Evaluation of Effective Class-Balancing Techniques for CNN-Based Assessment of Aphanomyces Root Rot Resistance in Pea (Pisum sativum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Data Collection
2.2. Dataset Pre-Processing and Class Balancing
2.2.1. Random Oversampling
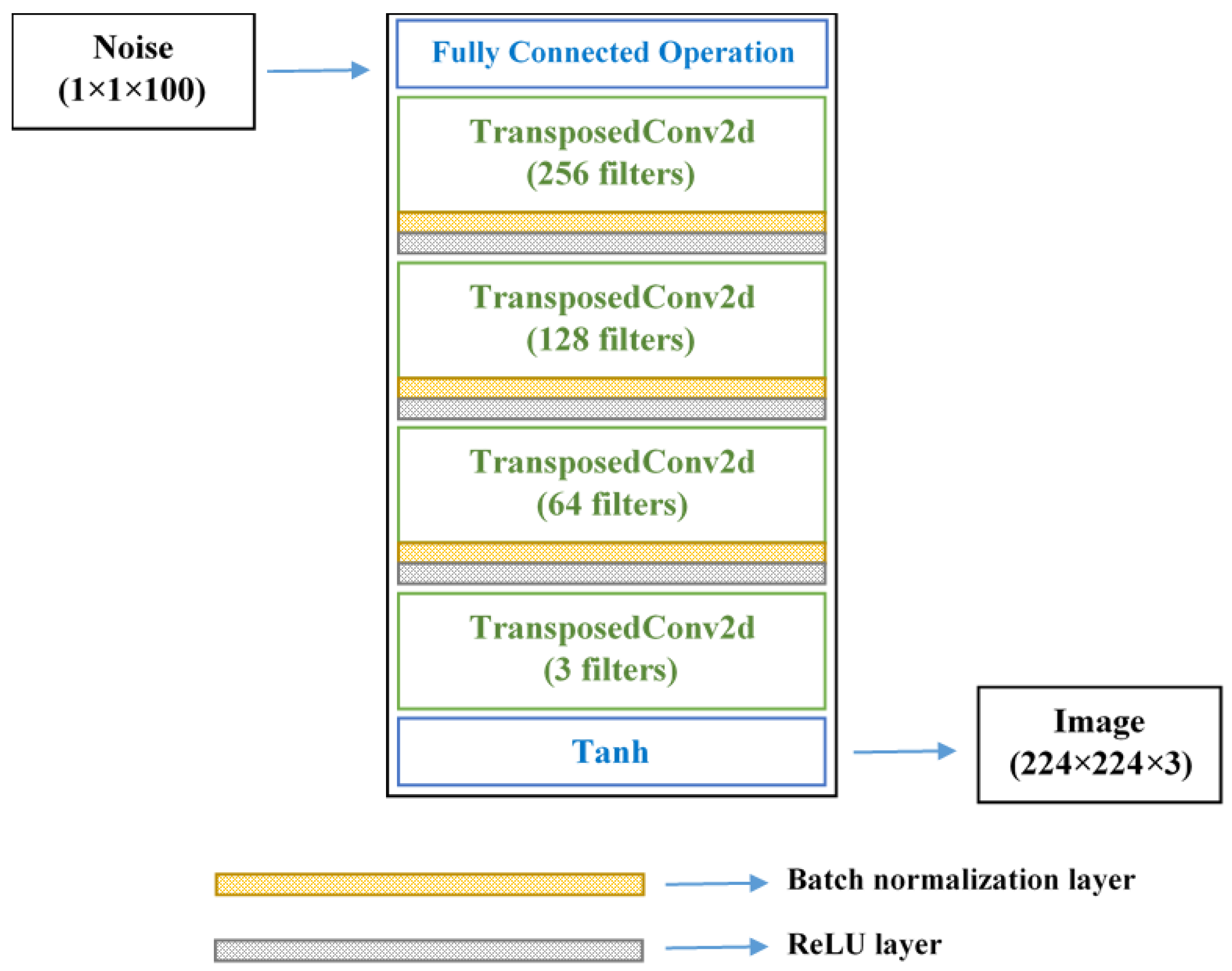
2.2.2. GAN-Based Image Augmentation
2.2.3. Loss Function with Weighted Ratio
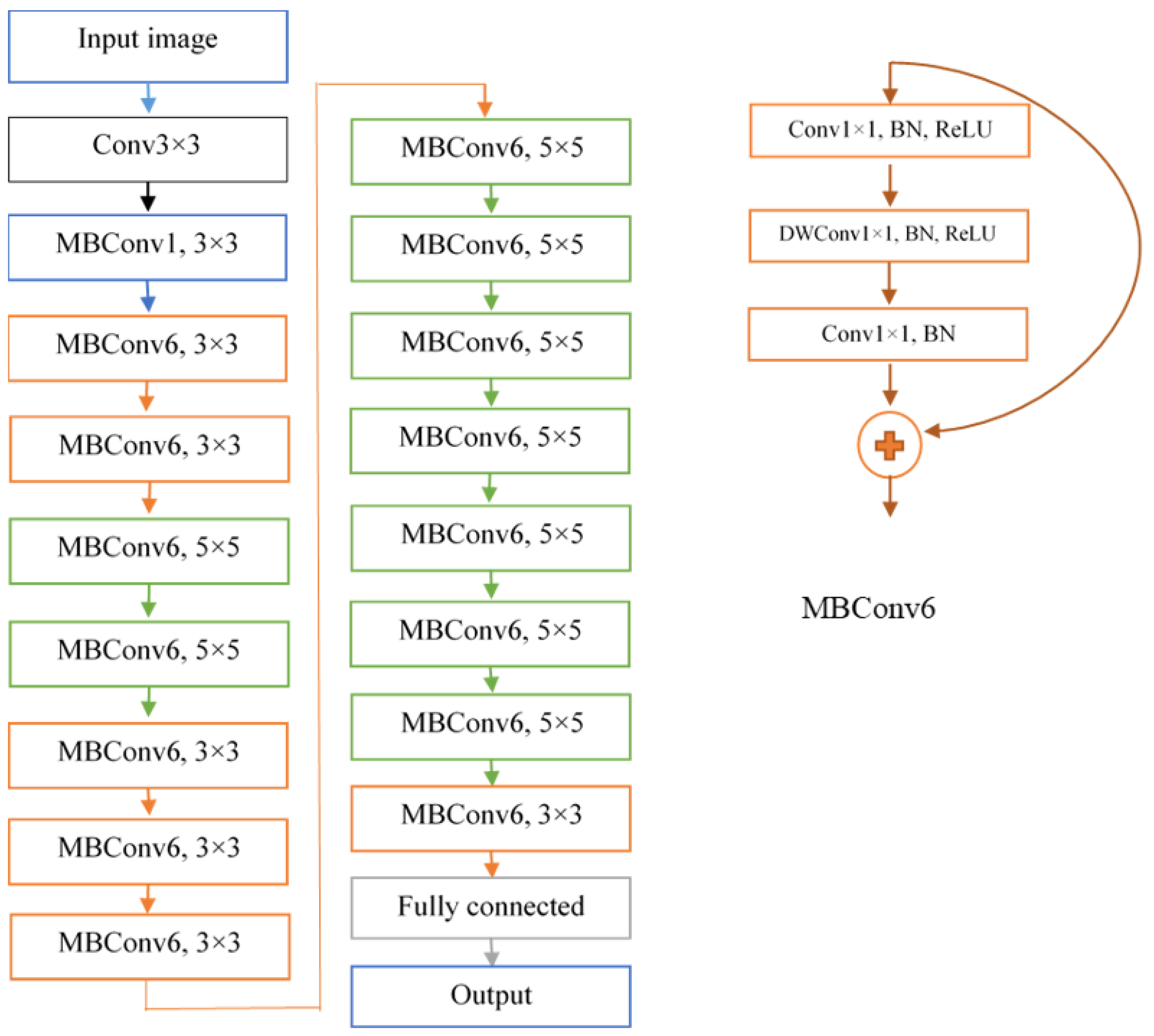
2.3. DeepARRNet Architecture
3. Results
3.1. Performance of the Model Using Original Images
3.2. Impact of Random Oversampling Method on Model Performance
3.3. Impact of Addition of GAN-Generated Images on Model Performance
3.4. Impact of Introducing Class-Weighted Ratio in Loss Function
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hossain, S.; Bergkvist, G.; Berglund, K.; Mårtensson, A.; Persson, P. Aphanomyces Pea Root Rot Disease and Control with Special Reference to Impact of Brassicaceae Cover Crops. Acta Agric. Scand. Sect. B Soil Plant Sci. 2012, 62, 477–487. [Google Scholar] [CrossRef]
- Wicker, E.; Rouxel, F. Specific Behaviour of French Aphanomyces euteiches Drechs. Populations For Virulence and Aggressiveness on Pea, Related to Isolates from Europe, America and New Zealand. Eur. J. Plant Pathol. 2001, 107, 919–929. [Google Scholar] [CrossRef]
- Chatterton, S.; Bowness, R.; Harding, M.W. First Report of Root Rot of Field Pea Caused by Aphanomyces euteiches in Alberta, Canada. Plant Dis. 2015, 99, 288. [Google Scholar] [CrossRef]
- Wu, L.; Chang, K.F.; Hwang, S.F.; Conner, R.; Fredua-Agyeman, R.; Feindel, D.; Strelkov, S.E. Evaluation of Host Resistance and Fungicide Application as Tools for the Management of Root Rot of Field Pea Caused by Aphanomyces euteiches. Crop J. 2019, 7, 38–48. [Google Scholar] [CrossRef]
- Pilet-Nayel, M.L.; Muehlbauer, F.J.; McGee, R.J.; Kraft, J.M.; Baranger, A.; Coyne, C.J. Quantitative Trait Loci for Partial Resistance to Aphanomyces Root Rot in Pea. Theor. Appl. Genet. 2002, 106, 28–39. [Google Scholar] [CrossRef]
