Quantitative Determination of Fusarium proliferatum Concentration in Intact Garlic Cloves Using Near-Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Quantification of F. proliferatum in Garlic Cloves
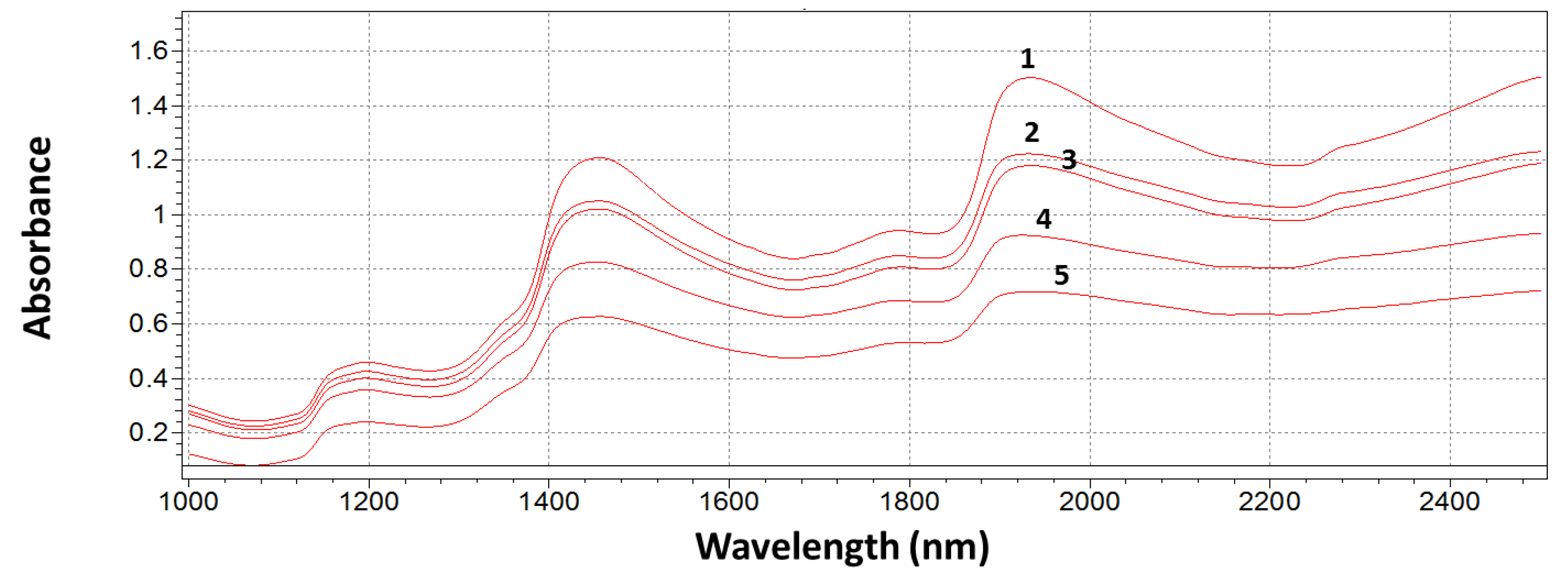
3.2. NIR Spectra Analysis
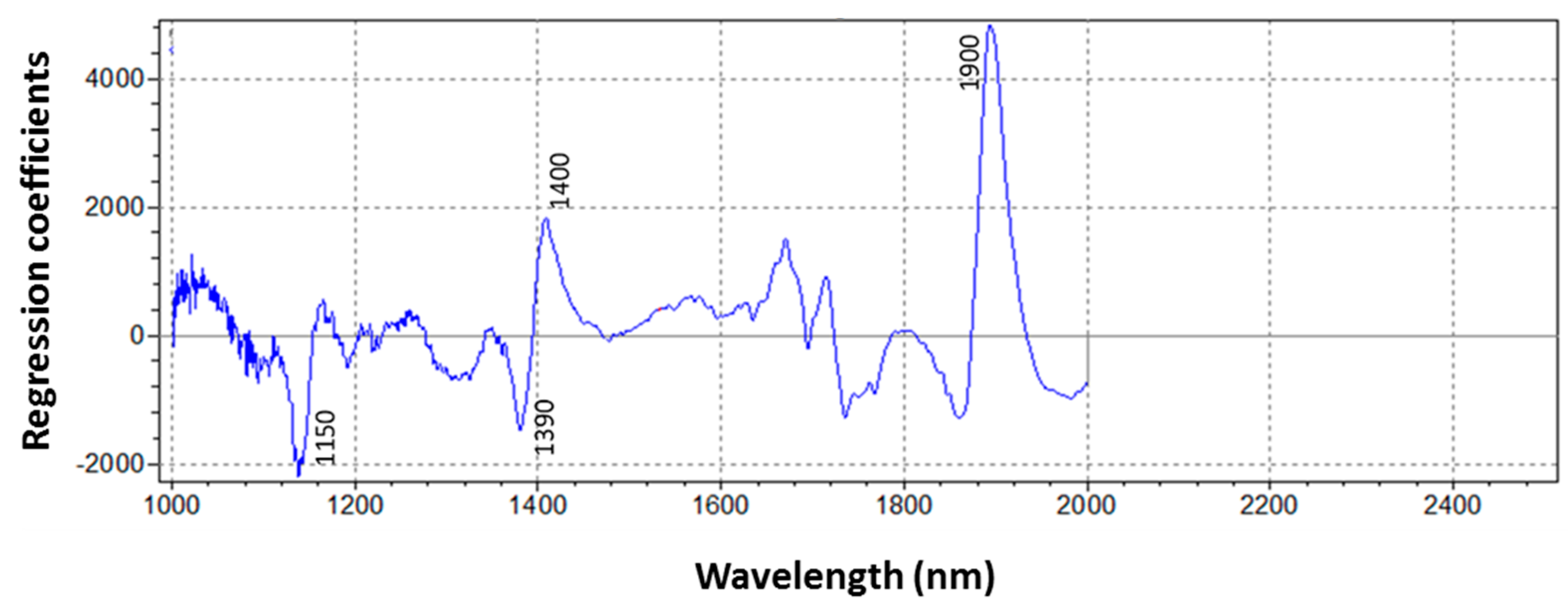
3.3. NIRS Model Development
3.4. NIRS Model External Validation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rheeder, J.P.; Marasas, W.F.O.; Vismer, H.F. Production of fumonisin analogs by Fusarium species. Appl. Environ. Microbiol. 2002, 68, 2101–2105. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha, M.E.B.; Freire, F.D.C.O.; Maia, F.E.F.; Guedes, M.I.F.; Rondina, D. Mycotoxins and their effects on human and animal health. Food Control 2014, 36, 159–165. [Google Scholar] [CrossRef]
- Waśkiewicz, A.; Beszterda, M.; Golinski, P. Occurrence of fumonisins in food—An interdisciplinary approach to the problem. Food Control 2012, 26, 491–499. [Google Scholar] [CrossRef]
- Jurado, M.; Marín, P.; Callejas, C.; Moretti, A.; Vázquez, C.; González-Jaén, M.T. Genetic variability and fumonisin production by Fusarium proliferatum. Food Microbiol. 2010, 27, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.; Burgess, L.W.; Summerell, B.A.; O’Keeffe, K. Fusarium species associated with cob rot of sweet corn and maize in New South Wales. Aust. Plant Dis. Notes 2014, 9, 1–4. [Google Scholar] [CrossRef]
- Logrieco, A.; Mule, G.; Moretti, A.; Bottalico, A. Toxigenic Fusarium species and mycotoxins associated with maize ear rot in Europe. Eur. J. Plant Pathol. 2002, 108, 597–609. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Busman, M.; Proctor, R.H.; Stessman, R. Wheat kernel black point and fumonisin contamination by Fusarium proliferatum. Food Addit. Contam. 