Simultaneous Detection of Multiple Fish Pathogens Using a Naked-Eye Readable DNA Microarray
Abstract
: We coupled 16S rDNA PCR and DNA hybridization technology to construct a microarray for simultaneous detection and discrimination of eight fish pathogens (Aeromonas hydrophila, Edwardsiella tarda, Flavobacterium columnare, Lactococcus garvieae, Photobacterium damselae, Pseudomonas anguilliseptica, Streptococcus iniae and Vibrio anguillarum) commonly encountered in aquaculture. The array comprised short oligonucleotide probes (30 mer) complementary to the polymorphic regions of 16S rRNA genes for the target pathogens. Targets annealed to the microarray probes were reacted with streptavidin-conjugated alkaline phosphatase and nitro blue tetrazolium/5-bromo-4-chloro-3′-indolylphosphate, p-toluidine salt (NBT/BCIP), resulting in blue spots that are easily visualized by the naked eye. Testing was performed against a total of 168 bacterial strains, i.e., 26 representative collection strains, 81 isolates of target fish pathogens, and 61 ecologically or phylogenetically related strains. The results showed that each probe consistently identified its corresponding target strain with 100% specificity. The detection limit of the microarray was estimated to be in the range of 1 pg for genomic DNA and 103 CFU/mL for pure pathogen cultures. These high specificity and sensitivity results demonstrate the feasibility of using DNA microarrays in the diagnostic detection of fish pathogens.1. Introduction
It may be assumed that fish are continually bathed in an aqueous suspension of microorganisms. Many of the members of the normal microflora of water can be bacterial fish pathogen candidates. Aeromonas hydrophila, an etiological agent of fish diseases, is considered as both a primary and secondary pathogen resulting in hemorrhagic septicemia [1]. Edwardsiella tarda causes the serious systemic septicemia commonly known as edwardsiellosis, which occurs in cultured Japanese eels [2,3], flounders [4], and tilapias [5]. Flavobacterium columnare, which causes gill damage or lesions on the fish body surface has been recognized as a universally occurring pathogen of numerous freshwater fish species, including both coldwater fish [6] and tropical aquarium fish [7]. Photobacterium damselae leads to pasteurellosis and infects a wide range of marine species such as white perch [8], yellowtail [9], gilthead seabream [10], sea bass [11], striped jack [12], Japanese flounder [13], and cobia [14]. Pseudomonas anguilliseptica causes red spot disease, which is one of the most destructive diseases of pond-cultured eels in Japan [15] and Taiwan [16]. This bacterium has also caused disease outbreaks in European eels in Scotland [17], France [18], and in other farmed fish such as black sea bream [19], salmonid [20], and ayu [21]. Outbreaks of disease caused by Vibrio anguillarum represent one of the most commonly occurring examples of vibriosis [22,23]. This pathogen usually produces hemorrhagic septicemia [24], is distributed worldwide, and affects a wide range of fish and shellfish [25–27]. The pathogenic gram-positive cocci Lactococcus garvieae and Streptococcus iniae usually cause hyperacute and hemorrhagic septicemia in both freshwater and marine aquaculture species such as catfish [28], tilapia [29], trout [30], and yellowtail [31]. These pathogens cause massive mortality and large economic losses in fish farming every year.
The above pathogens that infect cultured species are phylogenetically diverse. Consequently, detection of these pathogens using conventional culture-based microbiological methods is technically demanding and time consuming. The wide diversity of assays combined with frequently fastidious growth conditions make molecular tools such as PCR and DNA microarray better options for detection of fish pathogens. PCR assays have been developed for the rapid detection and identification of microorganisms in clinical samples without the need for further isolation [32,33]. A multiplex PCR (m-PCR) approach that can simultaneously identify several pathogens by the PCR amplicon size using gel electrophoresis has successfully been applied to detect fish and shellfish pathogens [34,35]. However, there are practical limits to PCR assays for detecting multiple pathogens at a time. It is not easy to incorporate more than six primer sets because of the cross-reaction in m-PCR, and the challenges inherent in size discrimination among PCR products by conventional electrophoresis [36]. Subsequent sequencing, which is a relatively costly and laborious process, is often needed to confirm product identity. The new developing method, three oligo (primers + probe) PCR (such as TaqMan® real-time PCR) may overcome the problems. However, it requires more expensive equipment and is suggested to be used in quantitative gene expression and allele discrimination research. Thus, to efficiently screen a complex mixture of sequences from different pathogens, DNA microarray is an excellent candidate.
