Actinomycetes from Red Sea Sponges: Sources for Chemical and Phylogenetic Diversity
Abstract
:1. Introduction
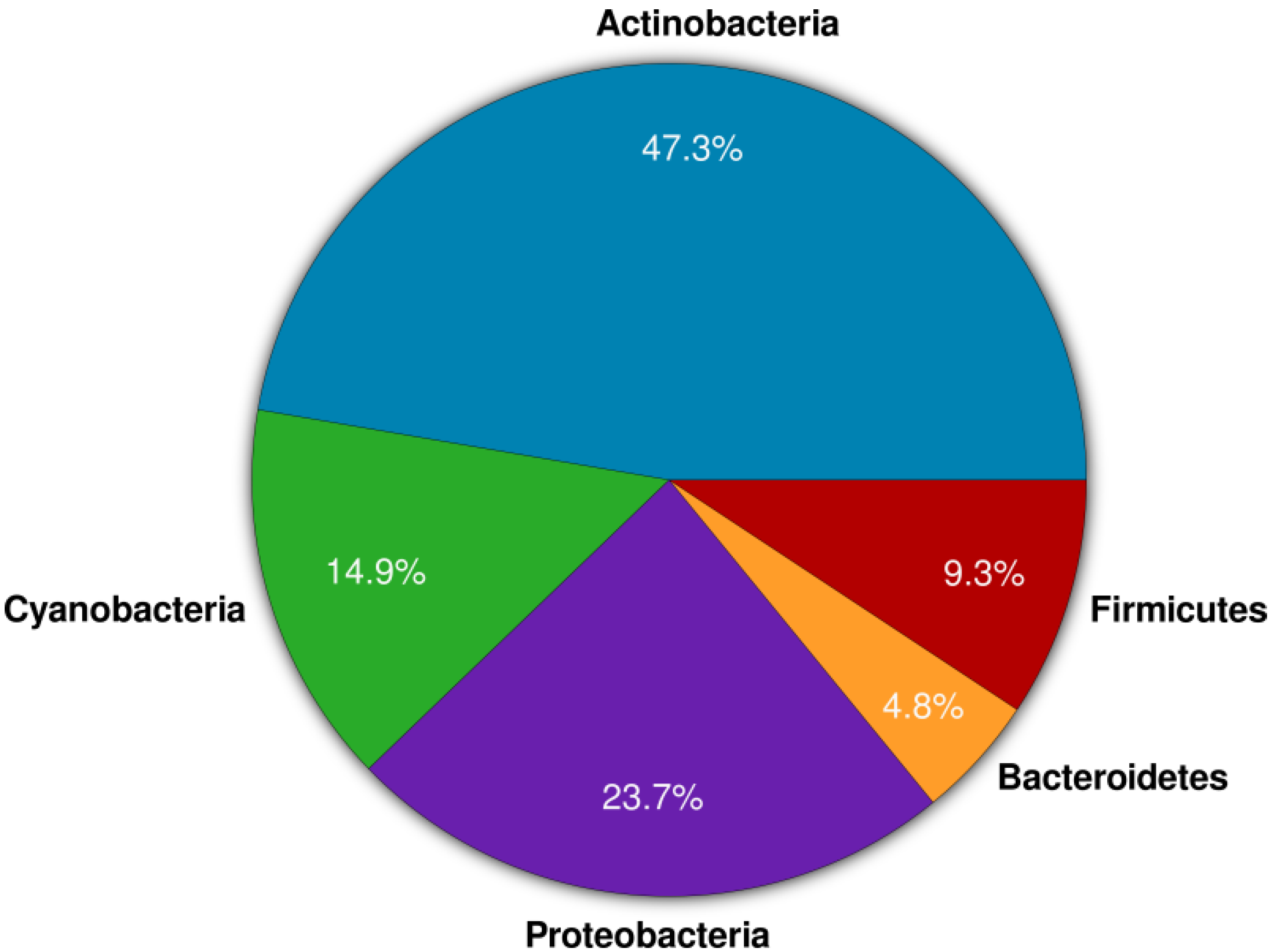
2. Results and Discussion
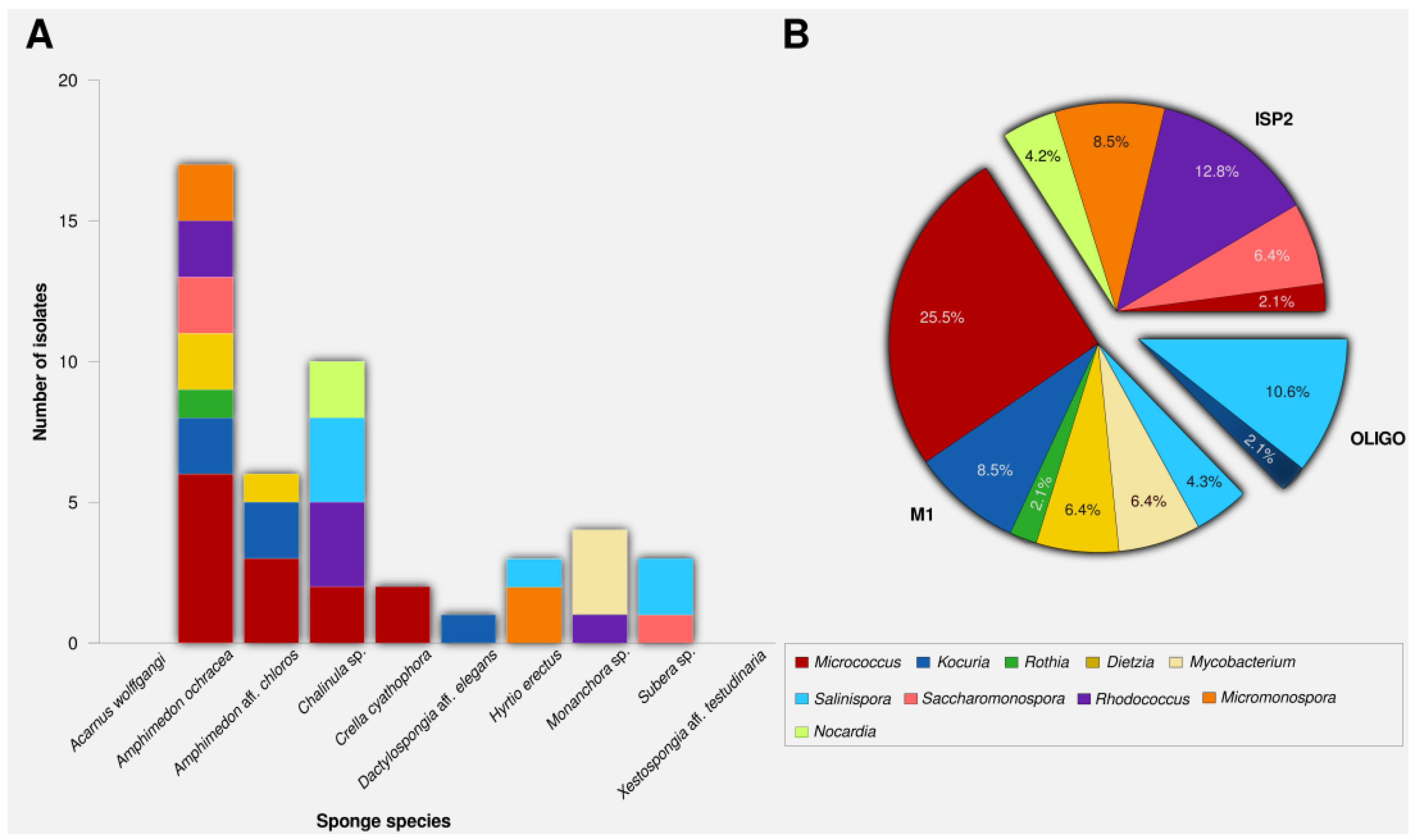
2.1. Actinomycete Isolation and Phylogenetic Identification
| Isolate Code | Isolation Medium | Sponge Source | Sequence Length (bp) | Closest Relative by BLAST | % Sequence Similarity |
|---|---|---|---|---|---|
| SA1 | M1 | Amphimedon ochracea | 1379 | Micrococcus sp. PN13 (KF554087) | 99.4 |
| SA2 | ISP2 | Amphimedon aff. chloros | 1435 | Micrococcus luteus strain DAG I (KC470045) | 99.7 |
| SA3 | ISP2 | Amphimedon aff. chloros | 1360 | Micrococcus sp. X2Bc2 (KF465977) | 99.5 |
| SA4 | ISP2 | Hyrtios erectus | 1423 | Micromonospora sp. S6 (HF674982) | 99.7 |
| SA5 | OLIGO | Chalinula sp. | 1395 | Salinispora arenicola CNS-205 (NR_074612) | 99.9 |
| SA6 | OLIGO | Chalinula sp. | 1372 | Salinispora arenicola strain SCSIOZ-SH19 (KC747487) | 100.0 |
| * SA7 | ISP2 | Chalinula sp. | 1338 | Nocardia sp. W9912 (GU992878) | 98.2 |
| * SA8 | ISP2 | Chalinula sp. | 1461 | Rhodococcus equi (AB738794) | 97.4 |
| * SA9 | ISP2 | Monanchora sp. | 1378 | Rhodococcus sp. L-15 (JN006270) | 97.9 |
| SA10 | M1 | Chalinula sp. | 1397 | Micrococcus sp. PA-E028 (FJ233852) | 99.8 |
| * SA11 | M1 | Monanchora sp. | 1416 | Mycobacterium sp. CNJ879 PL04 (DQ448780) | 97.6 |
| * SA12 | ISP2 | Chalinula sp. | 1374 | Rhodococcus sp. HL-3 (JF734314) | 97.9 |
| * SA13 | M1 | Monanchora sp. | 1419 | Mycobacterium sp. CNJ879 PL04 (DQ448780) | 97.8 |
| * SA14 | M1 | Amphimedon ochracea | 1341 | Kocuria sp. PN5 (KF554079) | 97.5 |
| * SA15 | M1 | Amphimedon aff. chloros | 1347 | Kocuria sp. SS263-23 (JX429815) | 97.3 |
| SA16 | M1 | Amphimedon aff. chloros | 1395 | Rothia terrae strain F77052 (HQ908743) | 99.7 |
| SA17 | ISP2 | Crella cyathophora | 1401 | Micrococcus sp. X-48(JX997909) | 99.9 |
| SA18 | M1 | Amphimedon aff. chloros | 1413 | Dietzia maris strain KMGL1309-AS3 (KF740541) | 100.0 |
| SA19 | OLIGO | Chalinula sp. | 1325 | Salinispora pacifica strain AMS365 (HQ873949) | 99.9 |
| SA20 | M1 | Subera sp. | 1330 | Salinispora arenicola strain SCSIOZ-SH19 (KC747487) | 100.0 |
| SA21 | ISP2 | Subera sp. | 1443 | Saccharomonospora azurea strain RR1 (KC855265) | 99.2 |
| SA22 | OLIGO | Subera sp. | 1337 | Salinispora pacifica strain S34 (JX007964) | 99.7 |
| SA23 | ISP2 | Amphimedon aff. chloros | 1396 | Saccharomonospora sp. G2Z21 (JF806667) | 99.8 |
| SA24 | M1 | Dactylospongia aff. elegans | 1494 | Kocuria palustris strain LJ27 (KF515677) | 99.1 |
| SA25 | OLIGO | Hyrtios erectus | 1453 | Salinispora arenicola strain SCSIOZ-SH11 (KC747479) | 99.9 |

2.2. PCR-Screening for PKS and NRPS Domains