- McGee, R.J.; Coyne, C.J.; Pilet-Nayel, M.-L.; Moussart, A.; Tivoli, B.; Baranger, A.; Hamon, C.; Vandemark, G.; McPhee, K. Registration of Pea Germplasm Lines Partially Resistant to Aphanomyces Root Rot for Breeding Fresh or Freezer Pea and Dry Pea Types. J. Plant Regist. 2012, 6, 203–207. [Google Scholar] [CrossRef]
- Walter, J.; Edwards, J.; Cai, J.; McDonald, G.; Miklavcic, S.J.; Kuchel, H. High-Throughput Field Imaging and Basic Image Analysis in a Wheat Breeding Programme. Front. Plant Sci. 2019, 10, 449. [Google Scholar] [CrossRef] [PubMed]
- Das Choudhury, S.; Samal, A.; Awada, T. Leveraging Image Analysis for High-Throughput Plant Phenotyping. Front. Plant Sci. 2019, 10, 508. [Google Scholar] [CrossRef]
- Jin, X.; Zarco-Tejada, P.J.; Schmidhalter, U.; Reynolds, M.P.; Hawkesford, M.J.; Varshney, R.K.; Yang, T.; Nie, C.; Li, Z.; Ming, B.; et al. High-Throughput Estimation of Crop Traits: A Review of Ground and Aerial Phenotyping Platforms. IEEE Geosci. Remote Sens. Mag. 2021, 9, 200–231. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics–Technologies to Relieve the Phenotyping Bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Jimenez-Berni, J.; Fischer, R.A.; Deery, D.M.; Smith, D.J. Review: High-Throughput Phenotyping to Enhance the Use of Crop Genetic Resources. Plant Sci. 2019, 282, 40–48. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Y.; Du, J.; Guo, X.; Wen, W.; Gu, S.; Wang, J.; Fan, J. Crop Phenomics: Current Status and Perspectives. Front. Plant Sci. 2019, 10, 714. [Google Scholar] [CrossRef]
- Song, P.; Wang, J.; Guo, X.; Yang, W.; Zhao, C. High-Throughput Phenotyping: Breaking through the Bottleneck in Future Crop Breeding. Crop J. 2021, 9, 633–645. [Google Scholar] [CrossRef]
- Jiao, L.; Zhang, F.; Liu, F.; Yang, S.; Li, L.; Feng, Z.; Qu, R. A Survey of Deep Learning-Based Object Detection. IEEE Access 2019, 7, 128837–128868. [Google Scholar] [CrossRef]
- Hao, S.; Zhou, Y.; Guo, Y. A Brief Survey on Semantic Segmentation with Deep Learning. Neurocomputing 2020, 406, 302–321. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Sapkota, B.; Singh, V.; Neely, C.; Rajan, N.; Bagavathiannan, M. Detection of Italian Ryegrass in Wheat and Prediction of Competitive Interactions Using Remote-Sensing and Machine-Learning Techniques. Remote Sens. 2020, 12, 2977. [Google Scholar] [CrossRef]