2007, 24, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Stępień, Ł.; Koczyk, G.; Waśkiewicz, A. Diversity of Fusarium species and mycotoxins contaminating pineapple. J. Appl. Genet. 2013, 54, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Von Bargen, S.; Martinez, O.; Schadock, I.; Eisold, A.M.; Gossmann, M.; Buttner, C. Genetic variability of phytopathogenic Fusarium proliferatum associated with crown rot in Asparagus officinalis. J. Phytopathol. 2009, 157, 446–456. [Google Scholar] [CrossRef]
- Waskiewicz, A.; Stepien, L.; Wilman, K.; Kachlicki, P. Diversity of Pea-Associated F. proliferatum and F. verticillioides Populations Revealed by FUM1 Sequence Analysis and Fumonisin Biosynthesis. Toxins 2013, 5, 488–503. [Google Scholar] [CrossRef] [PubMed]
- Bhale, U.N.; Chatage, V.S.; Ambuse, M.G. First Report of Fusarium proliferatum Inciting Wilt of Rumex acetosa L. in Maharashtra, India. J. Plant Pathol. Microbiol. 2012, 3, 116. [Google Scholar] [CrossRef]
- Medina, Á.; Rodríguez, A.; Magan, N. Climate change and mycotoxigenic fungi: Impacts on mycotoxin production. Curr. Opin. Food Sci. 2015, 5, 99–104. [Google Scholar] [CrossRef]
- Spanjer, M.C.; Rensen, P.M.; Scholten, J.M. LC–MS/MS multi-method for mycotoxins after single extraction, with validation data for peanut, pistachio, wheat, maize, cornflakes, raisins and figs. Food Addit. Contam. 2008, 25, 472–489. [Google Scholar] [CrossRef] [PubMed]
- Yörük, E.; Albayrak, G. Chemotyping of Fusarium graminearum and F. culmorum isolates from Turkey by PCR assay. Mycopathologia 2012, 173, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Fraga, M.E.; Kozakiewicz, Z.; Lima, N. Fourier transform infrared as a powerful technique for the identification and characterization of filamentous fungi and yeasts. Res. Microbiol. 2010, 161, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Siesler, H.W.; Ozaki, Y.; Kawata, S.; Heise, H.M. Near-Infrared Spectroscopy: Principles, Instruments, Applications; Wiley-VCH Verlag GmbH: Weinheim, Germany, 2008. [Google Scholar]
