Genotyping of KRAS Mutational Status by the In-Check Lab-on-Chip Platform
Abstract
:1. Introduction
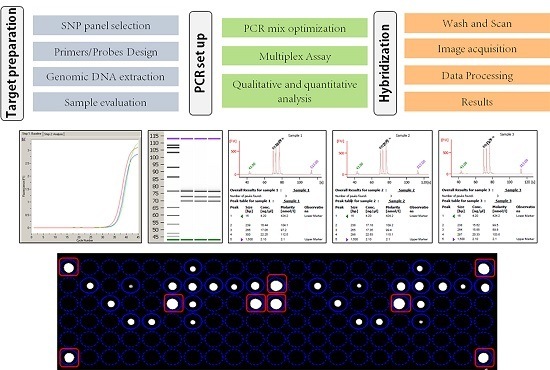
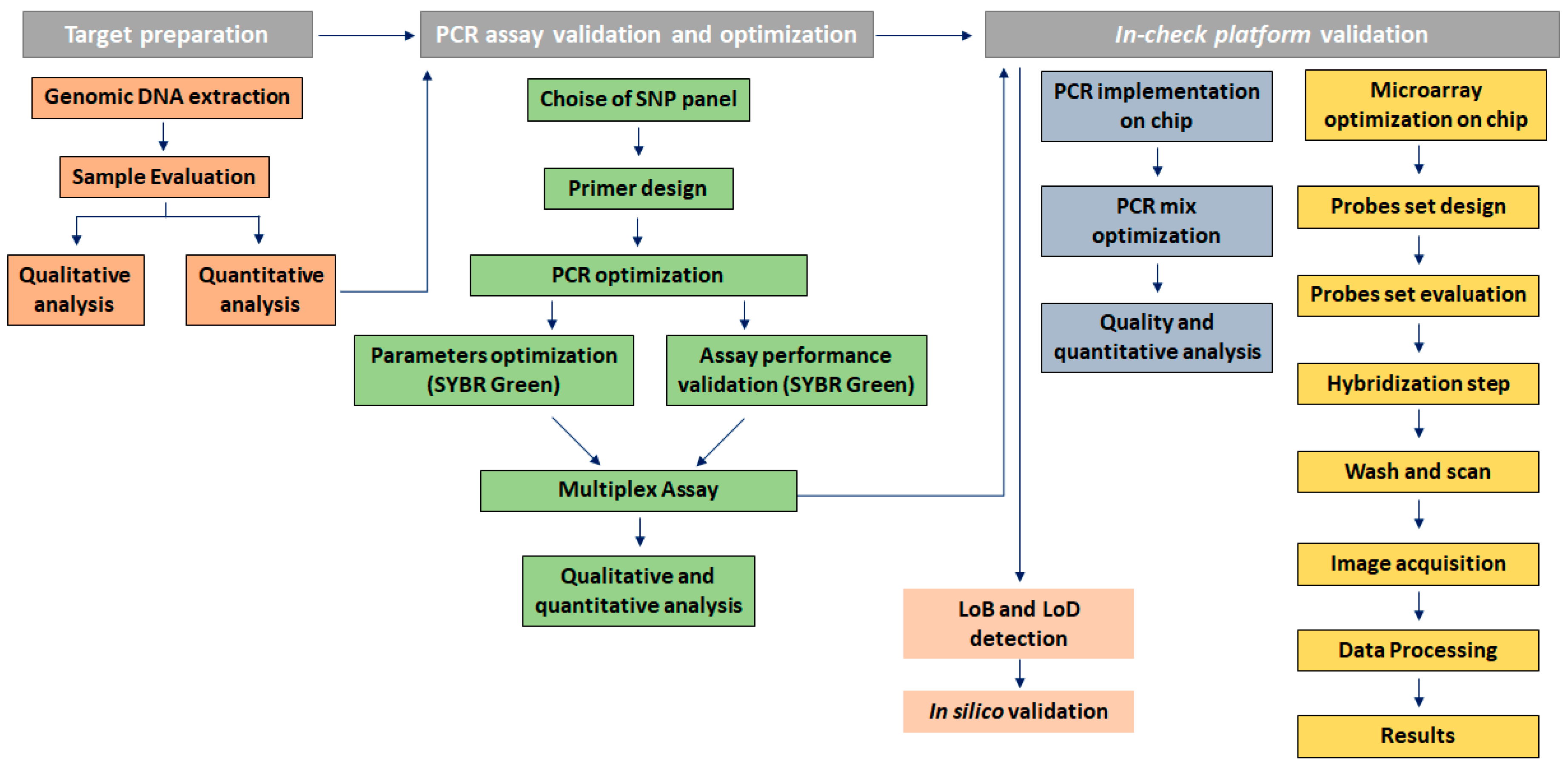
2. Materials and Methods
2.1. Assay Design
2.2. Samples Processing
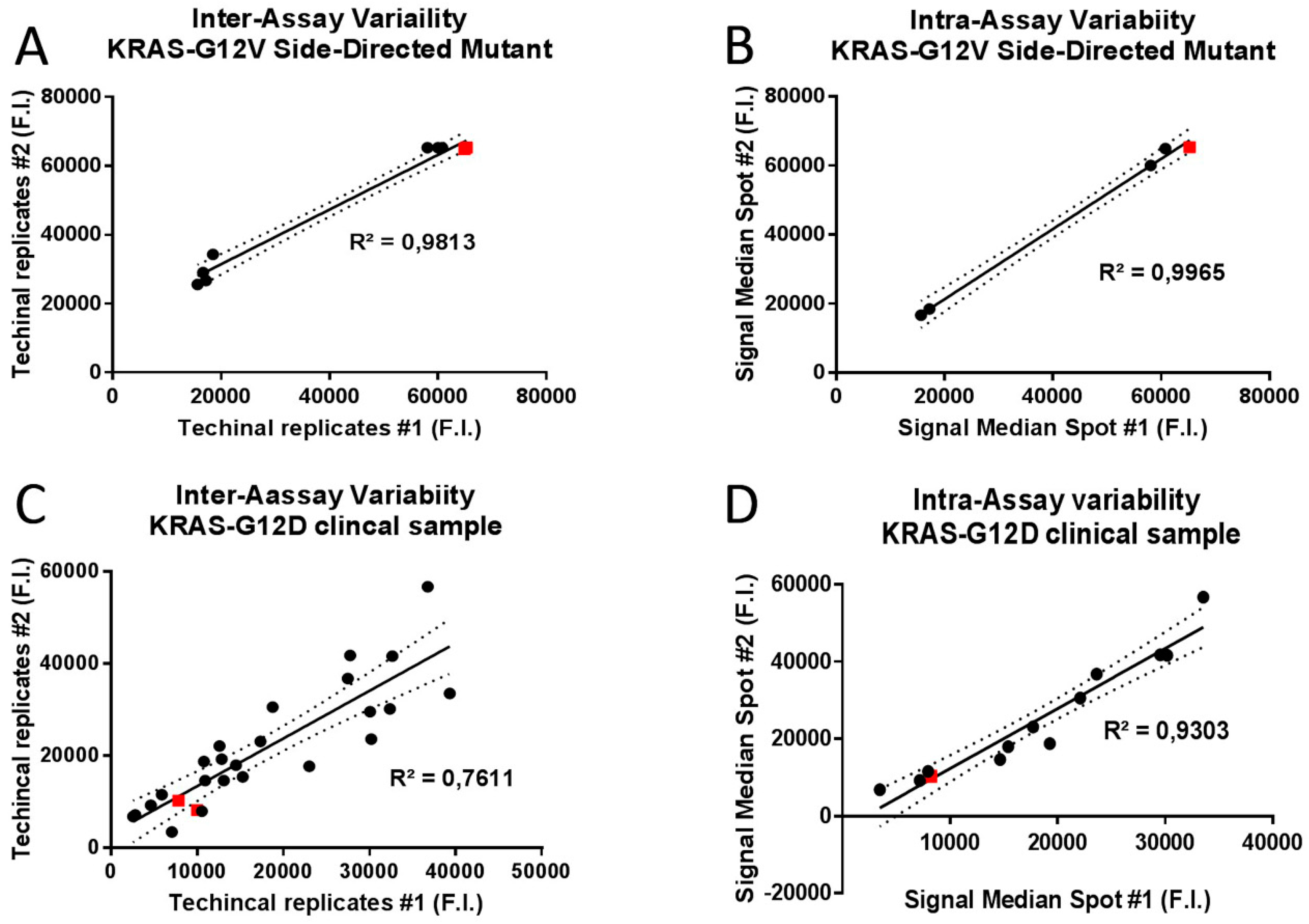
2.3. Analytic Performance Studies
3. Results
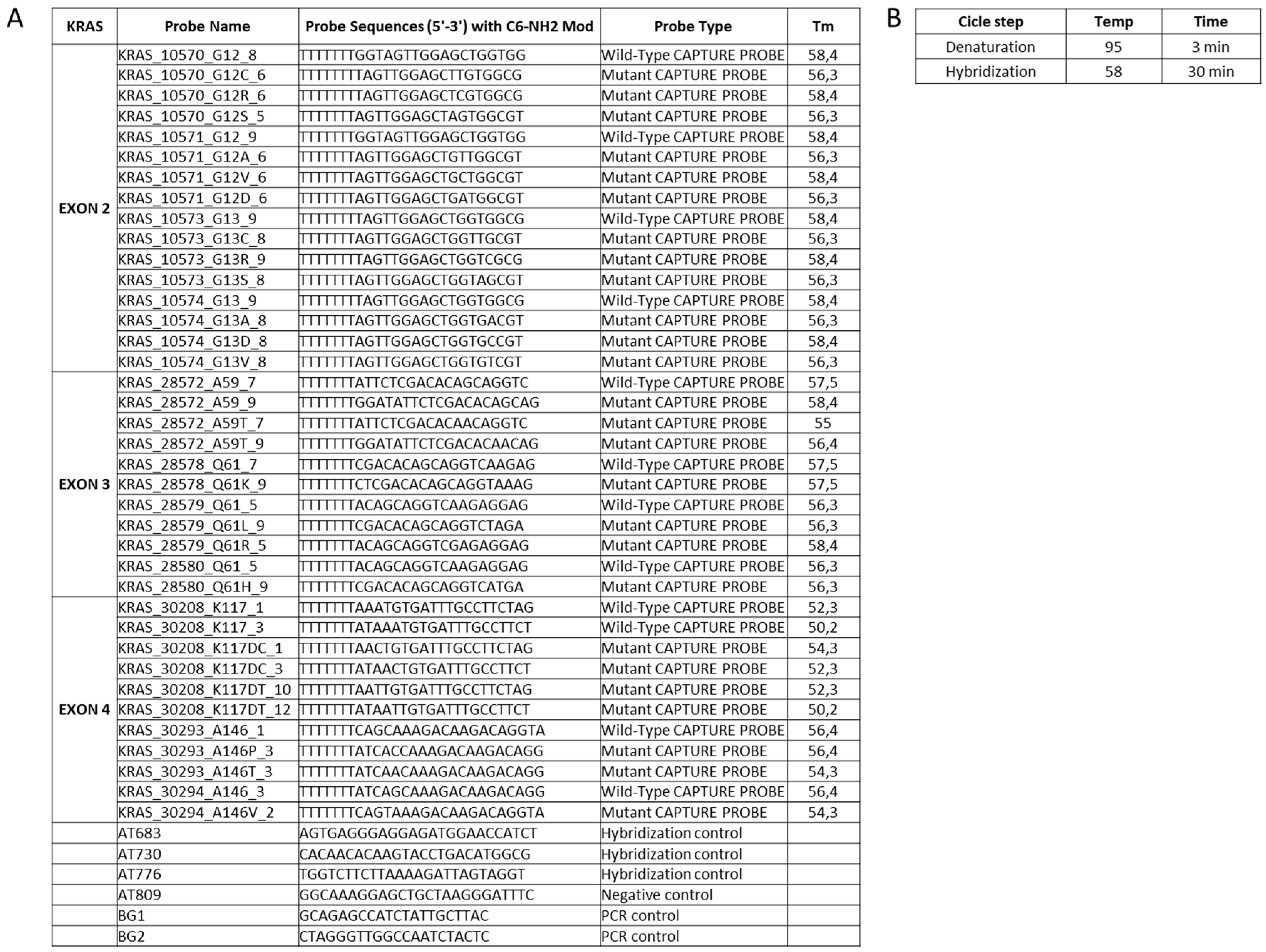
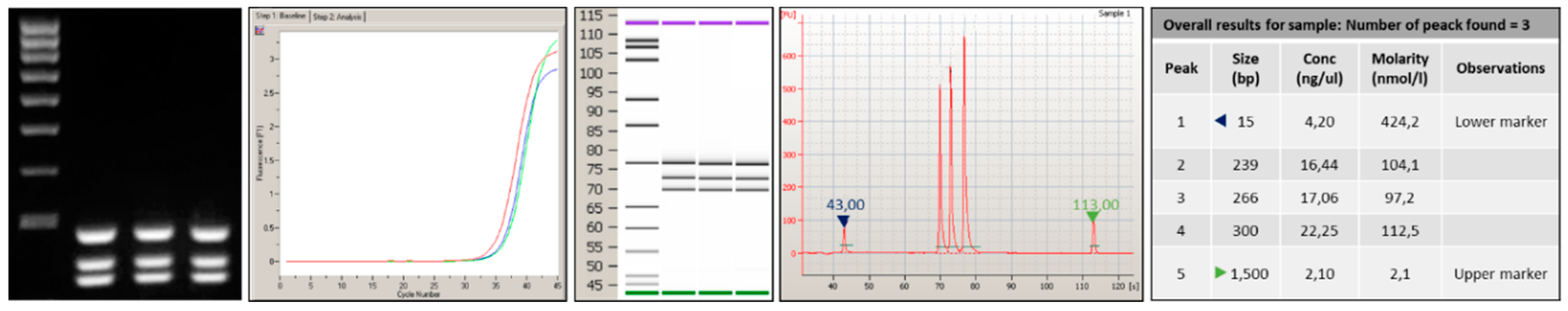
3.1. Primers Selection and PCR Optimization
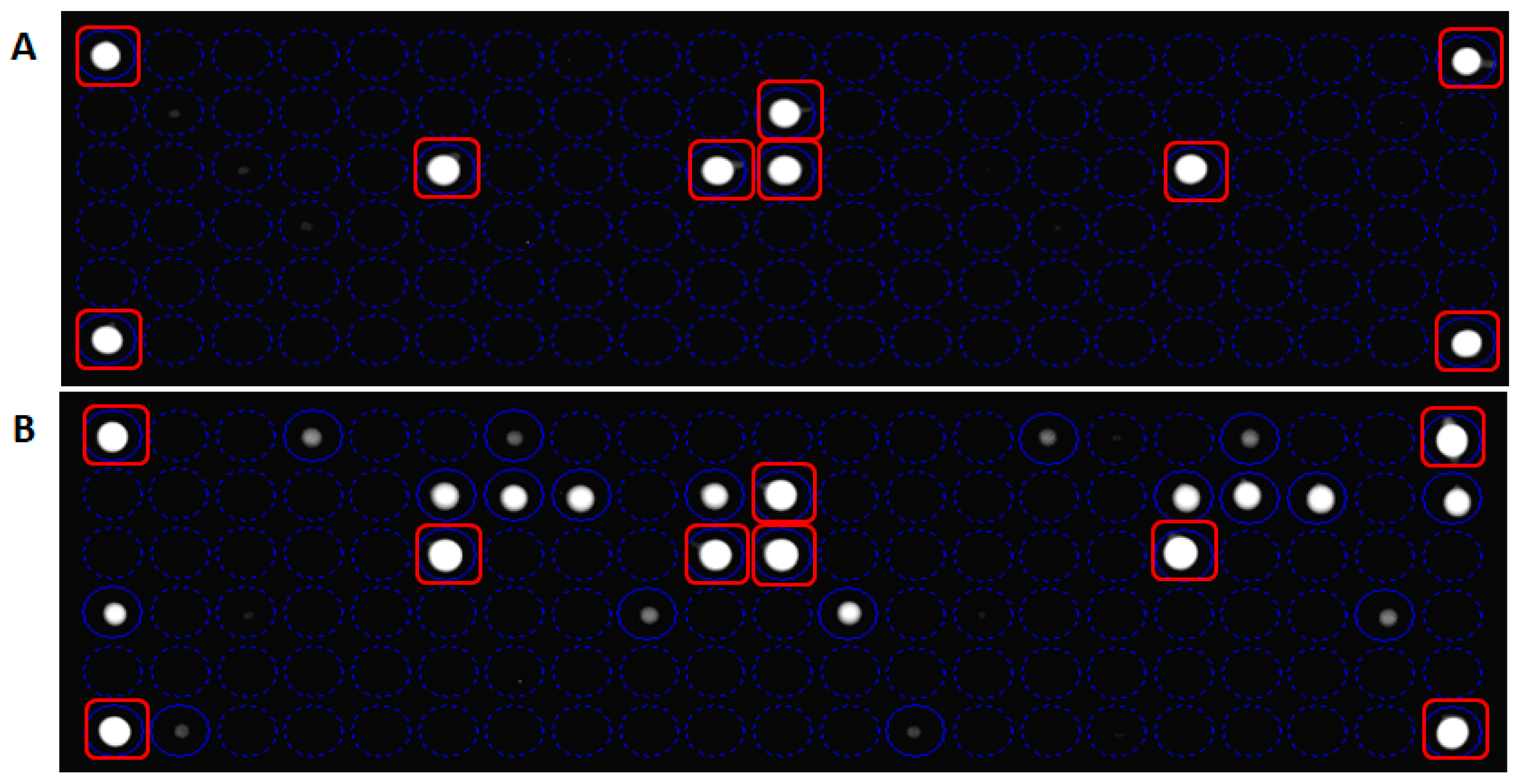
3.2. Probes Set Selection and Hybridization Assay Evaluation
3.3. Analytical Sensitivity and Specificity
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Domagala, P.; Hybiak, J.; Sulzyc-Bielicka, V.; Cybulski, C.; Rys, J.; Domagala, W. Kras mutation testing in colorectal cancer as an example of the pathologist’s role in personalized targeted therapy: A practical approach. Pol. J. Pathol. Off. J. Polish Soc. Pathol. 2012, 63, 145–164. [Google Scholar] [CrossRef]
- Phipps, A.I.; Buchanan, D.D.; Makar, K.W.; Win, A.K.; Baron, J.A.; Lindor, N.M.; Potter, J.D.; Newcomb, P.A. Kras-mutation status in relation to colorectal cancer survival: The joint impact of correlated tumour markers. Br. J. Cancer 2013, 108, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Du, X. Kras mutation testing in metastatic colorectal cancer. World J. Gastroenterol. WJG 2012, 18, 5171–5180. [Google Scholar] [PubMed]
- Yamauchi, M.; Morikawa, T.; Kuchiba, A.; Imamura, Y.; Qian, Z.R.; Nishihara, R.; Liao, X.; Waldron, L.; Hoshida, Y.; Huttenhower, C.; et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012, 61, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153–173. [Google Scholar] [PubMed]
