Quorum Quenching Bacillus sonorensis Isolated from Soya Sauce Fermentation Brine
Abstract
: An N-acylhomoserine lactone (AHL)-degrading bacterial strain, L62, was isolated from a sample of fermentation brine of Chinese soya sauce by using rich medium agar supplemented with soya sauce (10% v/v). L62, a rod-shaped Gram positive bacterium with amylolytic activity, was phylogentically related to Bacillus sonorensis by 16S ribosomal DNA and rpoB sequence analyses. B. sonorensis L62 efficiently degraded N-3-oxohexanoyl homoserine lactone and N-octanoylhomoserine lactone. However, the aiiA homologue, encoding an autoinducer inactivation enzyme catalyzing the degradation of AHLs, was not detected in L62, suggesting the presence of a different AHL-degrading gene in L62. To the best of our knowledge, this is the first report of AHL-degrading B. sonorensis from soya sauce liquid state fermentation.1. Introduction
Gram negative bacterial communication achieved via production and sensing of freely diffusible N-acylhomoserine lactone (AHL) molecules is known as quorum sensing (QS) [1]. The N-acyl side chains of different AHLs may vary in length, saturation, and substitution [2]. AHLs are synthesised by LuxI homologue synthase. Upon reaching threshold level, AHLs will bind to their cognate receptor (LuxR homologue) and affect several QS-dependent phenotypes. QS regulates a broad range of biological activities, including luminescence, antibiotic production, plasmid transfer, and virulence [3–5]. It has been shown that QS also plays significant role in food related bacteria [6].
Quorum quenching (QQ) refers to the process of interrupting QS and has been regarded as a promising novel approach to attenuate bacterial pathogens [3]. Enzymatic inactivation of AHL can be through either the opening of the lactone ring moiety by AHL lactonase or detachment of N-acyl side chain from the lactone ring via acylase [7–9].
Chinese soya sauce is widely used as a flavour enhancer in Asia, including China, Japan, and Southeast Asia. It is a liquid condiment employed in most oriental cuisine owing to its unique characteristic taste and it can be served plain as a sauce or form part of the cooking ingredients. Chinese soya sauce is produced by a two-stage fermentation process, namely solid state and liquid fermentations. In solid state fermentation, cooked soya beans are mixed with an arbitrary amount of flour to allow Aspergillus oryzae or Aspergillus sojae to grow on the materials at room temperature. However, brine fermentation involves the concurrent fermentation of soya mash in sea salt brine (17–20% w/v NaCl) with added bacteria and yeasts [10]. After a specific period of aging, the final product is made by pressing the soya mash and then filtered, before sugar is added to taste. Soya sauce is then sold as extra virgin for the first extraction. A subsequent extraction is achieved via extracting the second liquid fermentation after addition of more sea salt brine to the extracted soya mash and fermentation in the similar manner as described. The second extract will be sold as lower grade soya sauce.
To date, no QQ bacteria have been reported from fermented brines used to make soya sauce. To address this issue, we isolated culturable bacterium from a commercial well-aged (three months) soya sauce mash during brine fermentation. Subsequently, we characterised a novel amylolytic, QQ bacterium Bacillus sonorensis L62 that efficiently degraded AHLs in vitro.
2. Experimental Section
In addition to the bacterial strains isolated from soya sauce fermentation brine, bacteria used in this study included Escherichia coli DH5α, Bacillus cereus [11], and the biosensor Chromobacterium violaceum CV026 which detects short chain AHLs [12].
Lysogeny broth (LB) rich medium (per 100 mL: 1.0 g tryptone, 0.5 g yeast extract, 0.5 g NaCl) was used. To prepare modified LB medium (LBm) supplemented with different soya sauce concentrations, the following compositions were used (per 100 mL): 1.0 g tryptone, 0.5 g yeast extract, 0.5 g NaCl, supplemented with appropriate volumes of soya sauce at 5, 10, and 15% (v/v). The media were autoclaved. Bacteriological agar (1.5% w/v) was used to solidify LB and LBm. For selection of transformants, LB agar was supplemented with ampicillin (100 μg/mL).