- McClure, W.F. 204 years of near infrared technology: 1800–2003. J. Near Infrared Spectros. 2003, 11, 487–518. [Google Scholar] [CrossRef]
- Pettersson, H.; Åberg, L. Near infrared spectroscopy for determination of mycotoxins in cereals. Food Control 2003, 14, 229–232. [Google Scholar] [CrossRef]
- Fernández-Ibañez, V.; Soldado, A.; Martínez-Fernández, A.; De la Roza-Delgado, B. Application of near infrared spectroscopy for rapid detection of aflatoxin B1 in maize and barley as analytical quality assessment. Food Chem. 2009, 113, 629–634. [Google Scholar] [CrossRef]
- Hossain, M.Z.; Goto, T. Near- and mid-infrared spectroscopy as efficient tools for detection of fungal and mycotoxin contamination in agricultural commodities. World Mycotoxin J. 2014, 7, 507–515. [Google Scholar] [CrossRef]
- Hernández-Hierro, J.M.; García-Villanova, R.J.; González-Martín, I. Potential of near infrared spectroscopy for the analysis of mycotoxins applied to naturally contaminated red paprika found in the Spanish market. Anal. Chim. Acta 2008, 622, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Mishra, H.N. A rapid FT-NIR method for estimation of aflatoxin B 1 in red chili powder. Food Control 2009, 20, 840–846. [Google Scholar] [CrossRef]
- Sirisomboon, C.D.; Putthang, R.; Sirisomboon, P. Application of near infrared spectroscopy to detect aflatoxigenic fungal contamination in rice. Food Control 2013, 33, 207–214. [Google Scholar] [CrossRef]
- Singh, C.B.; Jayas, D.S.; Paliwal, J.; White, N.D.G. Fungal damage detection in wheat using short-wave near-infrared hyperspectral and digital colour imaging. Int. J. Food Prop. 2012, 15, 11–24. [Google Scholar] [CrossRef]
- FAOSTAT. 2014. Available online: http://faostat.fao.org/site/291/default.aspx (accessed on 21 April 2016).
