Multivalent Anchoring and Oriented Display of Single-Domain Antibodies on Cellulose
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Plasmids
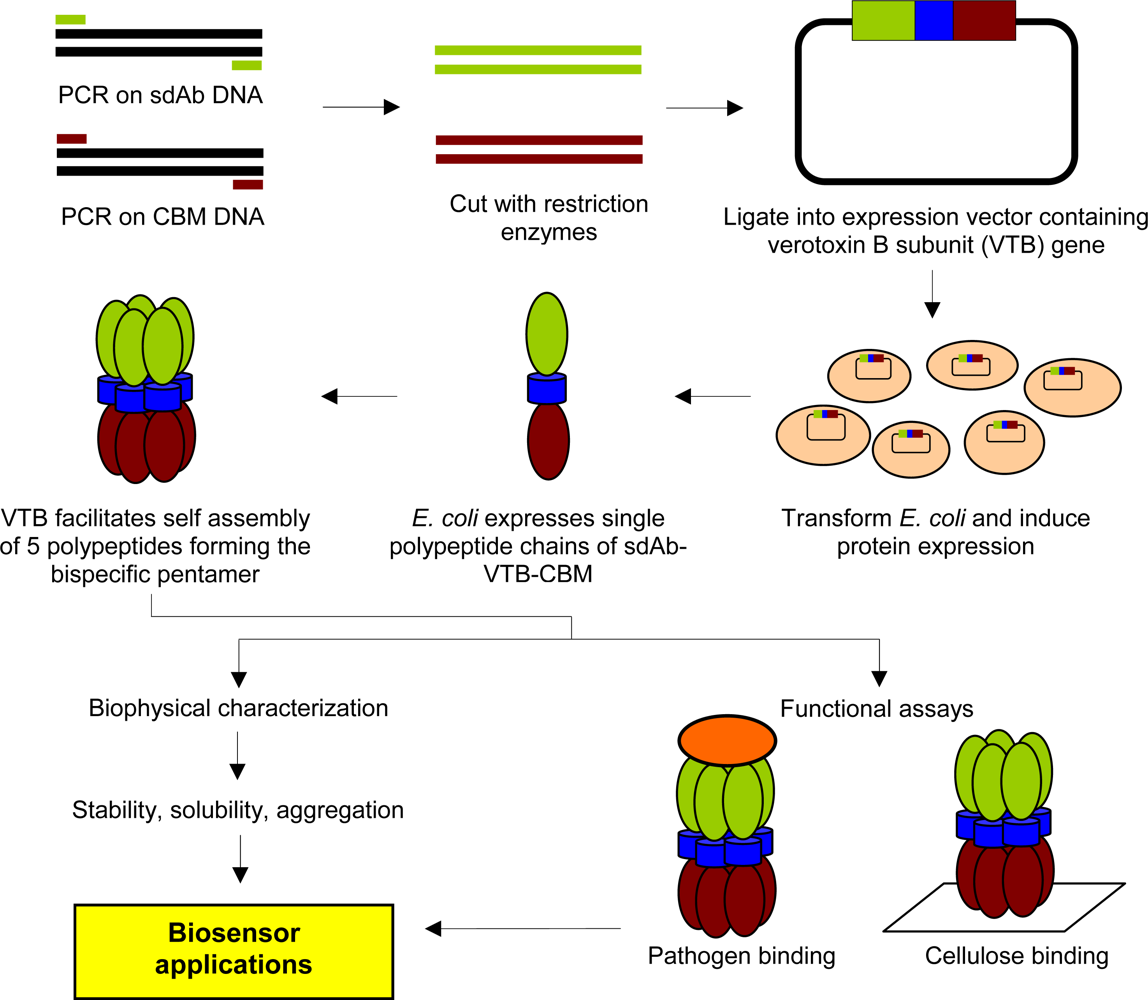
2.2. Cloning and Expression of sdAb-CBM Fusions
2.3. Purification of Recombinant Proteins and Size Exclusion Chromatography
2.4. Microagglutination Assays
2.5. Cellulose Binding Assays
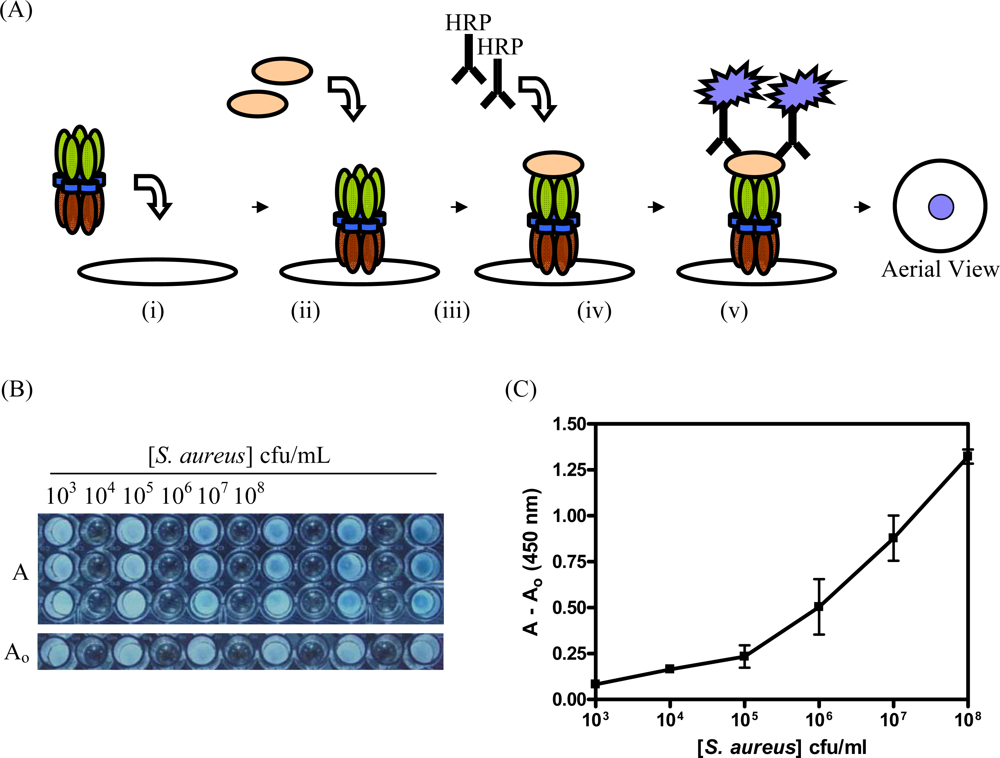
2.6 Detection of S. aureus with CBM9-VTB-HVHP428
3. Results
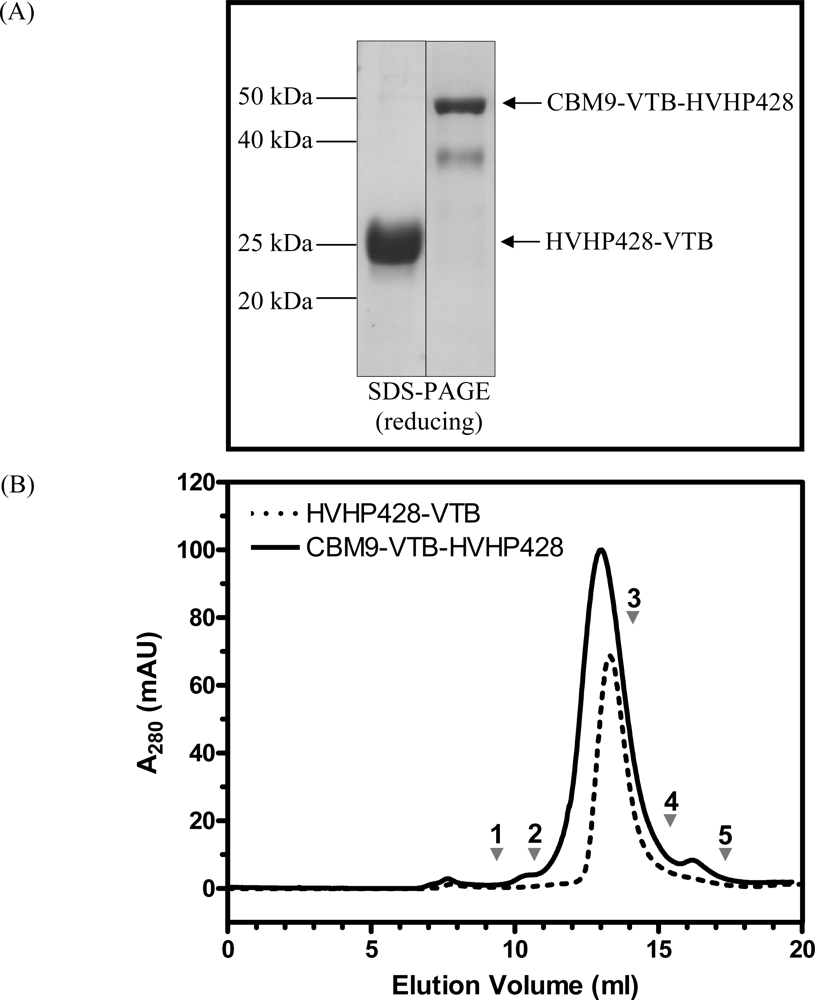
3.1. Expression and Characterization of CBM-sdAb Fusion Proteins
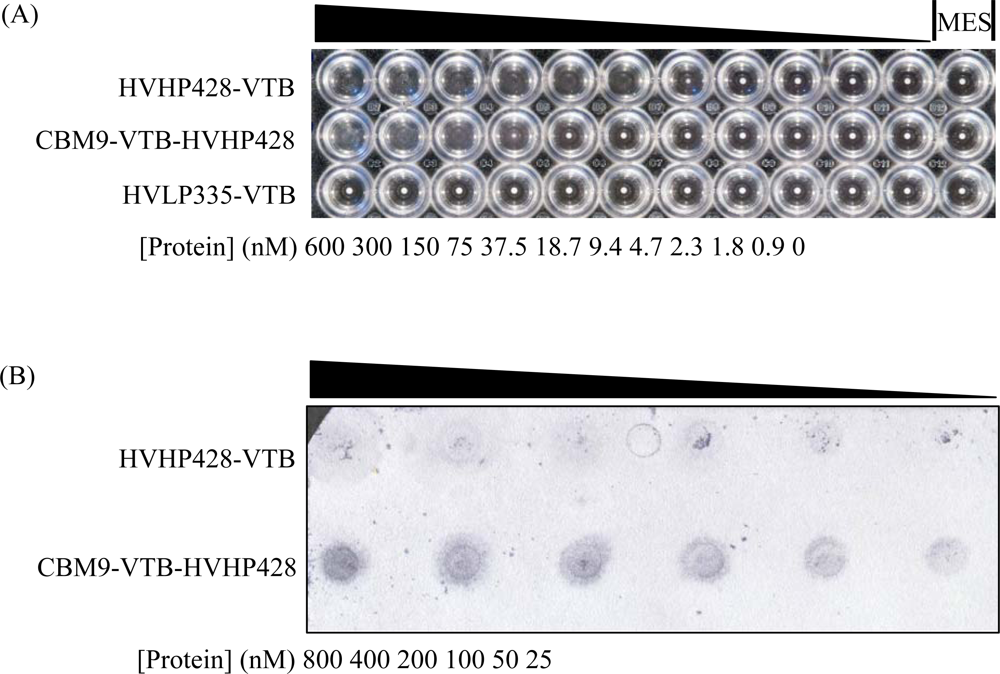
3.2. Functional Characterization of CBM9-VTB-HVHP428 Bispecific Pentamer
3.3. Detection of S. aureus with CBM9-VTB-HVHP428 Impregnated Filters
4. Discussion
5. Conclusions
Acknowledgments
References and Notes
- Ward, E.S.; Güssow, D.; Griffiths, A.D.; Jones, P.T.; Winter, G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature 1989, 341, 544–546. [Google Scholar]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar]
- Arbabi-Ghahroudi, M.; Desmyter, A.; Wyns, L.; Hamers, R.; Muyldermans, S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett 1997, 414, 521–526. [Google Scholar]
- Nuttall, S.D.; Krishnan, U.V.; Hattarki, M.; De Gori, R.; Irving, R.A.; Hudson, P.J. Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol. Immunol 2001, 38, 313–326. [Google Scholar]
- Goldman, E.R.; Liu, J.L.; Bernstein, R.D.; Swain, M.D.; Mitchell, S.Q.; Anderson, G.P. Ricin detection using phage displayed single domain antibodies. Sensors 2009, 9, 542–555. [Google Scholar]