- Beyer, K.; Altendorf-Hofmann, A.; Chen, Y.; Bickel, K.; Petersen, I. Kras and aneusomy of chromosomes 4, 10 and 12 in colorectal carcinomas. Pathol. Res. Pract. 2015, 211, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Knickelbein, K.; Zhang, L. Mutant kras as a critical determinant of the therapeutic response of colorectal cancer. Genes Dis. 2015, 2, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Barcelo, C.; Paco, N.; Beckett, A.J.; Alvarez-Moya, B.; Garrido, E.; Gelabert, M.; Tebar, F.; Jaumot, M.; Prior, I.; Agell, N. Oncogenic k-ras segregates at spatially distinct plasma membrane signaling platforms according to its phosphorylation status. J. Cell Sci. 2013, 126, 4553–4559. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Matsusaka, S.; Hirai, M.; Shibata, H.; Takagi, K.; Mizunuma, N.; Hatake, K. A novel approach to detect kras/braf mutation for colon cancer: Highly sensitive simultaneous detection of mutations and simple pre-treatment without DNA extraction. Int. J. Oncol. 2015, 47, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Zinsky, R.; Bolukbas, S.; Bartsch, H.; Schirren, J.; Fisseler-Eckhoff, A. Analysis of kras mutations of exon 2 codons 12 and 13 by snapshot analysis in comparison to common DNA sequencing. Gastroenterol. Res. Pract. 2010, 2010, 789363. [Google Scholar] [CrossRef] [PubMed]