- Palmero, D.; de Cara, M.; Nosir, W.; Galvez Patón, L.; Cruz, A.; Woodward, S.; González-Jaén, M.T.; Tello, J.C. Fusarium proliferatum isolated from garlic in Spain: Identification, toxigenic potential and pathogenicity on related Allium species. Phytopathol. Mediter. 2013, 51, 207–218. [Google Scholar]
- Clewer, A.G.; Scarisbrick, D.H. Practical Statistics and Experimental Design for Plant and Crop Science; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Isaksson, T.; Næs, T. The effect of multiplicative scatter correction (MSC) and linearity improvement in NIR spectroscopy. Appl. Spectrosc. 1988, 42, 1273–1284. [Google Scholar] [CrossRef]
- Martens, H.; Naes, T. Multivariate Calibration; John Wiley & Sons: Chichester, UK, 1988. [Google Scholar]
- Williams, P.C. Near-Infrared Technology in the Agricultural and Food Industries, 2nd ed.; American Association of Cereal Chemists: Saint Paul, MN, USA, 2001. [Google Scholar]
- Wold, S.; Antti, H.; Lindgren, F.; Öhman, J. Orthogonal signal correction of near-infrared spectra. Chemom. Intell. Lab. Syst. 1998, 44, 175–185. [Google Scholar] [CrossRef]
- Rocke, D.M.; Woodruff, D.L. Identification of Outliers in Multivariate Data. J. Am. Stat. Assoc. 1996, 91, 1047–1061. [Google Scholar] [CrossRef]
- Durbin, J.; Watson, G.S. Testing for serial correlation in least squares regression. Biometrika 1950, 37, 409–428. [Google Scholar] [PubMed]
- Tamburini, E.; Marchetti, M.G.; Pedrini, P. Monitoring Key Parameters in Bioprocesses Using Near-Infrared Technology. Sensors 2014, 14, 18941–18959. [Google Scholar] [CrossRef] [PubMed]
- Dhanoa, M.S.; Lister, S.J.; Sanderson, R.; Barnes, R.J. The link between multiplicative scatter correction (MSC) and standard normal variate (SNV) transformations of NIR spectra. J. Near Infrared Spectrosc. 1994, 2, 43–47. [Google Scholar] [CrossRef]
- Vivó-Truyols, G.; Schoenmakers, P.J. Automatic selection of optimal Savitzky-Golay smoothing. Anal. Chem. 2006, 78, 4598–4608. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.W.; Kellogg, C.A.; Peak, K.K.; Shinn, E.A. A rapid and efficient assay for extracting DNA from fungi. Lett. Appl. Microbiol. 2002, 34, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.M.; Jiménez-Gasco, M.; Kang, S.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’donnell, K. FUSARIUM-ID v.1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- Kennedy, S.; Oswald, N. PCR Troubleshooting and Optimization: The Essential Guide; Caister Academic: Edinburgh, UK, 2011. [Google Scholar]
- Martínez, N.; Martín, M.C.; Herrero, A.; Fernández, M.; Alvarez, M.A.; Ladero, V. qPCR as a powerful tool for microbial food spoilage quantification: Significance for food quality. Trends Food Sci. Technol. 2011, 22, 367–376. [Google Scholar] [CrossRef]
- Nam, H.-M.; Srinivasan, V.; Gillespie, B.E.; Murinda, S.E.; Oliver, S.P. Application of SYBR green real-time PCR assay for specific detection of Salmonella spp. in dairy farm environmental samples. Int. J. Food Microbiol. 2005, 102, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Postollec, F.; Falentin, H.; Pavan, S.; Combrisson, J.; Sohier, D. Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol. 2011, 28, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Levin, R.E. Rapid Detection and Characterization of Foodborne Pathogens by Molecular Techniques; CRC Press: Boca Raton, FL, USA, 2010; pp. 8–20. [Google Scholar]