- Saerens, D.; Huang, L.; Bonroy, K.; Muyldermans, S. Antibody fragments as probe in biosensor development. Sensors 2008, 8, 4669–4686. [Google Scholar]
- Tanha, J.; Xu, P.; Chen, Z.; Ni, F.; Kaplan, H.; Narang, S.A.; MacKenzie, C.R. Optimal design features of camelized human single-domain antibody libraries. J. Biol. Chem 2001, 276, 24774–24780. [Google Scholar]
- van der Linden, R.H.; Frenken, L.G.; de Geus, B.; Harmsen, M.M.; Ruuls, R.C.; Stok, W.; de Ron, L.; Wilson, S.; Davis, P.; Verrips, C.T. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta 1999, 1431, 37–46. [Google Scholar]
- Dumoulin, M.; Conrath, K.; van Meirhaeghe, A.; Meersman, F.; Heremans, K.; Frenken, L.G.; Muyldermans, S.; Wyns, L.; Matagne, A. Single-domain antibody fragments with high conformational stability. Protein Sci 2002, 11, 500–515. [Google Scholar]
- Harmsen, M.M.; van Solt, C.B.; van Bemmel, A.M.; Niewold, T.A.; van Zijderveld, F.G. Selection and optimization of proteolytically stable llama single-domain antibody fragments for oral immunotherapy. Appl. Microbiol. Biotechnol 2006, 72, 544–551. [Google Scholar]
- Koide, A.; Tereshko, V.; Uysal, S.; Margalef, K.; Kossiakoff, A.A.; Koide, S. Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope. J. Mol. Biol 2007, 373, 941–953. [Google Scholar]
- Stijlemans, B.; Conrath, K.; Cortez-Retamozo, V.; van Xong, H.; Wyns, L.; Senter, P.; Revets, H.; De Baetselier, P.; Muyldermans, S.; Magez, S. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J. Biol. Chem 2004, 279, 1256–1261. [Google Scholar]
- Arbabi-Ghahroudi, M.; Tanha, J.; MacKenzie, R. Prokaryotic expression of antibodies. Cancer Metastasis Rev 2005, 24, 501–519. [Google Scholar]