- Vigil, D.; Cherfils, J.; Rossman, K.L.; Der, C.J. Ras superfamily gefs and gaps: Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 2010, 10, 842–857. [Google Scholar] [CrossRef] [PubMed]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of kras mutations and acquired resistance to anti-egfr therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Prenen, H.; Tejpar, S.; Van Cutsem, E. New strategies for treatment of kras mutant metastatic colorectal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 2921–2926. [Google Scholar] [CrossRef] [PubMed]
- Van Krieken, J.H.; Jung, A.; Kirchner, T.; Carneiro, F.; Seruca, R.; Bosman, F.T.; Quirke, P.; Flejou, J.F.; Plato Hansen, T.; de Hertogh, G.; et al. Kras mutation testing for predicting response to anti-egfr therapy for colorectal carcinoma: Proposal for an european quality assurance program. Virchows Arch. Int. J. Pathol. 2008, 453, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Giovanna, M.; Giulia, G.; Sebastiano, C. Biomarkers: Towards the dream of personalized medicine. Available online: https://www.researchgate.net/publication/273703395_Biomarkers_Towards_the_Dream_of_Personalized_Medicine (accessed on 1 January 2018).
- Franklin, W.A.; Haney, J.; Sugita, M.; Bemis, L.; Jimeno, A.; Messersmith, W.A. Kras mutation: Comparison of testing methods and tissue sampling techniques in colon cancer. J. Mol. Diagn. JMD 2010, 12, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Simi, L.; Pratesi, N.; Vignoli, M.; Sestini, R.; Cianchi, F.; Valanzano, R.; Nobili, S.; Mini, E.; Pazzagli, M.; Orlando, C. High-resolution melting analysis for rapid detection of kras, braf, and pik3ca gene mutations in colorectal cancer. Am. J. Clin. Pathol. 