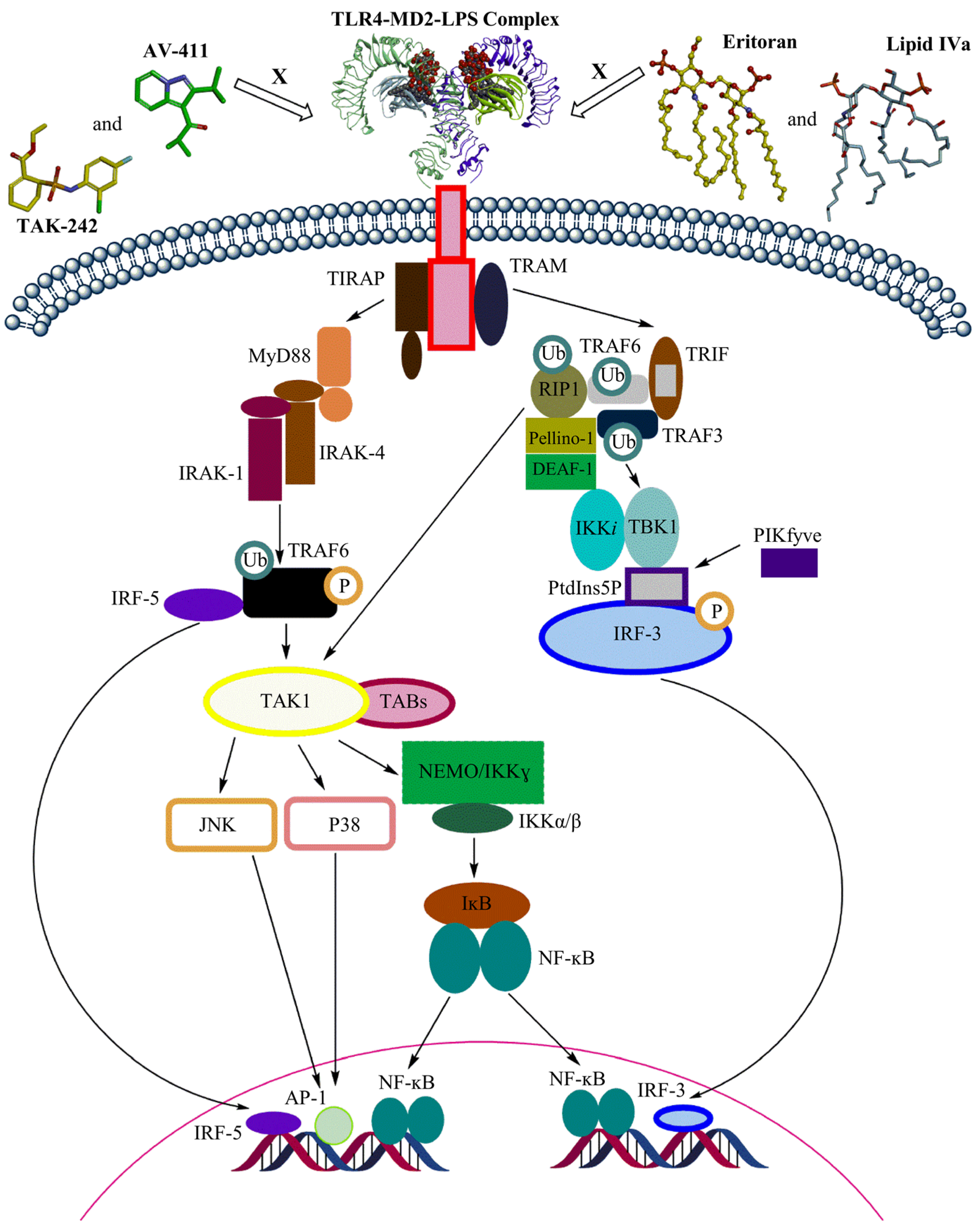
6. Cyanobacterial LPSs in TLR4 Signaling
7. Biological Activity of Cyanobacterial LPSs
8. Effects of Cyanobacterial LPSs on Humans
9. Effects of Cyanobacterial LPSs on Marine Organisms
Acknowledgements
Author Contributions
Conflict of Interest
References
- Berg, K.A.; Lyra, C.; Sivonen, K.; Paulin, L.; Suomalainen, S.; Tuomi, P.; Rapala, J. High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J. 2009, 3, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Santos, R.B.; Silva, C.R.; Verani, N.F.; Nonaka, K.O.; Rocha, O. Toxicity of a cyanobacteria bloom in barra bonita reservoir (middle tiete river, Sao Paulo, Brazil). Ecotoxicol. Environ. Saf. 2006, 64, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 2007, 68, 954–979. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.B.; Suseela, M.R. Cyanobacteria: Potential candidates for drug discovery. Antonie Van Leeuwenhoek 2013, 103, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Chorus, I.; Falconer, I.R.; Salas, H.J.; Bartram, J. Health risks caused by freshwater cyanobacteria in recreational waters. J. Toxicol. Environ. Health B Crit. Rev. 2000, 3, 323–347. [Google Scholar] [PubMed]
- Wang, L.; Liang, X.F.; Liao, W.Q.; Lei, L.M.; Han, B.P. Structural and functional characterization of microcystin detoxification-related liver genes in a phytoplanktivorous fish, nile tilapia (Oreochromis niloticus). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 144, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, C.; Pflugmacher, S. Ecotoxicological effects of selected cyanobacterial secondary metabolites: A short review. Toxicol. Appl. Pharmacol. 2005, 203, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.A.; Choi, S. Gram-negative marine bacteria: Structural features of lipopolysaccharides and their relevance for economically important diseases. Mar. Drugs 2014, 12, 2485–2514. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Holst, O.; Di Lorenzo, F.; Callaghan, M.; Nurisso, A.; D’Errico, G.; Zamyatina, A.; Peri, F.; Berisio, R.; Jerala, R.; et al. Chemistry of lipid a: At the heart of innate immunity. Chemistry 2015, 21, 500–519. [Google Scholar] [CrossRef] [PubMed]
- Hoiczyk, E.; Hansel, A. Cyanobacterial cell walls: News from an unusual prokaryotic envelope. J. Bacteriol. 2000, 182, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Leone, S.; Silipo, A.; Nazarenko, E.L.; Lanzetta, R.; Parrilli, M.; Molinaro, A. Molecular structure of endotoxins from Gram-negative marine bacteria: An update. Mar. Drugs 2007, 5, 85–112. [Google Scholar] [CrossRef] [PubMed]
- Snyder, D.S.; Brahamsha, B.; Azadi, P.; Palenik, B. Structure of compositionally simple lipopolysaccharide from marine synechococcus. J. Bacteriol. 2009, 191, 5499–5509. [Google Scholar] [CrossRef] [PubMed]
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003, 338, 2431–2447. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Quinn, P.J. Lipopolysaccharide: Biosynthetic pathway and structure modification. Prog. Lipid Res. 2010, 49, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Sperandeo, P.; Deho, G.; Polissi, A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta 2009, 1791, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Stewart, I.; Schluter, P.J.; Shaw, G.R. Cyanobacterial lipopolysaccharides and human health—A review. Environ. Health 2006, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.R. Cyanobacterial toxins and human health. Symp. Ser. Soc. Appl. Microbiol. 1998, 27, 35S–40S. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.G. Bacterial lipopolysaccharides—Themes and variations. Prog. Lipid Res. 1996, 35, 283–343. [Google Scholar] [CrossRef]