- Frenken, L.G.; van der Linden, R.H.; Hermans, P.W.; Bos, J.W.; Ruuls, R.C.; de Geus, B.; Verrips, C.T. Isolation of antigen specific llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae. J. Biotechnol 2000, 78, 11–21. [Google Scholar]
- Holliger, P.; Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol 2005, 23, 1126–1136. [Google Scholar]
- Zhang, J.; Tanha, J.; Hirama, T.; Khieu, N.H.; To, R.; Tong-Sevinc, H.; Stone, E.; Brisson, J.-R.; MacKenzie, C.R. Pentamerization of single-domain antibodies from phage libraries: a novel strategy for the rapid generation of high-avidity antibody reagents. J. Mol. Biol 2004, 335, 49–56. [Google Scholar]
- De Jonge, J.; Heirman, C.; de Veerman, M.; van Meirvenne, S.; Moser, M.; Leo, O.; Thielemans, K. In vivo retargeting of T cell effector function by recombinant bispecific single chain Fv (anti-CD3 × anti-idiotype) induces long-term survival in the murine BCL1 lymphoma model. J. Immunol 1998, 161, 1454–1461. [Google Scholar]
- Rodriguez, B.; Kavoosi, M.; Koska, J.; Creagh, A.L.; Kilburn, D.G.; Haynes, C.A. Inexpensive and generic affinity purification of recombinant proteins using a family 2a CBM fusion tag. Biotechnol. Prog 2004, 20, 1479–1489. [Google Scholar]
- Martinez, A.W.; Phillips, S.T.; Carrilho, E.; Thomas, S.W., III; Sindi, H.; Whitesides, G.M. Simple telemedicine for developing regions: camera phones and paper-based microfluidic devices for real-time, off-site diagnosis. Anal. Chem 2008, 80, 3699–3707. [Google Scholar]