| Isolate Code | Closest Relative by BLASTX (Sequence Length bp, % Sequence Similarity) | ||
|---|---|---|---|
| NRPS | PKS I | PKS II | |
| SA1 | Amino acid adenylation domain-containing protein [Micrococcus luteus (CP001628)] (347, 51) | - | - |
| SA2 | Non-ribosomal peptide synthetase [Micrococcus luteus (EFD52022)] (417, 56) | - | - |
| SA3 | Amino acid adenylation domain-containing protein [Micrococcus luteus (CP001628)] (511, 62) | - | - |
| SA4 | Amino acid adenylation domain-containing protein [Micromonospora aurantiaca (WP_013285021)] (501, 79) | Polyketide synthase [Micromonospora sp. CNB394 (WP_018787726)] (473, 67) | Type II PKS ketosynthase, partial [Micromonospora sp. SCSIO11524 (AHB18630)] (541, 63) |
| SA5 | Amino acid adenylation domain-containing protein [Salinispora arenicola (WP_012184557)] (521, 72) | Polyketide synthase [Salinispora arenicola (WP_018795623)] (541, 64) | Beta-ACP synthase, partial [Salinispora arenicola (WP_020608853)] (573, 70) |
| SA6 | Amino acid adenylation domain-containing protein [Salinispora arenicola (YP_001535283)] (513, 77) | Polyketide synthase [Salinispora arenicola (WP_018795623)] (516, 71) | KAS II [Salinispora arenicola (WP_020608853)] (581, 67) |
| * SA7 | - | - | Polyketide synthase [Nocardia nova SH22a (AHH16328)] (614, 85) |
| * SA8 | Non-ribosomal peptide synthetase [Rhodococcus equi (CBH48735)] (570, 69) | Putative polyketide synthase [Rhodococcus equi (WP_022593366)] (518, 65) | - |
| * SA9 | Non-ribosomal peptide synthetase, partial [Rhodococcus qingshengii (WP_007730195)] (609, 68) | Putative polyketide synthase [Rhodococcus opacus B4 (BAH55256)] (428, 75) | - |
| SA10 | - | - | - |
| * SA11 | - | - | - |
| * SA12 | Non-ribosomal peptide synthetase [Rhodococcus opacus (WP_005253470)] (622, 58) | Putative polyketide synthase [Rhodococcus opacus (BAH55256)] (428, 75) | - |
| * SA13 | - | - | - |
| * SA14 | Non-ribosomal peptide synthetase [Kocuria rhizophila (BAG29492)] (532, 74) | - | - |
| *SA15 | Non-ribosomal peptide synthetase [Kocuria rhizophila (BAG29492)] (462, 68) | - | - |
| SA16 | - | - | - |
| SA17 | Non-ribosomal peptide synthetase [Micrococcus luteus (EFD52022) (423, 66) | - | - |
| SA18 | - | - | - |
| SA19 | Adenylation domain of nonribosomal peptide synthetase [Salinispora pacifica (WP_018724218)] (465, 58) | Polyketide synthase [Salinispora pacifica (WP_018824659)] (421, 59) | Beta-ACP synthase [Salinispora pacifica (WP_018720155)] (541, 61) |
| SA20 | Amino acid adenylation domain-containing protein [Salinispora arenicola (WP_012184557)] (465, 58) | Polyketide synthase [Salinispora arenicola (WP_019032802)] (506, 61) | KAS II [Salinispora arenicola (WP_020608853)] (529, 57) |
| SA21 | Nonribosomal peptide synthetase [Saccharopolyspora spinosa (WP_010314019)] (665, 74) | Polyketide synthase [Saccharopolyspora spinosa (WP_010311945)] (565, 69) | - |