Starch agar was prepared according to Stark et al. [13], with the following composition (per 100 mL): 0.5 g soluble starch (BDH Chemical Ltd., Poole, UK), 0.2 g yeast extract, 0.5 g tryptone, 0.5 g NaCl, and 1.5 g bacteriological agar. Bacterial colonies were streaked onto the starch agar and incubated for 24 to 72 h at 37 °C. Amylolytic activity was detected by the formation of halos around bacterial colonies after flooding the agar with iodine solution.
A well-aged (three months) sample of soya sauce fermentation brine (100 mL) was collected at 0.5 cm below the surface of a liquid state fermentation in a fermentation tank of a local factory. Aliquots of the sample (100 μL) were spread on LB and LBm plates, which were subsequently incubated at 37 °C for 24 to 48 h. Pure colonies were obtained by repeated dilution streaking on LB agar. For routine maintenance, bacterial isolates were kept on LB agar slants and in glycerol (80% v/v) at −80 °C.
Microbiological and molecular techniques, as described previously [14], were performed to identify one of the bacterial isolates, L62. Gram staining was performed and bacterial cell morphology was observed using light microscope (Olympus, Japan) at 1,000× magnification. To obtain the 16S ribosomal DNA (rDNA) of L62, an internal fragment of ≈1.5 kb was amplified by the polymerase chain reaction (PCR) with bacterial genomic DNA as template. The 16S rDNA PCR primers 27F (5′-AGAGTTTGATC(M)TGGC-TCAG-3′) and 1525R (5′-AAGGAGGTG(W)TCCA(R)-CC-3′) were used as forward and reverse primers, respectively. The rpoB gene, encoding the β-subunit of RNA polymerase, of L62 was amplified with reported primers rpoBF (5′-AGGTCAACTAGTTCAGTA TGGACG-3′) and rpoBR (5′-ACCGTAACCGGCAACTTAC-3′) as described by Palmisano et al. [15]. Purification, ligation, transformation, and sequencing of PCR products were carried out essentially as previously described [16]. Phylogenetic analysis was done by the Neighbor-Joining method using MEGA version 4.0 [17] as reported elsewhere [16].
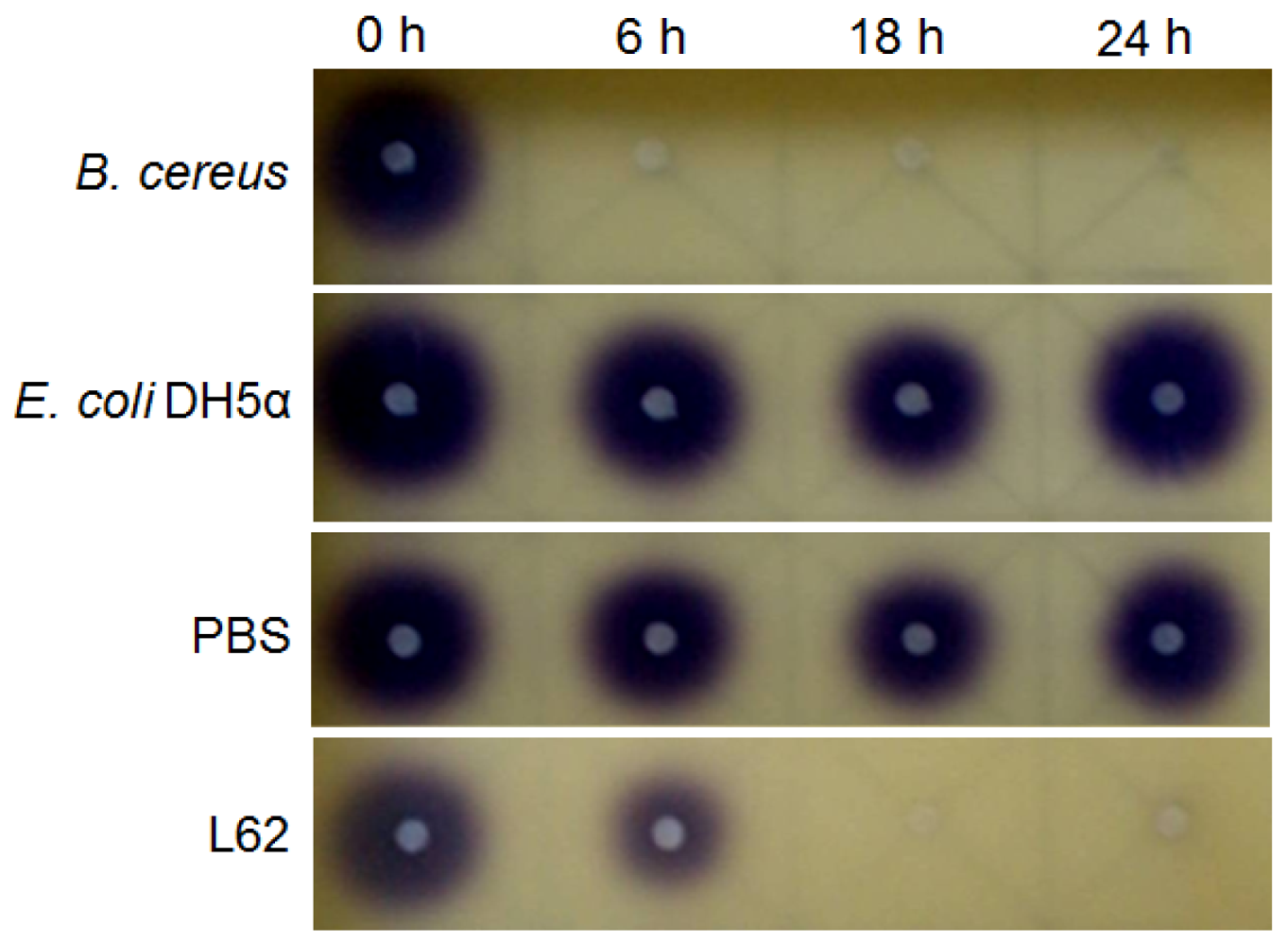