- Li, X.; Tian, J.; Nguyen, T.; Shen, W. Paper-based microfluidic devices by plasma treatment. Anal. Chem 2008, 80, 9131–9134. [Google Scholar]
- Zhao, W.; Ali, M.M.; Aguirre, S.D.; Brook, M.A.; Li, Y. Paper-based bioassays using gold nanoparticle colorimetric probes. Anal. Chem 2008, 80, 8431–8437. [Google Scholar]
- van Tilbeurgh, H.; Tomme, P.; Claeyssens, M.; Bhikhabhai, R.; Pettersson, G. Limited proteolysis of the cellobiohydrolase I from Trichoderma reesei. Separation of functional domains. FEBS Lett 1986, 204, 223–227. [Google Scholar]
- Gilkes, N.R.; Warren, R.A.; Miller, R.C., Jr.; Kilburn, D.G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J. Biol. Chem 1988, 263, 10401–10407. [Google Scholar]
- Shoseyov, O.; Shani, Z.; Levy, I. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol. Mol. Biol. Rev 2006, 70, 283–295. [Google Scholar]
- Stone, E.; Hirama, T.; Tanha, J.; Tong-Sevinc, H.; Li, S.; MacKenzie, C.R.; Zhang, J. The assembly of single domain antibodies into bispecific decavalent molecules. J. Immunol. Methods 2007, 318, 88–94. [Google Scholar]
- To, R.; Hirama, T.; Arbabi-Ghahroudi, M.; MacKenzie, R.; Wang, P.; Xu, P.; Ni, F.; Tanha, J. Isolation of monomeric human V(H)s by a phage selection. J. Biol. Chem 2005, 280, 41395–41403. [Google Scholar]
- McLean, B.W.; Bray, M.R.; Boraston, A.B.; Gilkes, N.R.; Haynes, C.A.; Kilburn, D.G. Analysis of binding of the family 2a carbohydrate-binding module from Cellulomonas fimi xylanase 10A to cellulose: specificity and identification of functionally important amino acid residues. Protein Eng 2000, 13, 801–809. [Google Scholar]