- Moran, A.P. Molecular structure, biosynthesis, and pathogenic roles of lipopolysaccharides. In Helicobacter pylori: Physiology and Genetics; Mobley, H.L.T., Mendz, G.L., Hazell, S.L., Eds.; ASM Press: Washington, DC, USA, 2001. [Google Scholar]
- Raziuddin, S.; Siegelman, H.W.; Tornabene, T.G. Lipopolysaccharides of the cyanobacterium Microcystis aeruginosa. Eur. J. Biochem. 1983, 137, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Codd, G.A.; Siegelman, H.W.; Weckesser, J. Lipopolysaccharides and polysaccharides of the cell envelope of toxic Microcystis aeruginosa strains. Arch. Microbiol. 1989, 152, 90–94. [Google Scholar] [CrossRef]
- Fujii, M.; Sato, Y.; Ito, H.; Masago, Y.; Omura, T. Monosaccharide composition of the outer membrane lipopolysaccharide and O-chain from the freshwater cyanobacterium Microcystis aeruginosa NIES-87. J. Appl. Microbiol. 2012, 113, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Weise, G.; Drews, G.; Jann, B.; Jann, K. Identification and analysis of a lipopolysaccharide in cell walls of the blue-green alga Anacystis nidulans. Arch. Mikrobiol. 1970, 71, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.; Weckesser, J.; Drews, G.; Mayer, H. Chemical and biological studies on the lipopolysaccharide (O-antigen) of Anacystis nidulans. Arch. Microbiol. 1977, 113, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Buttke, T.M.; Ingram, L.O. Comparison of lipopolysaccharides from Agmenellum quadruplicatum to Escherichia coli and Salmonella typhimurium by using thin-layer chromatography. J. Bacteriol. 1975, 124, 1566–1573. [Google Scholar] [PubMed]
- Keleti, G.; Sykora, J.L.; Lippy, E.C.; Shapiro, M.A. Composition and biological properties of lipopolysaccharides isolated from Schizothrix calcicola (Ag.) Gomont (Cyanobacteria). Appl. Environ. Microbiol. 1979, 38, 471–477. [Google Scholar] [PubMed]
- Keleti, G.; Sykora, J.L. Production and properties of cyanobacterial endotoxins. Appl. Environ. Microbiol. 1982, 43, 104–109. [Google Scholar] [PubMed]
- Tornabene, T.; Bourne, T.; Raziuddin, S.; Ben-Amotz, A. Lipid and lipopolysaccharide constituents of cyanobacterium Spirulina platensis (Cyanophyceae, Nostocales). Mar. Ecol. Prog. Ser. 1985, 22, 121–125. [Google Scholar] [CrossRef]
- Carillo, S.; Pieretti, G.; Bedini, E.; Parrilli, M.; Lanzetta, R.; Corsaro, M.M. Structural investigation of the antagonist LPS from the cyanobacterium Oscillatoria planktothrix FP1. Carbohydr. Res. 2014, 388, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Mikheyskaya, L.V.; Ovodova, R.G.; Ovodov, Y.S. Isolation and characterization of lipopolysaccharides from cell walls of blue-green algae of the genus Phormidium. J. Bacteriol. 1977, 130, 1–3. [Google Scholar] [PubMed]