Bacterial cells (5 mL) were grown in LB broth at 37 °C (220 rpm) to stationary phase. Cells were collected by centrifugation, washed twice, and suspended in phosphate buffered saline (PBS, 150 mM, pH 6.5). The resulting concentrated cell suspension was used as the source of resting cells for in vitro AHL inactivation assays, as previously described [14]. The AHLs tested were N-3-oxohexanoyl homoserine lactone (3-oxo-C6-HSL), N-heptanoylhomoserine lactone (C7-HSL) and N-octanoylhomoserine lactone (C8-HSL) (Sigma-Aldrich, St. Louis, MO, USA). For the whole-cell assay, 5 μL of AHL in absolute ethanol was dispensed into a sterile tube and the solvent evaporated to dryness. The dried AHL was then rehydrated with 100 μL of a bacterial cell suspension to a final AHL concentration of 0.5 μg/μL. The resting cell suspension was incubated at 37 °C for up to 24 h and at regular intervals (0, 6, 18, and 24 h), aliquots (15 μL) were withdrawn and heat inactivated (95 °C, 3 min). Experiments involving E. coli DH5α and PBS served as negative controls and B. cereus was included as a positive control for AHL-degradation. Residual AHLs were detected by the formation of purple pigmentation on C. violaceum CV026 lawn [14].
Rapid Resolution Liquid Chromatography (RRLC) analysis was carried out to further confirm the QQ activity of L62. To analyze the degradation of AHLs over a period of time, we used an RRLC instrument (Agilent Technologies 1200 series) equipped with an Agilent ZORBAX Eclipse® XDB-C18 column (4.6 × 50 mm, 1.8 μm particle size) as reported [14]. The elution procedure consisted of an isocratic profile of acetonitrile-water (35:65, v/v) for 3 min at a constant flow rate of 0.7 mL/min and monitored at 210 nm. Both the retention time and spectral properties were compared to those of synthetic AHL standards. AHLs incubated with washed E. coli DH5α cells and PBS were used as negative controls.
The 16S rDNA and rpoB gene sequences of L62 (GenBank accession numbers of HM191249 and HQ108343, respectively) have been deposited at GenBank.