- Boraston, A.B.; Warren, R.A.J.; Kilburn, D.G. Glycosylation by Pichia pastoris decreases the affinity of a family 2a carbohydrate-binding module from Cellulomonas fimi: a functional and mutational analysis. Biochem. J 2001, 358, 423–430. [Google Scholar]
- Kavoosi, E.; Meijer, J.; Kwan, E.; Creagh, A.L.; Kilburn, D.G.; Haynes, C.A. Inexpensive one-step purification of polypeptides expressed in Escherichia coli as fusions with the family 9 carbohydrate-binding module of xylanase 10A from T. maritima. J. Chromatogr. B 2004, 807, 87–94. [Google Scholar]
- Wimalasena, R.L; Wilson, G.S. Factors affecting the specific activity of immobilized antibodies and their biologically active fragments. J. Chromatogr 1991, 572, 85–102. [Google Scholar]
- Kavoosi, M.; Sanaie, N.; Dismer, F.; Hubbuch, J.; Kilburn, D.G.; Haynes, C.A. A novel two-zone protein uptake model for affinity chromatography and its application to the description of elution band profiles fused to a family 9 cellulose binding module affinity tag. J. Chromatogr. A 2007, 1160, 137–149. [Google Scholar]
- Ramirez, C.; Fung, J.; Miller, R.C., Jr.; Antony, R.; Warren, J.; Kilburn, D.G. A bifunctional affinity linker to couple antibodies to cellulose. Biotechnology 1993, 11, 1570–1573. [Google Scholar]
- Linder, M.; Salovuori, I.; Ruohonen, L.; Teeri, T.T. Characterization of a double cellulose-binding domain. Synergistic high affinity binding to crystalline cellulose. J. Biol. Chem 1996, 271, 21268–21272. [Google Scholar]
- Pangu, G.; Johnston, E.; Petkov, J.; Parry, N.; Leach, M.; Hammer, D.A. Targeted particulate adhesion to cellulose surfaces mediated by bifunctional fusion proteins. Langmuir 2007, 23, 10682–10693. [Google Scholar]