- Divyanth, L.G.; Ahmad, A.; Saraswat, D. A Two-Stage Deep-Learning Based Segmentation Model for Crop Disease Quantification Based on Corn Field Imagery. Smart Agric. Technol. 2022, 3, 100108. [Google Scholar] [CrossRef]
- Fu, L.; Majeed, Y.; Zhang, X.; Karkee, M.; Zhang, Q. Faster R–CNN–Based Apple Detection in Dense-Foliage Fruiting-Wall Trees Using RGB and Depth Features for Robotic Harvesting. Biosyst. Eng. 2020, 197, 245–256. [Google Scholar] [CrossRef]
- Divyanth, L.G.; Chelladurai, V.; Loganathan, M.; Jayas, D.S.; Soni, P. Identification of Green Gram (Vigna radiata) Grains Infested by Callosobruchus maculatus Through X-Ray Imaging and GAN-Based Image Augmentation. J. Biosyst. Eng. 2022, 47, 302–317. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine Learning Approaches for Crop Yield Prediction and Nitrogen Status Estimation in Precision Agriculture: A Review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- Gao, J.; Westergaard, J.C.; Sundmark, E.H.R.; Bagge, M.; Liljeroth, E.; Alexandersson, E. Automatic Late Blight Lesion Recognition and Severity Quantification Based on Field Imagery of Diverse Potato Genotypes by Deep Learning. Knowl.-Based Syst. 2021, 214, 106723. [Google Scholar] [CrossRef]
- Ghosal, S.; Blystone, D.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. An Explainable Deep Machine Vision Framework for Plant Stress Phenotyping. Proc. Natl. Acad. Sci. USA 2018, 115, 4613–4618. [Google Scholar] [CrossRef] [PubMed]
- Ubbens, J.R.; Stavness, I. Deep Plant Phenomics: A Deep Learning Platform for Complex Plant Phenotyping Tasks. Front. Plant Sci. 2017, 8, 1190. [Google Scholar] [CrossRef]
- Azimi, S.; Kaur, T.; Gandhi, T.K. A Deep Learning Approach to Measure Stress Level in Plants Due to Nitrogen Deficiency. Measurement 2021, 173, 108650. [Google Scholar] [CrossRef]
- Mishra, P.; Sadeh, R.; Ryckewaert, M.; Bino, E.; Polder, G.; Boer, M.P.; Rutledge, D.N.; Herrmann, I. A Generic Workflow Combining Deep Learning and Chemometrics for Processing Close-Range Spectral Images to Detect Drought Stress in Arabidopsis Thaliana to Support Digital Phenotyping. Chemom. Intell. Lab. Syst. 2021, 216, 104373. [Google Scholar] [CrossRef]