2008, 130, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Guarnaccia, M.; Gentile, G.; Alessi, E.; Schneider, C.; Petralia, S.; Cavallaro, S. Is this the real time for genomics? Genomics 2014, 103, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Heller, M.J. DNA microarray technology: Devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002, 4, 129–153. [Google Scholar] [CrossRef] [PubMed]
- Ziober, B.L.; Mauk, M.G.; Falls, E.M.; Chen, Z.; Ziober, A.F.; Bau, H.H. Lab-on-a-chip for oral cancer screening and diagnosis. Head Neck 2008, 30, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Petralia, S.; Verardo, R.; Klaric, E.; Cavallaro, S.; Alessi, E.; Schneider, C. In-check system: A highly integrated silicon lab-on-chip for sample preparation, pcr amplification and microarray detection of nucleic acids directly from biological samples. Sens. Actuators B Chem. 2013, 187, 99–105. [Google Scholar] [CrossRef]
- Teo, J.; Di Pietro, P.; San Biagio, F.; Capozzoli, M.; Deng, Y.M.; Barr, I.; Caldwell, N.; Ong, K.L.; Sato, M.; Tan, R.; et al. Vereflu: An integrated multiplex rt-pcr and microarray assay for rapid detection and identification of human influenza a and b viruses using lab-on-chip technology. Arch. Virol. 2011, 156, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.; Alessi, E.; Conoci, S.; Marchi, M.; Panvini, G. Develop the “in-check” platform for diagnostic applications. Int. Soc. Opt. Photonics 2008. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, D. Miniaturized pcr chips for nucleic acid amplification and analysis: Latest advances and future trends. Nucleic Acids Res. 2007, 35, 4223–4237. [Google Scholar] [CrossRef] [PubMed]
- Pemov, A.; Modi, H.; Chandler, D.P.; Bavykin, S. DNA analysis with multiplex microarray-enhanced pcr. Nucleic Acids Res. 2005, 33, e11. [Google Scholar] [CrossRef] [PubMed]