- Lewis, W.; Keshavarz-Moore, E.; Windust, J.; Bushell, D.; Parry, N. Construction and evaluation of novel fusion proteins for targeted delivery of micro particles to cellulose surfaces. Biotechnol. Bioeng 2006, 94, 625–632. [Google Scholar]
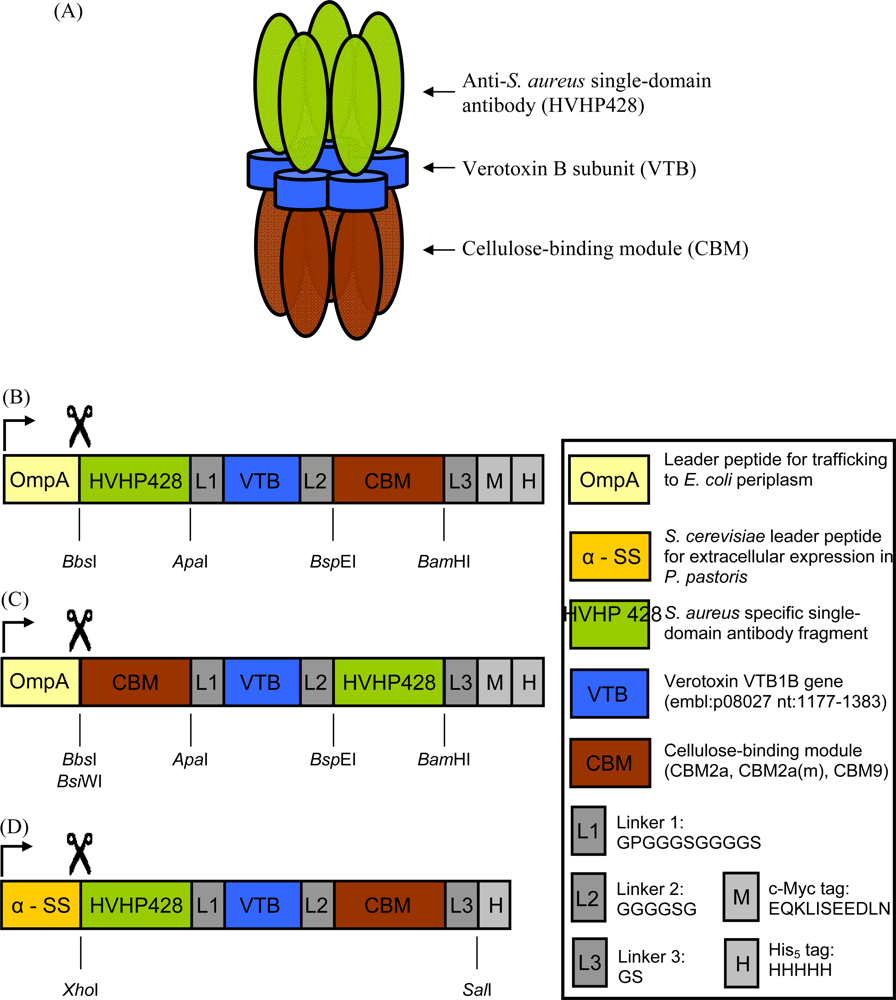
| Construct | Oligonucleotide sequence (5′ → 3′, enzyme sites underlined) | RE |
|---|---|---|
| 1-For | GTTGCGCAGGCCGTCTTCCAGCTGCAGCTGCAGGAGT | BbsI |
| 1-Rev | GGAGCCGCCGCCGGGCCCTGAGGAGACGGTGACCA | ApaI |
| 2-For | GGTGGGTCCGGACAGCTGCAGCTGCAGGAG | BspEI |
| 2-Rev | AGTTTTTGTTCGGATCCTGAGGAGACGGTGACCA | BamHI |
| 3-For | GGTGGGTCCGGATCTGGTCCGGCGGGTTGTC | BspEI |
| 3-Rev | AGTTTTTGTTCGGATCCACCAACGGTACACGGGGCAC | BamHI |
| 4-For | GGTGGGTCCGGAAGCGGCCCGGCCGGGTG | BspEI |
| 4-Rev | AGTTTTTGTTCGGATCCGCCGACCGTGCAGGGCGTG | BamHI |
| 5-For | GGTGGGTCCGGAATGACTAGCGGAATAATGGTAG | BspEI |
| 5-Rev | AGTTTTTGTTCGGATCCAAAGCTTGATGAGCCTGAGG | BamHI |
| 6-For | GTTGCGCAGGCCGTCTTCTCTGGTCCGGCGGGTTGTC | BbsI |
| 6-Rev | GGAGCCGCCGCCGGGCCCACCAACGGTACACGGGGCAC | ApaI |
| 7-For | GTTGCGCAGGCCGTCTTCAGCGGCCCGGCCGGGTG | BbsI |
| 7-Rev | GGAGCCGCCGCCGGGCCCGCCGACCGTGCAGGGCGTG | ApaI |
| 8-For | GGCCGTCTTCGTACGAATGACTAGCGGAATAATGGTAG | BsiWI |
| 8-Rev | GGAGCCGCCGCCGGGCCCAAAGCTTGATGAGCCTGAG | ApaI |
| 9-For (pPICZαA) | TTTTGTCTCGAGAAAAGAGAGGCTGAAGCTCAGCTGCAGCTGCAGGAGTCG | XhoI |
| 9-Rev (pPICZαA) | TTTTGTGTCGACCTATCAATGGTGATGGTGATGGTTCAGATC | SalI |
| Construct (No./Description) | Expression | Purified Protein | |||
|---|---|---|---|---|---|