3. Results and Discussion
3.1. Isolation of L62 from Soya Sauce Fermentation Brine
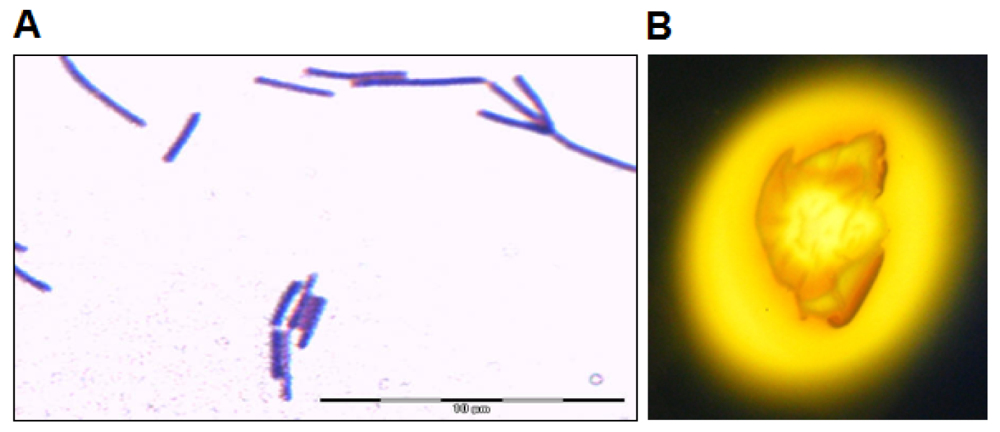
Isolate L62 was isolated from a sample of soya sauce fermentation brine plated on LBm (10% v/v soya sauce). Bacterial isolates were then streaked on LBm agar repeatedly to obtain pure colonies, which appeared after incubation for 24 to 48 h at 37 °C. The bacterial colonies were morphologically homogeneous, suggesting that limited types of bacteria were enriched in LBm. When grown on LB agar, its colonies appeared brown yellowish and irregular with undulate margin. Its rod-shaped cells were Gram stain positive (Figure 1(A)). L62 showed amylolytic activity after incubation for 72 h (Figure 1(B)).
3.2. Molecular Characterisation of L62
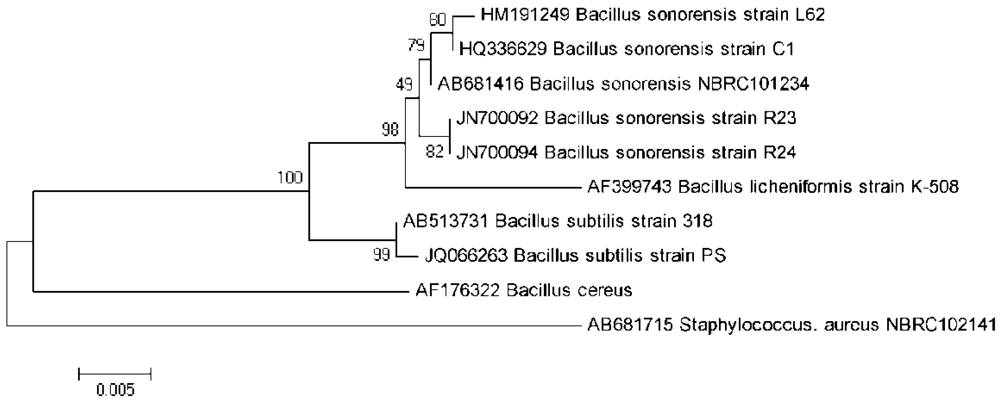
The 16S rDNA gene of isolate L62 was PCR amplified, sequenced, edited, aligned, and matched against similar sequences in the GenBank. Its 1,542 nucleotides showed 99.8% similarity with the 16S rDNA of B. sonorensis strain C1 (GenBank accession number HQ336629). A phylogenetic tree was constructed based on the 16S rDNA gene sequences of L62, other related bacterial species, and an outgroup (Figure 2). Strain L62 is phylogenetically related to B. sonorensis as compared to other bacilli. Furthermore, the amplification and analysis of the rpoB gene confirmed our identification of isolate L62 to be B. sonorensis (data not shown). Hence, L62 was named B. sonorensis L62.