- Dyrmann, M.; Karstoft, H.; Midtiby, H.S. Plant Species Classification Using Deep Convolutional Neural Network. Biosyst. Eng. 2016, 151, 72–80. [Google Scholar] [CrossRef]
- Bae, S.Y.; Lee, J.; Jeong, J.; Lim, C.; Choi, J. Effective Data-Balancing Methods for Class-Imbalanced Genotoxicity Datasets Using Machine Learning Algorithms and Molecular Fingerprints. Comput. Toxicol. 2021, 20, 100178. [Google Scholar] [CrossRef]
- Reddy, S.T.; Georgiou, G. Systems Analysis of Adaptive Immunity by Utilization of High-Throughput Technologies. Curr. Opin. Biotechnol. 2011, 22, 584–589. [Google Scholar] [CrossRef]
- Korkmaz, S. Deep Learning-Based Imbalanced Data Classification for Drug Discovery. J. Chem. Inf. Model. 2020, 60, 4180–4190. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Jain, S.; Gour, M.; Vankudothu, S. Tomato Plant Disease Detection Using Transfer Learning with C-GAN Synthetic Images. Comput. Electron. Agric. 2021, 187, 106279. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Giuffrida, M.V.; Scharr, H.; Tsaftaris, S.A. ARIGAN: Synthetic Arabidopsis Plants Using Generative Adversarial Network. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2064–2071. [Google Scholar] [CrossRef]
- Espejo-Garcia, B.; Mylonas, N.; Athanasakos, L.; Vali, E.; Fountas, S. Combining Generative Adversarial Networks and Agricultural Transfer Learning for Weeds Identification. Biosyst. Eng. 2021, 204, 79–89. [Google Scholar] [CrossRef]
- Arsenovic, M.; Karanovic, M.; Sladojevic, S.; Anderla, A.; Stefanovic, D. Solving Current Limitations of Deep Learning Based Approaches for Plant Disease Detection. Symmetry 2019, 11, 939. [Google Scholar] [CrossRef]
- Madsen, S.L.; Dyrmann, M.; Jørgensen, R.N.; Karstoft, H. Generating Artificial Images of Plant Seedlings Using Generative Adversarial Networks. Biosyst. Eng. 2019, 187, 147–159. [Google Scholar] [CrossRef]
- Wicker, E.; Hullé, M.; Rouxel, F. Pathogenic Characteristics of Isolates of Aphanomyces euteiches from Pea in France. Plant Pathol. 2001, 50, 433–442. [Google Scholar] [CrossRef]