| SA22 | Amino acid adenylation domain of nonribosomal peptide synthetase [Salinispora pacifica (WP_018724218)] (476, 59) | Polyketide synthase [Salinispora pacifica (WP_018824659)] (501, 63) | Beta-ACP synthase [Salinispora pacifica (WP_018823591)] (531, 71) |
| SA23 | Amino acid adenylation domain-containing protein [Saccharopolyspora spinosa (WP_010694383)] (643, 71) | Polyketide synthase [Saccharopolyspora erythraea (WP_011873765)] (533, 61) | - |
| SA24 | Nonribosomal peptide synthetase [Kocuria rhizophila (WP_019309050)] (592, 73) | - | - |
| SA25 | Amino acid adenylation domain-containing protein [Salinispora arenicola (YP_001539321)] (453, 68) | Rifamycin polyketide synthase [Salinispora arenicola (WP_020217874)] (578, 71) | Ketosynthase, partial [Salinispora arenicola AFO70123] (548, 63) |
2.3. Anti-Infective Screening
| Isolate Code | Inhibition Zone Diameter (mm) | IC50 (μg/mL, 72 h) | Growth Inhibition (%) | ||
|---|---|---|---|---|---|
| Bacillus sp. P25 | Escherichia coli K12 | Fusarium sp. P21 | Trypanosoma brucei TC 221 | West Nile Virus Protease | |
| Micrococcus sp. SA1E | 8 | 12 | - | <10 | - |
| Micrococcus sp. SA3E | - | - | 14 | - | - |
| Salinispora sp. SA6E | 20 | 7 | 22 | <10 | 84 |
| Salinispora sp. SA22E | 18 | 9 | 15 | <10 | 79 |
| * Rhodococcus sp. SA9E | - | - | 13 | - | - |
| * Rhodococcus sp. SA12E | - | - | 16 | - | 93 |
| Mycobacterium sp. SA11E | 14 | - | - | <10 | - |
| Saccharomonospora sp. SA21 E | 10 | 12 | - | - | - |
| Saccharomonospora sp. SA23 E | 11 | 13 | 17 | <10 | - |
3. Experimental Section
3.1. Sponge Collection
3.2. Actinomycete Isolation and Identification
3.3. PCR Screening of NRPS and PKS-II Gene Fragments
3.4. Secondary Metabolites Extraction and Bioactivity Testing
3.4.1. Antibacterial and Antifungal Activities Testing
3.4.2. Anti-Trypanosomal Activity
3.4.3. Anti-Leishmanial Activity
3.4.4. West Nile Virus NS3 Protease Inhibition Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hentschel, U.; Piel, J.; Degnan, S.M.; Taylor, M.W. Genomic insights into the marine sponge microbiome. Nature 2012, 10, 641–654. [Google Scholar]
- Van Soest, R.W.; Boury-Esnault, N.; Vacelet, J.; Dohrmann, M.; Erpenbeck, D.; de Voogd, N.J.; Santodomingo, N.; Vanhoorne, B.; Kelly, M.; Hooper, J.N. Global diversity of sponges (Porifera). PLoS One 2012, 7, e35105. [Google Scholar] [CrossRef]
- Schmitt, S.; Tsai, P.; Bell, J.; Fromont, J.; Ilan, M.; Lindquist, N.; Perez, T.; Rodrigo, A.; Schupp, P.J.; Vacelet, J.; et al. Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012, 6, 564–576. [Google Scholar] [CrossRef]
- Belarbi el, H.; Contreras Gomez, A.; Chisti, Y.; Garcia Camacho, F.; Molina Grima, E. Producing drugs from marine sponges. Biotechnol. Adv. 2003, 21, 585–598. [Google Scholar] [CrossRef]