- Dugan, F.M.; Hellier, B.C.; Lupien, S.L. Pathogenic fungi in garlic seed cloves from the United States and China, and efficacy of fungicides against pathogens in garlic germplasm in Washington State. J. Phytopathol. 2007, 155, 437–445. [Google Scholar] [CrossRef]
- Sun, J. Statistical analysis of NIR data: Data pretreatment. J. Chemom. 1997, 11, 525–532. [Google Scholar] [CrossRef]
- Workman, J., Jr.; Weyer, L. Practical Guide to Interpretive Near-Infrared Spectroscopy; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Kodera, Y.; Ayabe, M.; Ogasawara, K.; Yoshida, S.; Hayashi, N.; Ono, K. Allixin accumulation with long-term storage of garlic. Chem. Pharm. Bull. 2002, 50, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Gregori, R.; Meriggi, P.; Pietri, A.; Formenti, S.; Baccarini, G.; Battilani, P. Dynamics of fungi and related mycotoxins during cereal storage in silo bags. Food Control 2013, 30, 280–287. [Google Scholar] [CrossRef]
- Lawson, L.D.; Gardner, C.D. Composition, Stability, and Bioavailability of Garlic Products Being Used in a Clinical Trial. J. Agric. Food. Chem. 2005, 53, 6254–6261. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Dowell, F.E.; Ram, M.S.; Schapaugh, W.T. Classification of fungal-damaged soybean seeds using near-infrared spectroscopy. Int. J. Food Prop. 2004, 7, 75–82. [Google Scholar] [CrossRef]


| Grade of Infection | Range (ppb) | Average Value (ppb) | # Samples | # Outliers |
|---|---|---|---|---|
| 1 | 0.34–20.68 | 10.51 | 20 | 6 |
| 2 | 18.31–79.88 | 49.10 | 20 | 4 |
| 3 | 59.76–322.13 | 190.94 | 20 | 2 |
| 4 | 345.27–1986.34 | 1165.80 | 15 | 2 |
| 5 | 2019.20–7231.15 | 4625.17 | 20 | 5 |
| Statistical Parameter | Calibration | Cross Validation |
|---|---|---|
| Units | ppb | ppb |
| SEL-reproducibility | 0.10 | 0.10 |
| # Samples | 190 | 95 |
| Outliers | 0 | 0 |
| Min | 0.34 | 0.95 |
| Max | 7231.15 | 6541.89 |
| SD | 1391.17 | 1467.98 |
| Segment (nm) | 4 | 4 |
| WL range/step (nm) | 1000–2500/8 | 1000–2500/8 |
| Pretreatments | SNV/D1 | SNV/D1 |
| Regression method | PLSR | blockwise CV |
| Number of factors | 4 | - |
| SEC | 615.17 | - |
| R2 | 0.829 | 0.774 |
| SECV | - | 717.41 |
| NIRS repeatability | 0.11 | 0.11 |
| DW | 2.09 | 1.84 |
| C-Set Durbin–Watson in range 1.5 to 2.5? | yes | yes |
| Q-value | 0.85 | - |
| RPD | 2.26 | 2.04 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamburini, E.; Mamolini, E.; De Bastiani, M.; Marchetti, M.G. Quantitative Determination of Fusarium proliferatum Concentration in Intact Garlic Cloves Using Near-Infrared Spectroscopy. Sensors 2016, 16, 1099. https://doi.org/10.3390/s16071099
Tamburini E, Mamolini E, De Bastiani M, Marchetti MG. Quantitative Determination of Fusarium proliferatum Concentration in Intact Garlic Cloves Using Near-Infrared Spectroscopy. Sensors. 2016; 16(7):1099. https://doi.org/10.3390/s16071099
Chicago/Turabian StyleTamburini, Elena, Elisabetta Mamolini, Morena De Bastiani, and Maria Gabriella Marchetti. 2016. "Quantitative Determination of Fusarium proliferatum Concentration in Intact Garlic Cloves Using Near-Infrared Spectroscopy" Sensors 16, no. 7: 1099. https://doi.org/10.3390/s16071099
APA StyleTamburini, E., Mamolini, E., De Bastiani, M., & Marchetti, M. G. (2016). Quantitative Determination of Fusarium proliferatum Concentration in Intact Garlic Cloves Using Near-Infrared Spectroscopy. Sensors, 16(7), 1099. https://doi.org/10.3390/s16071099