- Marzougui, A.; Ma, Y.; Zhang, C.; McGee, R.J.; Coyne, C.J.; Main, D.; Sankaran, S. Advanced Imaging for Quantitative Evaluation of Aphanomyces Root Rot Resistance in Lentil. Front. Plant Sci. 2019, 10, 383. [Google Scholar] [CrossRef]
- Marzougui, A.; Ma, Y.; McGee, R.J.; Khot, L.R.; Sankaran, S. Generalized Linear Model with Elastic Net Regularization and Convolutional Neural Network for Evaluating Aphanomyces Root Rot Severity in Lentil. Plant Phenomics 2020, 2020, 11. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning PMLR, Long Beach, CA, USA, 9–15 June 2019; Volume 97, pp. 10691–10700. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, J.; Yang, G.; Zhang, H.; He, Y. Localization and Classification of Paddy Field Pests Using a Saliency Map and Deep Convolutional Neural Network. Sci. Rep. 2016, 6, 20410. [Google Scholar] [CrossRef]
- Stewart, E.L.; Wiesner-Hanks, T.; Kaczmar, N.; DeChant, C.; Wu, H.; Lipson, H.; Nelson, R.J.; Gore, M.A. Quantitative Phenotyping of Northern Leaf Blight in UAV Images Using Deep Learning. Remote Sens. 2019, 11, 2209. [Google Scholar] [CrossRef]
- Gao, J.; French, A.P.; Pound, M.P.; He, Y.; Pridmore, T.P.; Pieters, J.G. Deep Convolutional Neural Networks for Image-Based Convolvulus Sepium Detection in Sugar Beet Fields. Plant Methods 2020, 16, 29. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B. Plant Disease Identification Using Explainable 3D Deep Learning on Hyperspectral Images. Plant Methods 2019, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Miao, H.; Li, J.; Jian, F.; Jayas, D.S. A Low-Resolution Image Restoration Classifier Network to Identify Stored-Grain Insects from Images of Sticky Boards. Comput. Electron. Agric. 2019, 162, 593–601. [Google Scholar] [CrossRef]
- Nazki, H.; Yoon, S.; Fuentes, A.; Park, D.S. Unsupervised Image Translation Using Adversarial Networks for Improved Plant Disease Recognition. Comput. Electron. Agric. 2020, 168, 105117. [Google Scholar] [CrossRef]




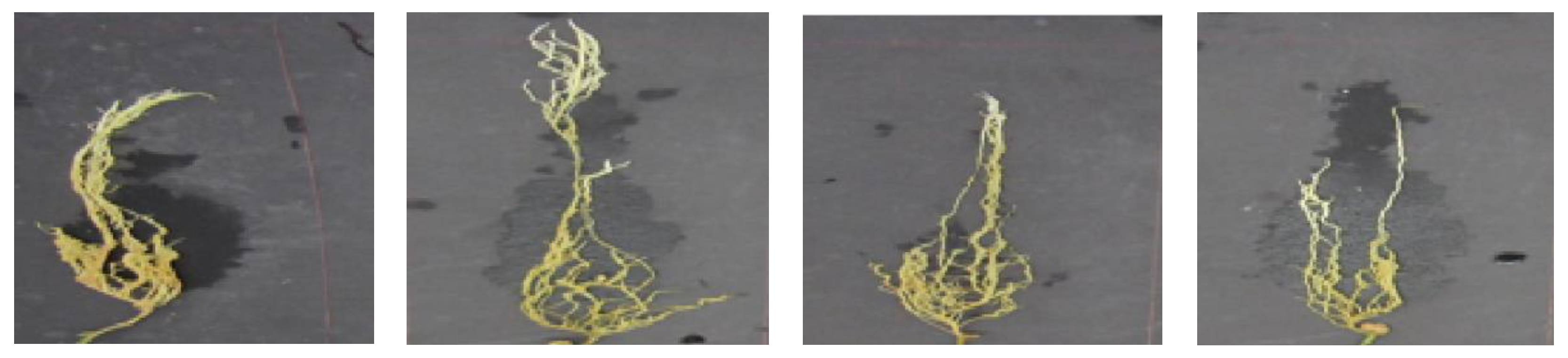
| Visual Disease Score | Symptoms | Class | Number of Image Samples |
|---|---|---|---|
| 0.0 | No discolored lesions on the entire root | Healthy/Resistant | 784 |