- Webster, N.S.; Taylor, M.W. Marine sponges and their microbial symbionts: Love and other relationships. Environ. Microbiol. 2012, 14, 335–346. [Google Scholar] [CrossRef]
- Hentschel, U.; Fieseler, L.; Wehrl, A.; Gernert, C.; Steinert, M.; Hacker, J.; Horn, M. Microbial diversity of marine sponges. In Sponges (Porifera); Müller, W.E.G., Ed.; Springer: Berlin, Germany, 2003; pp. 59–88. [Google Scholar]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Goodfellow, M.; Williams, S.T. Ecology of actinomycetes. Annu. Rev. Microbiol. 1983, 37, 189–216. [Google Scholar] [CrossRef]
- Garrity, G.M.; Bell, J.A.; Lilburn, T.G. Taxonomic outline of the prokaryotes. In Bergey’s Manual of Systematic Bacteriology; Gray, M.W., Burger, G., Lang, B.F., Eds.; Springer: New York, NY, USA, 2004; pp. 204–301. [Google Scholar]
- Adegboye, M.F.; Babalola, O.O. Taxonomy and ecology of antibiotic producing actinomycetes. Afr. J. Agric. Res. 2012, 15, 2255–2261. [Google Scholar]
- Andrade, M.R.M.; Amaral, E.P.; Ribeiro, S.C.M.; Almeida, F.M.; Peres, T.V.; Lanes, V.; D’Imperio-Lima, M.R.; Lasunskaia, E.B. Pathogenic Mycobacterium bovis strains differ in their ability to modulate the proinflammatory activation phenotype of macrophages. BMC Microbiol. 2012, 12, 166. [Google Scholar] [CrossRef] [Green Version]
- Salem, H.; Kreutzer, E.; Sudakaran, S.; Kaltenpoth, M. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ. Microbiol. 2013, 15, 1956–1968. [Google Scholar] [CrossRef]
- Sun, W.; Dai, S.; Jiang, S.; Wang, G.; Liu, G.; Wu, H.; Li, X. Culture-dependent and culture-independent diversity of Actinobacteria associated with the marine sponge Hymeniacidon perleve from the South China Sea. Anton Van Leeuwenhoek 2010, 98, 65–75. [Google Scholar] [CrossRef]
- Zhang, L.M.; Xi, L.J.; Ruan, J.S.; Huang, Y. Microbacterium marinum sp nov., isolated from deep-sea water. Syst. Appl. Microbiol. 2012, 35, 81–85. [Google Scholar] [CrossRef]
- Becerril-Espinosa, A.; Freel, K.C.; Jensen, P.R.; Soria-Mercado, I.E. Marine actinobacteria from the Gulf of California: Diversity, abundance and secondary metabolite biosynthetic potential. Antonie Van Leeuwenhoek 2013, 103, 809–819. [Google Scholar] [CrossRef]
- Sun, W.; Peng, C.S.; Zhao, Y.Y.; Li, Z.Y. Functional gene-guided discovery of type II polyketides from culturable actinomycetes associated with soft coral Scleronephthya sp. PLoS One 2012, 7, e42847. [Google Scholar]
- Abdelmohsen, U.R.; Pimentel-Elardo, S.M.; Hanora, A.; Radwan, M.; Abou-El-Ela, S.H.; Ahmed, S.; Hentschel, U. Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated actinomycetes. Mar. Drugs 2010, 8, 399–412. [Google Scholar] [CrossRef]