3.3. AHL-Degradation by Isolate L62
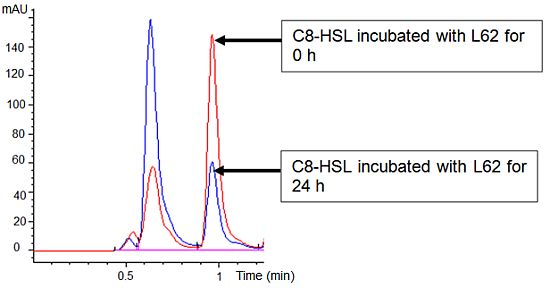
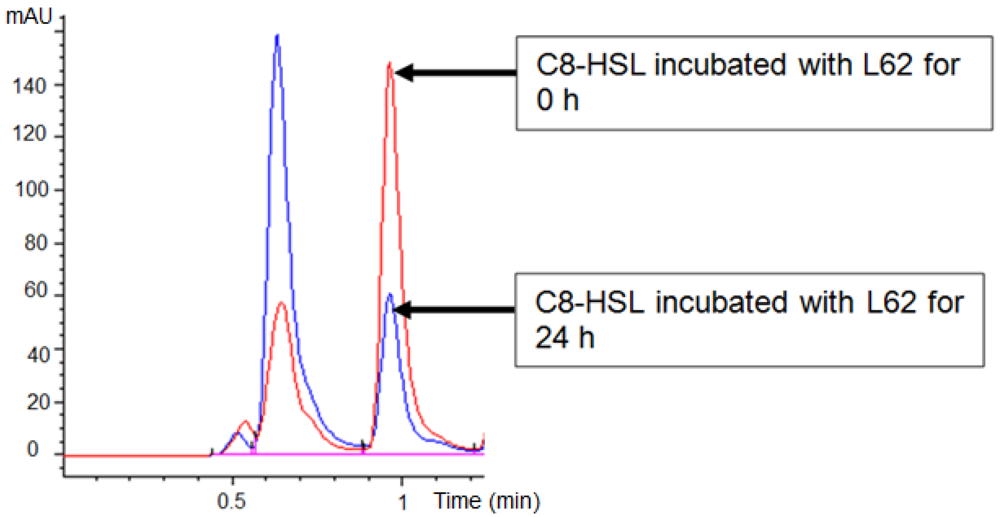
Isolate L62 degraded 3-oxo-C6-HSL efficiently within 6 h and no detectable AHL molecules were found after further incubation to 18 h (Figure 3). No significant C7-HSL degradation by isolate L62 was observed (data not shown). To further investigate AHL-degradation specificity, we incubated PBS-washed L62 cells with C8-HSL and used RRLC to analyze the resulting growth medium. Based on a published criterion [16], Figure 4 shows that L62 degraded about 60% of C8-HSL within 24 h, suggesting that L62 could inactivate AHL molecules with different substitutions at C3 position and with C6 and C8 N-acyl side chains. No apparent C8-HSL degradation was observed when the same experiment was repeated with the incubation buffer (PBS) and E. coli DH5α cells serving as negative controls (data not shown).
Recently, a number of food-related bacteria have been reported to show QS activity [6]. However, no report has indicated the presence of AHL-degrading bacteria in the unique fermentation of soya sauce. Here, we describe a QQ positive bacterial isolate, B. sonorensis L62, from soya sauce fermentation brine. L62 grew well on LBm (10% v/v soya sauce) and LB agar.
B. sonorensis, first isolated from desert soil, has been reported by Palmisano et al. [15] to withstand highly saline soils and dehydration. Later, B. sonorensis has also been isolated from a sample of ropy bread [18] and semifinal gelatin extracts [19]. Recently, Park et al. [20] reported the isolation of B. sonorensis under anaerobic conditions from a traditionally fermented Korean soya bean paste called eoyukjang. In contrast to Park et al. [20], we isolated QQ B. sonorensis L62 under aerobic conditions from traditional Chinese soya sauce liquid fermentation.
Palmisano et al. [15] described B. sonorensis as a novel species that could only be phenotypically distinguished from B. licheniformis by salt tolerance and pigmentation. Isolate L62 was salt tolerant and grew optimally in rich media supplemented with 3% (w/v) NaCl (data not shown). Palmisano et al. [15] also reported that B. sonorensis is an active heterofermenter and a spore-forming bacillus with casein and starch hydrolysis activities. However, isolate L62 did not show positive proteolytic and lipolytic activities (data not shown) but possessed starch hydrolysis activity. It is speculated that B. sonorensis L62 may play a role in hydrolysis of the flour introduced during the solid-state (koji) phase fermentation with its amylase.
Palmisano et al. [15] suggested using the rpoB sequence to distinguish between B. licheniformis and B. sonorensis. Indeed, the rpoB gene sequence of L62 showed closer similarity with that of B. sonorensis. Phylogenetic analysis revealed that L62 was closely related to B. sonorensis.