| 0.5 | Up to 5% of discolored lesions on the entire root | Resistant | 4 |
| 1.0 | 5–15% of discolored lesions on the entire root | ||
| 1.5 | 15–25% of discolored lesions on the entire root | ||
| 2.0 | 25–50% minor discoloration on the entire root | Intermediate | 727 |
| 2.5 | 50–75% major discoloration on the entire root | ||
| 3.0 | More than 75% of brown discoloration on the entire root | ||
| 3.5 | More than 75% of brown discoloration on entire root system with some symptoms on hypocotyl | Susceptible | 70 |
| 4.0 | Brown discoloration on entire root system with shriveled and brown hypocotyl | ||
| 4.5 | Brown discoloration on entire root system with a shriveled, brown, and soft hypocotyl | ||
| 5.0 | Dead plant |
| Dataset and Class-Balancing Technique Implemented | 1st Seed (Sia) | 2nd Seed (Sib) | 3rd Seed (Sic) |
|---|---|---|---|
| S1—Without class balancing (original dataset) | Evaluate on S1a (training with R1a and test on T1a) | Evaluate on S1b (training with R1b and test on T1b) | Evaluate on S1c (training with R1c and test on T1c) |
| S2—Random oversampling | Evaluate on S2a (training with R2a and test on T2a) | Evaluate on S2b (training with R2b and test on T2b) | Evaluate on S2c (training with R2c and test on T2c) |
| S3—GAN-based image synthesis | Evaluate on S3a (training with R3a and test on T3a) | Evaluate on S3b (training with R3b and test on T3b) | Evaluate on S3c (training with R3c and test on T3c) |
| S4—Loss function with weighted ratio | Evaluate on S4a (training with R4a and test on T4a) | Evaluate on S4b (training with R4b and test on T4b) | Evaluate on S4c (training with R4c and test on T4c) |
| Class | Precision | Recall | F1-Score |
|---|---|---|---|
| Resistant | 0.99 ± 0.02 | 0.92 ± 0.03 | 0.95 ± 0.03 |
| Intermediate | 0.80 ± 0.03 | 0.99 ± 0.03 | 0.88 ± 0.03 |
| Susceptible | 0.97 ± 0.05 | 0.06 ± 0.05 | 0.09 ± 0.05 |
| Overall | 0.93 ± 0.03 | 0.72 ± 0.03 | 0.83 ± 0.03 |
| Class | Precision | Recall | F1-Score |
|---|---|---|---|
| Resistant | 0.99 ± 0.02 | 0.92 ± 0.03 | 0.96 ± 0.03 |
| Intermediate | 0.86 ± 0.04 | 0.98 ± 0.04 | 0.91 ± 0.04 |
| Susceptible | 0.91 ± 0.06 | 0.68 ± 0.06 | 0.78 ± 0.06 |