- Wyche, T.P.; Hou, Y.P.; Vazquez-Rivera, E.; Braun, D.; Bugni, T.S. Peptidolipins B–F, antibacterial lipopeptides from an ascidian-derived Nocardia sp. J. Nat. Prod. 2012, 75, 735–740. [Google Scholar] [CrossRef]
- Zhang, H.; Lee, Y.K.; Zhang, W.; Lee, H.K. Culturable actinobacteria from the marine sponge Hymeniacidon perleve: Isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Anton Van Leeuwenhoek 2006, 90, 159–169. [Google Scholar] [CrossRef]
- Xi, L.; Ruan, J.; Huang, Y. Diversity and biosynthetic potential of culturable actinomycetes associated with marine sponges in the china seas. Int. J. Mol. Sci. 2012, 13, 5917–5932. [Google Scholar] [CrossRef]
- Lam, K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 2006, 9, 245–251. [Google Scholar] [CrossRef]
- Thomas, T.R.A.; Kavlekar, D.P.; LokaBharathi, P.A. Marine drugs from sponge-microbe association-A Review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar] [CrossRef]
- Pimentel-Elardo, S.M.; Kozytska, S.; Bugni, T.S.; Ireland, C.M.; Moll, H.; Hentschel, U. Anti-parasitic compounds from Streptomyces sp. strains isolated from Mediterranean sponges. Mar. Drugs 2010, 8, 373–380. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2013, 30, 237–323. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Szesny, M.; Othman, E.M.; Schirmeister, T.; Grond, S.; Stopper, H.; Hentschel, U. Antioxidant and anti-Protease activities of diazepinomicin from the sponge-associated Micromonospora strain RV115. Mar. Drugs 2012, 10, 2208–2221. [Google Scholar] [CrossRef]
- Bull, A.T.; Stach, J.E. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef]
- Solanki, R.; Khanna, M.; Lal, R. Bioactive compounds from marine actinomycetes. Indian J. Microbiol. 2008, 48, 410–431. [Google Scholar] [CrossRef]
- Subramani, R.; Aalbersberg, W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Tiwari, K.; Gupta, R.K. Rare actinomycetes: A potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 2012, 32, 108–132. [Google Scholar] [CrossRef]
- Donadio, S.; Monciardini, P.; Sosio, M. Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Nat. Prod. Rep. 2007, 24, 1073–1109. [Google Scholar] [CrossRef]
- Fieseler, L.; Hentschel, U.; Grozdanov, L.; Schirmer, A.; Wen, G.; Platzer, M.; Hrvatin, S.; Butzke, D.; Zimmermann, K.; Piel, J. Widespread occurrence and genomic context of unusually small polyketide synthase genes in microbial consortia associated with marine sponges. Appl. Environ. Microbiol. 2007, 73, 2144–2155. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Ntai, I.; Ju, K.S.; Unger, M.; Zamdborg, L.; Robinson, S.J.; Doroghazi, J.R.; Labeda, D.P.; Metcalf, W.W.; Kelleher, N.L. A proteomic survey of nonribosomal peptide and polyketide biosynthesis in Actinobacteria. J. Proteome Res. 2012, 11, 85–94. [Google Scholar] [CrossRef]