- Tsarfati-BarAd, I.; Gheber, L. Recent and future developments of microarrays: Miniaturization and lab-on-chip approaches. In Microarrays in Diagnostics and Biomarker Development; Jordan, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 153–168. [Google Scholar]
- Pernagallo, S.; Ventimiglia, G.; Cavalluzzo, C.; Alessi, E.; Ilyine, H.; Bradley, M.; Diaz-Mochon, J.J. Novel biochip platform for nucleic acid analysis. Sensors 2012, 12, 8100–8111. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-blast: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Rogan, P.K.; Cazcarro, P.M.; Knoll, J.H. Sequence-based design of single-copy genomic DNA probes for fluorescence in situ hybridization. Genome Res. 2001, 11, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Krause, A.; Krautner, M.; Meier, H. Accurate method for fast design of diagnostic oligonucleotide probe sets for DNA microarrays. In Proceedings of the International Parallel and Distributed Processing Symposium, Nice, France, 22–26 April 2003. [Google Scholar]
- Chou, C.C.; Chen, C.H.; Lee, T.T.; Peck, K. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 2004, 32, e99. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Gupta, V. Methods for the Determination of Limit of Detection and Limit of Quantitation of the Analytical Methods. Chron. Young Sci. 2011, 2, 21–25. [Google Scholar] [CrossRef]