Bacilli possessing QQ AHL lactonase (AiiA), similar to that of Bacillus sp. 240B1 [7], have been readily isolated from Malaysian rain forest soil by simple heat-treatment of the soil and enrichment with AHL-supplemented KG medium [11,14]. We used published primers [7,11] to amplify the aiiA gene. However, no aiiA homologue was detected in B. sonorensis L62 by PCR (data not shown), suggesting the possibility of a different QQ gene in B. sonorensis L62.
B. sonorensis L62 showed rapid degradation of AHLs with different C-3 substituents and acyl chain lengths, suggesting broad substrate activity against AHLs. This may represent a nutritional symbiosis in which products of degraded AHLs can be used by other species living in proximity or other well adapted bacteria may act in concert with B. sonorensis L62 in the soya sauce fermentation brine to further metabolise AHLs. Moreover, AHLs exist in D- and L-isomers, but our biosensors can only detect the latter. Hence, further work should be done to clone the QQ gene and determine whether the AHLase of B. sonorensis L62 is stereospecific against the two isomers of AHLs.
Many AHLs are quite stable in acidic and neutral pH environments [21]. Soya sauce liquid state fermentation is typically characterised by its low pH values (<5) owing largely to the activities of lactobacilli. This low pH environment would not cause AHLs present in the soya sauce fermentation brine to degrade. Therefore, they are most likely degraded by the QQ activities of bacteria such as B. sonorensis L62, preventing them from accumulating in soy sauce.
Little is known about the microbial QS and QQ physiotypes during soya sauce liquid state fermentation. Hence, further work should be directed on the role of this QQ B. sonorensis L62 in soya sauce fermentation. In addition, we need to isolate QS bacteria from this unique niche and to study them in order to understand the production of AHLs in soya sauce fermentation brine. We also need to determine the concentration of AHLs in soya sauce (if any) as there is no report on the effect of AHLs on humans after consumption. The degradation of AHLs by B. sonorensis L62 in the soya sauce mash may provide insights to the role of AHL quenching species, metabolic pathways of AHL turnover in soya sauce mash, and the microbial ecological significance of AHL degradation in soya sauce liquid state fermentation. It is plausible that B. sonorensis L62 uses its QQ ability to gain competitive advantage over AHL-dependent signalling bacteria, or B. sonorensis L62 is an important bacterium that has vital role in the soya sauce liquid fermentation process.
4. Conclusions
Not much is known about the presence of QQ bacteria in the traditional soya sauce fermentation except reports on the presence of halophilic bacteria in this unique fermentation process. This is the first report of a bacterial isolate with broad QQ activity, B. sonorensis strain L62, from traditional Chinese soya sauce brine.
Acknowledgments
This research was supported by the University of Malaya HIR Grant (A-000001-50001) and FRGS grant (FP022/2009) to KG Chan. We thank Paul Williams (University of Nottingham, UK) for providing us the biosensor.
References
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar]
- Chhabra, S.R.; Philipp, B.; Eberl, L.; Givskov, M.; Williams, P. Extracellular communication in bacteria. Top. Curr. Chem. 2005, 240, 279–315. [Google Scholar]
- Williams, P. Compromising bacterial communication skills. J. Pharm. Pharmacol. 1994, 46, 252–260. [Google Scholar]
- Williams, P. Quorum sensing, communication and cross-kingdom signaling in the bacterial world. Microbiology 2007, 153, 3923–3938. [Google Scholar]
- Chan, K.G.; Puthucheary, S.D.; Chan, X.Y.; Yin, W.F.; Wong, C.S.; Too, W.S.; Chua, K.H. Quorum sensing in Aeromonas species isolated from patients in Malaysia. Curr. Microbiol. 2011, 62, 167–172. [Google Scholar]