- Plahn, O.; Baschek, B.; Badewien, T.H.; Walter, M.; Rhein, M. Importance of the Gulf of Aqaba for the formation of bottom water in the Red Sea. J. Geophys. Res. Ocean. 2002, 107, 22/1–22/18. [Google Scholar]
- Ilan, M.; Gugel, J.; van Soest, R.W.M. Taxonomy, reproduction and ecology of new and known Red Sea sponges. Sarsia 2004, 89, 388–410. [Google Scholar] [CrossRef]
- Ngugi, D.K.; Antunes, A.; Brune, A.; Stingl, U. Biogeography of pelagic bacterioplankton across an antagonistic temperature-salinity gradient in the Red Sea. Mol. Ecol. 2012, 21, 388–405. [Google Scholar] [CrossRef]
- Radwan, M.; Hanora, A.; Zan, J.; Mohamed, N.M.; Abo-Elmatty, D.M.; Abou-El-Ela, S.H.; Hill, R.T. Bacterial community analyses of two Red Sea sponges. Mar. Biotechnol. 2010, 12, 350–360. [Google Scholar] [CrossRef]
- Vicente, J.; Stewart, A.; Song, B.; Hill, R.; Wright, J. Biodiversity of actinomycetes associated with Caribbean sponges and their potential for natural product discovery. Mar. Biotechnol. (N. Y.) 2013, 15, 413–424. [Google Scholar] [CrossRef]
- Montalvo, N.F.; Mohamed, N.M.; Enticknap, J.J.; Hill, R.T. Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 2005, 87, 29–36. [Google Scholar] [CrossRef]
- Xin, Y.; Wu, P.; Deng, M.; Zhang, W. Phylogenetic diversity of the culturable rare actinomycetes in marine sponge Hymeniacidon perlevis by improved isolation media. Wei Sheng Wu Xue Bao 2009, 49, 859–866. [Google Scholar]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381–399. [Google Scholar] [CrossRef]
- Webster, N.S.; Wilson, K.J.; Blackall, L.L.; Hill, R.T. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 2001, 67, 434–444. [Google Scholar] [CrossRef]
- Mincer, T.J.; Fenical, W.; Jensen, P.R. Culture-dependent and culture-independent diversity within the obligate marine actinomycete genus Salinispora. Appl. Environ. Microbiol. 2005, 71, 7019–7028. [Google Scholar] [CrossRef]
- Prieto-Davo, A.; Villarreal-Gomez, L.J.; Forschner-Dancause, S.; Bull, A.T.; Stach, J.E.; Smith, D.C.; Rowley, D.C.; Jensen, P.R. Targeted search for actinomycetes from near-shore and deep-sea marine sediments. FEMS Microbiol. Ecol. 2013, 84, 510–518. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Kirkland, T.N.; Jensen, P.R.; Fenical, W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot. (Tokyo) 2010, 63, 37–39. [Google Scholar] [CrossRef]
- Hodges, T.W.; Slattery, M.; Olson, J.B. Unique actinomycetes from marine caves and coral reef sediments provide novel PKS and NRPS biosynthetic gene clusters. Mar. Biotechnol. 2012, 14, 270–280. [Google Scholar] [CrossRef]
- Schneemann, I.; Nagel, K.; Kajahn, I.; Labes, A.; Wiese, J.; Imhoff, J.F. Comprehensive investigation of marine actinobacteria associated with the sponge Halichondria panicea. Appl. Environ. Microbiol. 2010, 76, 3702–3714. [Google Scholar] [CrossRef] [Green Version]