- Ianaro, A.; Tersigni, M.; D’Acquisto, F. New insight in LPS antagonist. Mini Rev. Med. Chem. 2009, 9, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Savva, A.; Roger, T. Targeting Toll-like receptors: Promising therapeutic strategies for the management of sepsis-associated pathology and infectious diseases. Front. Immunol. 2013, 4, 387. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Roach, J.C.; Glusman, G.; Rowen, L.; Kaur, A.; Purcell, M.K.; Smith, K.D.; Hood, L.E.; Aderem, A. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 9577–9582. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Segal, D.M.; Davies, D.R. The structural biology of Toll-like receptors. Structure 2011, 19, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Durai, P.; Govindaraj, R.G.; Choi, S. Structure and dynamic behavior of Toll-like receptor 2 subfamily triggered by malarial glycosylphosphatidylinositols of Plasmodium falciparum. FEBS J. 2013, 280, 6196–6212. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Lee, J.O. Structural biology of the Toll-like receptor family. Annu. Rev. Biochem. 2011, 80, 917–941. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.S.; Lee, H.; Lee, J.O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Kawai, T.; Akira, S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012, 33, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Lee, J.O. Structures of the Toll-like receptor family and its ligand complexes. Immunity 2008, 29, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Nijland, R.; Hofland, T.; van Strijp, J.A. Recognition of LPS by TLR4: Potential for anti-inflammatory therapies. Mar. Drugs 2014, 12, 4260–4273. [Google Scholar] [CrossRef] [PubMed]
- Macagno, A.; Molteni, M.; Rinaldi, A.; Bertoni, F.; Lanzavecchia, A.; Rossetti, C.; Sallusto, F. A cyanobacterial LPS antagonist prevents endotoxin shock and blocks sustained TLR4 stimulation required for cytokine expression. J. Exp. Med. 2006, 203, 1481–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorgersen, E.B.; Macagno, A.; Rossetti, C.; Mollnes, T.E. Cyanobacterial LPS antagonist (CyP)—A novel and efficient inhibitor of Escherichia coli LPS-induced cytokine response in the pig. Mol. Immunol. 2008, 45, 3553–3557. [Google Scholar] [CrossRef] [PubMed]
- Jemmett, K.; Macagno, A.; Molteni, M.; Heckels, J.E.; Rossetti, C.; Christodoulides, M. A cyanobacterial lipopolysaccharide antagonist inhibits cytokine production induced by Neisseria meningitidis in a human whole-blood model of septicemia. Infect. Immun. 2008, 76, 3156–3163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, R.; Staples, K.J.; Heckels, J.; Rossetti, C.; Molteni, M.; Christodoulides, M. Coadministration of the cyanobacterial lipopolysaccharide antagonist CyP with antibiotic inhibits cytokine production by an in vitro meningitis model infected with Neisseria meningitidis. J. Antimicrob. Chemother. 2012, 67, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- De Paola, M.; Mariani, A.; Bigini, P.; Peviani, M.; Ferrara, G.; Molteni, M.; Gemma, S.; Veglianese, P.; Castellaneta, V.; Boldrin, V.; et al. Neuroprotective effects of Toll-like receptor 4 antagonism in spinal cord cultures and in a mouse model of motor neuron degeneration. Mol. Med. 2012, 18, 971–981. [Google Scholar] [PubMed]
- Raetz, C.R.; Reynolds, C.M.; Trent, M.S.; Bishop, R.E. Lipid a modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 2007, 76, 295–329. [Google Scholar] [CrossRef] [PubMed]