- Gobbetti, M.; Angelis, M.D.; Cagno, R.D.; Minervini, F.; Limitone, A. Cell-cell communication in food related bacteria. Int. J. Food. Microbiol. 2007, 120, 34–45. [Google Scholar]
- Dong, Y.H.; Xu, J.L.; Li, X.Z.; Zhang, L.H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 2000, 97, 3526–3531. [Google Scholar]
- Lin, Y.H.; Xu, J.L.; Hu, J.; Wang, L.H.; Ong, S.L.; Leadbetter, J.R.; Zhang, L.H. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 2003, 47, 849–860. [Google Scholar]
- Chan, K.G.; Atkinson, S.; Mathee, K.; Sam, C.K.; Chhabra, S.R.; Koh, C.L.; Williams, P. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber offinale (ginger) rhizosphere: Co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol 2011. [Google Scholar] [CrossRef]
- Whitaker, J.R. Biochemical changes occurring during the fermentation of high-protein foods. Food Tech. 1978, 32, 175–180. [Google Scholar]
- Chan, K.G.; Tiew, S.Z.; Ng, C.C. Rapid isolation method of soil bacilli and screening of their quorum quenching activity. As. Pac. J. Mol. Biol. Biotechnol. 2007, 15, 153–156. [Google Scholar]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Cámara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar]
- Stark, E.; Wellerson, R., Jr.; Tetrault, P.A.; Kossack, C.F. Bacterial alpha amylase paper disc tests on starch agar. Appl. Microbiol. 1953, 1, 236–243. [Google Scholar]
- Chan, K.G.; Yin, W.F.; Sam, C.K.; Koh, C.L. A novel medium for the isolation of N-acylhomoserine lactone-degrading bacteria. J. Ind. Microbiol. Biotechnol. 2009, 36, 247–251. [Google Scholar]
- Palmisano, M.M.; Nakamura, L.K.; Duncan, K.E.; Istock, C.A.; Cohan, F.M. Bacillus sonorensis sp. nov., a close relative of Bacillus licheniformis, isolated from soil in the Sonoran Desert, Arizona. Int. Syst. Evol. Microbiol. 2001, 51, 1671–1679. [Google Scholar]
- Chan, K.G.; Wong, C.S.; Yin, W.F.; Sam, C.K.; Koh, C.L. Rapid degradation of N-3-oxo-acylhomoserine lactones by a Bacillus cereus isolate from Malaysian rainforest soil. Antonie van Leeuwenhoek 2010, 98, 299–305. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar]
- Sorokulova, I.B.; Reva, O.N.; Smirnov, V.V.; Pinchuk, I.V.; Lapa, S.V.; Urdaci, M.C. Genetic diversity and involvement in bread spoilage of Bacillus strains isolated from flour and ropy bread. Lett. Appl. Microbiol. 2003, 37, 169–173. [Google Scholar]
- De Clerck, E.; Vanhoutte, T.; Hebb, T.; Geerinck, J.; Devos, J.; de Vos, P. Isolation, characterization, and identification of bacterial contaminants in semifinal gelatin extracts. Appl. Environ. Microbiol. 2004, 70, 3664–3672. [Google Scholar]
- Park, J.W.; Lee, N.K.; Kim, B.Y.; Kim, H.K.; Kwon, K.O.; Hahm, Y.T. Characterization of traditionally fermented Korean soybean paste, eoyukjang, and isolation of its microorganisms. Food Sci. Biotechnol. 2010, 19, 425–430. [Google Scholar]
- Yates, E.A.; Philipp, B.; Buckley, C.; Atkinson, S.; Chhabra, S.R.; Sockett, R.E.; Goldner, M.; Dessaux, Y.; Cámara, M.; Smith, H.; Williams, P. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 2002, 70, 5635–5646. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yin, W.-F.; Tung, H.-J.; Sam, C.-K.; Koh, C.-L.; Chan, K.-G. Quorum Quenching Bacillus sonorensis Isolated from Soya Sauce Fermentation Brine. Sensors 2012, 12, 4065-4073. https://doi.org/10.3390/s120404065
Yin W-F, Tung H-J, Sam C-K, Koh C-L, Chan K-G. Quorum Quenching Bacillus sonorensis Isolated from Soya Sauce Fermentation Brine. Sensors. 2012; 12(4):4065-4073. https://doi.org/10.3390/s120404065
Chicago/Turabian StyleYin, Wai-Fong, Hun-Jiat Tung, Choon-Kook Sam, Chong-Lek Koh, and Kok-Gan Chan. 2012. "Quorum Quenching Bacillus sonorensis Isolated from Soya Sauce Fermentation Brine" Sensors 12, no. 4: 4065-4073. https://doi.org/10.3390/s120404065