- Bultel-Ponce, V.V.; Debitus, C.; Berge, J.P.; Cerceau, C.; Guyot, M. Metabolites from the sponge-associated bacterium Micrococcus luteus. J. Mar. Biotechnol. 1998, 6, 233–236. [Google Scholar]
- Palomo, S.; Gonzalez, I.; de la Cruz, M.; Martin, J.; Tormo, J.R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef]
- Forner, D.; Berrue, F.; Correa, H.; Duncan, K.; Kerr, R.G. Chemical dereplication of marine actinomycetes by liquid chromatography-high resolution mass spectrometry profiling and statistical analysis. Anal. Chim. Acta 2013, 805, 70–79. [Google Scholar] [CrossRef]
- Chauhan, D.; Catley, L.; Li, G.; Podar, K.; Hideshima, T.; Velankar, M.; Mitsiades, C.; Mitsiades, N.; Yasui, H.; Letai, A.; et al. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell 2005, 8, 407–419. [Google Scholar] [CrossRef]
- Kim, T.K.; Hewavitharana, A.K.; Shaw, P.N.; Fuerst, J.A. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl. Environ. Microbiol. 2006, 72, 2118–2125. [Google Scholar] [CrossRef]
- Jensen, P.R.; Williams, P.G.; Oh, D.C.; Zeigler, L.; Fenical, W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 2007, 73, 1146–1152. [Google Scholar] [CrossRef]
- Williams, P.G.; Asolkar, R.N.; Kondratyuk, T.; Pezzuto, J.M.; Jensen, P.R.; Fenical, W. Saliniketals A and B, bicyclic polyketides from the marine actinomycete Salinispora arenicola. J. Nat. Prod. 2007, 70, 83–88. [Google Scholar] [CrossRef]
- Oh, D.C.; Gontang, E.A.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinipyrones and pacificanones, mixed-precursor polyketides from the marine actinomycete Salinispora pacifica. J. Nat. Prod. 2008, 71, 570–575. [Google Scholar] [CrossRef]
- May, F.J.; Davis, C.T.; Tesh, R.B.; Barrett, A.D. Phylogeography of West Nile virus: From the cradle of evolution in Africa to Eurasia, Australia, and the Americas. J. Virol. 2011, 85, 2964–2974. [Google Scholar] [CrossRef]
- De Filette, M.; Soehle, S.; Ulbert, S.; Richner, J.; Diamond, M.S.; Sinigaglia, A.; Barzon, L.; Roels, S.; Lisziewicz, J.; Lorincz, O.; Sanders, N.N. Vaccination of mice using the West Nile Virus E-Protein in a DNA prime-protein boost strategy stimulates cell-mediated immunity and protects mice against a lethal challenge. PLoS One 2014, 9, e87837. [Google Scholar] [CrossRef] [Green Version]
- Samanta, S.; Cui, T.; Lam, Y. Discovery, synthesis, and in vitro in vitro evaluation of West Nile virus protease inhibitors based on the 9,10-dihydro-3H,4aH-1,3,9,10a-tetraazaphenanthren-4-one scaffold. ChemMedChem 2012, 7, 1210–1216. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Hofs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Zhang, G.L.; Philippe, A.; Schmitz, W.; Pimentel-Elardo, S.M.; Hertlein-Amslinger, B.; Hentschel, U.; Bringmann, G. Cyclodysidins A7#x2013;D, cyclic lipopeptides from the marine sponge-derived Streptomyces strain RV15. Tetrahedron Lett. 2012, 53, 23–29. [Google Scholar] [CrossRef]