| Overall | 0.93 ± 0.03 | 0.85 ± 0.04 | 0.91 ± 0.04 |
| Class | Precision | Recall | F1-Score |
|---|---|---|---|
| Resistant | 0.99 ± 0.01 | 0.93± 0.01 | 0.96 ± 0.01 |
| Intermediate | 0.90 ± 0.05 | 0.99 ± 0.05 | 0.91 ± 0.05 |
| Susceptible | 0.91 ± 0.07 | 0.75 ± 0.04 | 0.81 ± 0.06 |
| Overall | 0.96 ± 0.03 | 0.87 ± 0.04 | 0.92 ± 0.033 |
| Weight Ratio | Class | Precision | Recall | F1-Score |
|---|---|---|---|---|
| INS | Resistant | 0.99 ± 0.01 | 0.93 ± 0.02 | 0.96 ± 0.02 |
| Intermediate | 0.88 ± 0.05 | 0.98 ± 0.07 | 0.94 ± 0.06 | |
| Susceptible | 0.90 ± 0.08 | 0.64 ± 0.06 | 0.78 ± 0.07 | |
| Overall | 0.94 ± 0.04 | 0.85 ± 0.05 | 0.88 ± 0.05 | |
| ISRNS | Resistant | 0.99 ± 0.03 | 0.93 ± 0.03 | 0.96 ± 0.03 |
| Intermediate | 0.87 ± 0.06 | 0.98 ± 0.06 | 0.92 ± 0.06 | |
| Susceptible | 0.85 ± 0.07 | 0.60 ± 0.08 | 0.79 ± 0.07 | |
| Overall | 0.92 ± 0.05 | 0.83 ± 0.04 | 0.87 ± 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Divyanth, L.G.; Marzougui, A.; González-Bernal, M.J.; McGee, R.J.; Rubiales, D.; Sankaran, S. Evaluation of Effective Class-Balancing Techniques for CNN-Based Assessment of Aphanomyces Root Rot Resistance in Pea (Pisum sativum L.). Sensors 2022, 22, 7237. https://doi.org/10.3390/s22197237
Divyanth LG, Marzougui A, González-Bernal MJ, McGee RJ, Rubiales D, Sankaran S. Evaluation of Effective Class-Balancing Techniques for CNN-Based Assessment of Aphanomyces Root Rot Resistance in Pea (Pisum sativum L.). Sensors. 2022; 22(19):7237. https://doi.org/10.3390/s22197237
Chicago/Turabian StyleDivyanth, L. G., Afef Marzougui, Maria Jose González-Bernal, Rebecca J. McGee, Diego Rubiales, and Sindhuja Sankaran. 2022. "Evaluation of Effective Class-Balancing Techniques for CNN-Based Assessment of Aphanomyces Root Rot Resistance in Pea (Pisum sativum L.)" Sensors 22, no. 19: 7237. https://doi.org/10.3390/s22197237
APA StyleDivyanth, L. G., Marzougui, A., González-Bernal, M. J., McGee, R. J., Rubiales, D., & Sankaran, S. (2022). Evaluation of Effective Class-Balancing Techniques for CNN-Based Assessment of Aphanomyces Root Rot Resistance in Pea (Pisum sativum L.). Sensors, 22(19), 7237. https://doi.org/10.3390/s22197237