- Beasley, A.S.; Cotter, R.J.; Vogel, S.N.; Inzana, T.J.; Qureshi, A.A.; Qureshi, N. A variety of novel lipid a structures obtained from Francisella tularensis live vaccine strain. Innate. Immun. 2012, 18, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Schilling, B.; McLendon, M.K.; Phillips, N.J.; Apicella, M.A.; Gibson, B.W. Characterization of lipid a acylation patterns in Francisella tularensis, Francisella novicida, and Francisella philomiragia using multiple-stage mass spectrometry and matrix-assisted laser desorption/ionization on an intermediate vacuum source linear ion trap. Anal. Chem. 2007, 79, 1034–1042. [Google Scholar] [PubMed]
- Rose, J.R.; Christ, W.J.; Bristol, J.R.; Kawata, T.; Rossignol, D.P. Agonistic and antagonistic activities of bacterially derived Rhodobacter sphaeroides lipid A: Comparison with activities of synthetic material of the proposed structure and analogs. Infect. Immun. 1995, 63, 833–839. [Google Scholar] [PubMed]
- Blahova, L.; Adamovsky, O.; Kubala, L.; Svihalkova Sindlerova, L.; Zounkova, R.; Blaha, L. The isolation and characterization of lipopolysaccharides from Microcystis aeruginosa, a prominent toxic water bloom forming cyanobacteria. Toxicon 2013, 76, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Zanchett, G.; Oliveira-Filho, E.C. Cyanobacteria and cyanotoxins: From impacts on aquatic ecosystems and human health to anticarcinogenic effects. Toxins (Basel) 2013, 5, 1896–1917. [Google Scholar] [CrossRef] [PubMed]
- Rapala, J.; Lahti, K.; Rasanen, L.A.; Esala, A.L.; Niemela, S.I.; Sivonen, K. Endotoxins associated with cyanobacteria and their removal during drinking water treatment. Water Res. 2002, 36, 2627–2635. [Google Scholar] [CrossRef]
- Jakubowska, N.; Szelag-Wasielewska, E. Toxic picoplanktonic cyanobacteria—Review. Mar. Drugs 2015, 13, 1497–1518. [Google Scholar] [CrossRef] [PubMed]
- Best, J.H.; Pflugmacher, S.; Wiegand, C.; Eddy, F.B.; Metcalf, J.S.; Codd, G.A. Effects of enteric bacterial and cyanobacterial lipopolysaccharides, and of microcystin-LR, on glutathione S-transferase activities in zebra fish (Danio rerio). Aquat. Toxicol. 2002, 60, 223–231. [Google Scholar] [CrossRef]
- Jaja-Chimedza, A.; Gantar, M.; Mayer, G.D.; Gibbs, P.D.; Berry, J.P. Effects of cyanobacterial lipopolysaccharides from microcystis on glutathione-based detoxification pathways in the zebrafish (Danio rerio) embryo. Toxins (Basel) 2012, 4, 390–404. [Google Scholar] [CrossRef] [PubMed]
- Notch, E.G.; Miniutti, D.M.; Berry, J.P.; Mayer, G.D. Cyanobacterial LPS potentiates cadmium toxicity in zebrafish (Danio rerio) embryos. Environ. Toxicol. 2011, 26, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Ferrao-Filho Ada, S.; Kozlowsky-Suzuki, B. Cyanotoxins: Bioaccumulation and effects on aquatic animals. Mar. Drugs 2011, 9, 2729–2772. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durai, P.; Batool, M.; Choi, S. Structure and Effects of Cyanobacterial Lipopolysaccharides. Mar. Drugs 2015, 13, 4217-4230. https://doi.org/10.3390/md13074217
Durai P, Batool M, Choi S. Structure and Effects of Cyanobacterial Lipopolysaccharides. Marine Drugs. 2015; 13(7):4217-4230. https://doi.org/10.3390/md13074217
Chicago/Turabian StyleDurai, Prasannavenkatesh, Maria Batool, and Sangdun Choi. 2015. "Structure and Effects of Cyanobacterial Lipopolysaccharides" Marine Drugs 13, no. 7: 4217-4230. https://doi.org/10.3390/md13074217