| E. coli | P. pastoris | Yield (mg/L) | Aggregationa | Degradationb | |
| Monospecifc | |||||
| 1 HVHP428-VTB | S | 15.7 | No | No | |
| 2 VTB-HVHP428 | |||||
| 3 VTB-CBM2a(m) | S | NA | NA | NA | |
| 4 VTB-CBM2a | |||||
| 5 VTB-CBM9 | |||||
| 6 CBM2a(m)-VTB | IB | ||||
| 7 CBM2a-VTB | S | NA | NA | NA | |
| 8 CBM9-VTB | |||||
| Bispecific | |||||
| 9 HVHP428-VTB-CBM2a(m) | NE | S | NA | NA | NA |
| 10 HVHP428-VTB-CBM2a | NE | ||||
| 11 HVHP428-VTB-CBM9 | NE | ||||
| 12 CBM2a(m)-VTB-VHV28 | S | Low | Yes | Yes | |
| 13 CBM2a-VTB-HVHP428 | S | 3.8 | Yes | Yes | |
| 14 CBM9-VTB-HVHP428 | S | 7.8 | No | Low | |
© 2009 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hussack, G.; Luo, Y.; Veldhuis, L.; Hall, J.C.; Tanha, J.; MacKenzie, R. Multivalent Anchoring and Oriented Display of Single-Domain Antibodies on Cellulose. Sensors 2009, 9, 5351-5367. https://doi.org/10.3390/s90705351
Hussack G, Luo Y, Veldhuis L, Hall JC, Tanha J, MacKenzie R. Multivalent Anchoring and Oriented Display of Single-Domain Antibodies on Cellulose. Sensors. 2009; 9(7):5351-5367. https://doi.org/10.3390/s90705351
Chicago/Turabian StyleHussack, Greg, Yan Luo, Linda Veldhuis, J. Christopher Hall, Jamshid Tanha, and Roger MacKenzie. 2009. "Multivalent Anchoring and Oriented Display of Single-Domain Antibodies on Cellulose" Sensors 9, no. 7: 5351-5367. https://doi.org/10.3390/s90705351
APA StyleHussack, G., Luo, Y., Veldhuis, L., Hall, J. C., Tanha, J., & MacKenzie, R. (2009). Multivalent Anchoring and Oriented Display of Single-Domain Antibodies on Cellulose. Sensors, 9(7), 5351-5367. https://doi.org/10.3390/s90705351