- Liang, X.; Peng, L.; Li, K.; Peterson, T.; Katzen, F. A method for multi-site-directed mutagenesis based on homologous recombination. Anal. Biochem. 2012, 427, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Ritari, J.; Paulin, L.; Hultman, J.; Auvinen, P. Application of hybridization control probe to increase accuracy on ligation detection or minisequencing diagnostic microarrays. BMC Res. Notes 2009, 2, 249. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.X.; Ai, L.; Chen, J.H.; Feng, X.Y.; Chen, S.H.; Cai, Y.C.; Lu, Y.; Zhou, X.N.; Chen, J.X.; Hu, W. DNA microarray detection of 18 important human blood protozoan species. PLoS Negl. Trop. Dis. 2016, 10, e0005160. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.J.; Capozzoli, M.; Sato, M.; Watthanaworawit, W.; Ling, C.L.; Mauduit, M.; Malleret, B.; Gruner, A.C.; Tan, R.; Nosten, F.H.; et al. An integrated lab-on-chip for rapid identification and simultaneous differentiation of tropical pathogens. PLoS Negl. Trop. Dis. 2014, 8, e3043. [Google Scholar] [CrossRef] [PubMed]
- Dutse, S.W.; Yusof, N.A. Microfluidics-based lab-on-chip systems in DNA-based biosensing: An overview. Sensors 2011, 11, 5754–5768. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Keum, N.; Nishihara, R.; Ogino, S. Molecular pathological epidemiology: New developing frontiers of big data science to study etiologies and pathogenesis. J. Gastroenterol. 2017, 52, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Chan, A.T.; Fuchs, C.S.; Giovannucci, E. Molecular pathological epidemiology of colorectal neoplasia: An emerging transdisciplinary and interdisciplinary field. Gut 2011, 60, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, M. Personalized medicine: Special treatment. Nature 2014, 513, S8–S9. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.U. Genetics: Big hopes for big data. Nature 2015, 527, S108–S109. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, U.H.; Inci, F.; Wang, S.; Toy, M.; Tekin, H.C.; Javaid, A.; Lau, D.T.; Demirci, U. Recent advances in micro/nanotechnologies for global control of hepatitis b infection. Biotechnol. Adv. 2015, 33, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Katsanis, S.H.; Katsanis, N. Molecular genetic testing and the future of clinical genomics. Nat. Rev. Genet. 2013, 14, 415–426. [Google Scholar] [CrossRef] [PubMed]

| Exon 2 | rs121913530 |
| 10570G>T/G>C/G>A-->Gly12Cys/GLY12ARG/Gly12Ser | |
| LUNG CANCER, SQUAMOUS CELL, SOMATIC | |
| BLADDER CANCER, SOMATIC, INCLUDED | |
| rs121913529 | |
| 10571G>C/G>T/G>A--> Gly12Ala/GLY12Val/Gly12Asp | |
| LUNG CANCER, SQUAMOUS CELL, SOMATIC | |
| BLADDER CANCER, SOMATIC, INCLUDED | |
| rs121913535 | |
| 10573G>T/G>C/G>A --> Gly13Cys/Gly13Arg/Gly13Ser | |
| BREAST ADENOCARCINOMA, SOMATIC | |
| rs112445441 | |
| 10574G>A/G>C/G>T --> Gly13Ala/Gly13Asp/Gly13Val | |
| BREAST ADENOCARCINOMA, SOMATIC | |
| Exon 3 | rs121913528 |
| 28572G>A/Ala59Thr | |
| BLADDER CANCER, TRANSITIONAL CELL, SOMATIC | |
| rs121913238 | |
| 28578C>A-->Gln61Lys | |
| COLORECTAL CANCER | |
| rs121913240 | |
| 28579A>T/A>G-->Gln61Leu/Gln>Arg | |
| NON-SMALL CELL LUNG CANCER | |
| rs17851045 | |
| 28580A>C/A>T-->Gln61His | |
| COLORECTAL CANCER | |
| Exon 4 | rs770248150 |
| 30208A>C/A>T-->Lys117Asp) | |
| COLORECTAL CANCER | |
| rs121913527 | |
| 30293G>C/G>A-->Ala146Pro/Ala146Thr | |
| COLORECTAL CANCER | |
| rs1057519725 | |
| 30294C>T-->Ala146Val | |
| COLORECTAL CANCER |