- Ashelford, K.E.; Chuzhanova, N.A.; Fry, J.C.; Jones, A.J.; Weightman, A.J. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 2005, 71, 7724–7736. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glockner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Keane, T.M.; Creevey, C.J.; Pentony, M.M.; Naughton, T.J.; McLnerney, J.O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006, 6, 29. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Stover, B.C.; Muller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef]
- Ayuso-Sacido, A.; Genilloud, O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: Detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005, 49, 10–24. [Google Scholar] [CrossRef]
- Ayuso, A.; Clark, D.; Gonzalez, I.; Salazar, O.; Anderson, A.; Genilloud, O. A novel actinomycete strain de-replication approach based on the diversity of polyketide synthase and nonribosomal peptide synthetase biosynthetic pathways. Appl. Microbiol. Biotechnol. 2005, 67, 795–806. [Google Scholar] [CrossRef]
- Huber, W.; Koella, J.C. A comparison of three methods of estimating EC50 in studies of drug resistance of malaria parasites. Acta Trop. 1993, 55, 257–261. [Google Scholar] [CrossRef]
- Ponte-Sucre, A.; Vicik, R.; Schultheis, M.; Schirmeister, T.; Moll, H. Aziridine-2,3-dicarboxylates, peptidomimetic cysteine protease inhibitors with antileishmanial activity. Antimicrob. Agents Chemother. 2006, 50, 2439–2447. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Abdelmohsen, U.R.; Yang, C.; Horn, H.; Hajjar, D.; Ravasi, T.; Hentschel, U. Actinomycetes from Red Sea Sponges: Sources for Chemical and Phylogenetic Diversity. Mar. Drugs 2014, 12, 2771-2789. https://doi.org/10.3390/md12052771
Abdelmohsen UR, Yang C, Horn H, Hajjar D, Ravasi T, Hentschel U. Actinomycetes from Red Sea Sponges: Sources for Chemical and Phylogenetic Diversity. Marine Drugs. 2014; 12(5):2771-2789. https://doi.org/10.3390/md12052771
Chicago/Turabian StyleAbdelmohsen, Usama Ramadan, Chen Yang, Hannes Horn, Dina Hajjar, Timothy Ravasi, and Ute Hentschel. 2014. "Actinomycetes from Red Sea Sponges: Sources for Chemical and Phylogenetic Diversity" Marine Drugs 12, no. 5: 2771-2789. https://doi.org/10.3390/md12052771
APA StyleAbdelmohsen, U. R., Yang, C., Horn, H., Hajjar, D., Ravasi, T., & Hentschel, U. (2014). Actinomycetes from Red Sea Sponges: Sources for Chemical and Phylogenetic Diversity. Marine Drugs, 12(5), 2771-2789. https://doi.org/10.3390/md12052771