| Name | Primer F | Primer R | Product Size | Basic Tm |
|---|---|---|---|---|
| KRAS exon 2 | ACTGGTGGAGTATTTGATAGTGTAT | AGAATGGTCCTGCACCAGTAA | 249 | 52 °C |
| KRAS exon 3 | AGGTGCACTGTAATAATCCAGACT | AACCCACCTATAATGGTGAATATCT | 228 | 53 °C |
| KRAS exon 4 | AAGGACTCTGAAGATGTACCTATG | AGAAGCAATGCCCTCTCAAG | 293 | 53 °C |
| Reagents | Volume (µL) | Final Concentration |
|---|---|---|
| Forward Primer 10 µM | 0.5 | 0.2 µM |
| Reverse Primer 10 µM | 0.5 | 0.2 µM |
| HotStart Taq plus DNA Polymerase | 0.4 | 2.5 U |
| Cl2Mg 25 mM | 0.2 | 0.5 mM |
| dNTPs Solution Mix 10 mM each | 0.5 | 4 mM each |
| Buffer 10X | 2.5 | |
| Genomic DNA | 5 | 20 ng/µL |
| DNase Free Water | 1 | / |
| Total Volume | 12.5 | |
| Cycle Steps | Temp. | Time | Number of Cycles |
|---|---|---|---|
| Initial Denaturation | 95° | 900 s | 1 |
| Denaturation | 94° | 60 s | 35 |
| Annealing | 61° | 60 s | |
| Extension | 72° | 60 s | |
| Final Extension | 72° | 600 s | 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guarnaccia, M.; Iemmolo, R.; San Biagio, F.; Alessi, E.; Cavallaro, S. Genotyping of KRAS Mutational Status by the In-Check Lab-on-Chip Platform. Sensors 2018, 18, 131. https://doi.org/10.3390/s18010131
Guarnaccia M, Iemmolo R, San Biagio F, Alessi E, Cavallaro S. Genotyping of KRAS Mutational Status by the In-Check Lab-on-Chip Platform. Sensors. 2018; 18(1):131. https://doi.org/10.3390/s18010131
Chicago/Turabian StyleGuarnaccia, Maria, Rosario Iemmolo, Floriana San Biagio, Enrico Alessi, and Sebastiano Cavallaro. 2018. "Genotyping of KRAS Mutational Status by the In-Check Lab-on-Chip Platform" Sensors 18, no. 1: 131. https://doi.org/10.3390/s18010131