DNA microarrays are miniaturized microsystems based on the ability of DNA to specifically bind to its complementary sequence in hybridization. Oligonucleotide probes for specific targets are stained at distinct sites on a solid support to which the PCR product is then hybridized and detected [37]. Recent developments in DNA microarray allow parallel hybridizations to occur on the same surface and permit multiple independent detections [38]. In most microarray formats, slides are stained with streptavidin-conjugated fluorophore, and the interaction of the target with specific probes is measured by epifluorescence confocal microscopy using an argon ion laser. On the other hand, precipitation staining methods based on the catalytically induced chromogenic precipitation were applied to the microarray technology. Some commercial products (such as TubeArray™ of Alere Technologies GmbH, Germany and LCD-Array kits of Chipron GmbH, Germany) were developed based on different platforms and chromogenic phosphatase substrates. In this study, the NBT/BCIP (nitro blue tetrazolium/5-bromo-4-chloro-3′-indolylphosphate, p-toluidine salt) microarray system was applied to detection of fish pathogens. In the system, biotin-labelled PCR amplicons are firstly captured on the microarray during hybridization. Then the streptavidin conjugated alkaline phosphatase (Strep-AP) in the staining reagent binds to the biotinylated site. The BCIP in the colorimetric developing reagent reacts to Strep-AP and produces a blue-colored precipitate at the site of enzymatic activity. NBT acts as a co-precipitant agent for the BCIP reaction, forming a dark blue, precisely localized precipitate thus helps to visualize positive spots on the microarray. Here we demonstrate a naked-eye reading microarray system targeting 16S rDNA to identify eight common fish pathogens, obviating the need for expensive fluorescence detection facilities.
2. Experimental Section
2.1. Bacterial Strains
The strains used in this study are listed in Table 1. These include 26 representative collection strains, 81 isolates of target fish pathogens (belonging to eight species: A. hydrophila, E. tarda, F. columnare, L. garvieae, P. damselae, P. anguilliseptica, S. iniae and V. anguillarum), and 61 other strains of bacterial species. Strains were grown and maintained following American Type Culture guidelines. In brief, the following organisms were cultured on nutrient agar (incubation temperature/time): A. hydrophila (30 °C/24 h), E. tarda (37 °C/24 h), P. aeruginosa (37 °C/24 h), P. anguilliseptica (20 °C/24–36 h), and Staphylococcus epidermidis (37 °C/24 h). A. sobria (30 °C/24 h) and A. salmonicida (26 °C/24–48 h) were cultured on trypticase soy agar. E. faecalis (37 °C/24 h), E. faecium (37 °C/24 h), L. garvieae (30 °C/24–36 h), and S. iniae (37 °C/24–48 h) were cultured on brain heart infusion agar. V. anguillarum (18 °C/24–48 h) was cultured on enriched nutrient broth. V. proteolyticus (26 °C/24–36 h) was cultured on nutrient agar with 3% NaCl. F. columnare (20–22 °C/72 h) was cultured on Anacker and Ordal medium. L. pelagia (26 °C/24 h), P. damselae (26 °C/24–48 h), V. aestuarianus (26 °C/24–48 h), V. alginolyticus (37 °C/24 h), V. marinus (18 °C/36–48 h), V. salmonicida (15 °C/48 h), and V. vulnificus (30 °C/24 h) were cultured on Marine agar 2216. V. parahaemolyticus (25 °C/24 h) was cultured on modified seawater yeast extract agar. V. harveyi (26 °C/24 h) was cultured on Photobacterium broth. Mycobacterium fortuitum (37 °C/3–5 days) and M. marinum (30 °C/5–10 days) were cultured on Middlebrook 7H10 agar with Middlebrook OADC enrichment.
2.2. Genomic DNA Preparation
Genomic DNA was extracted from pure cultures using the UltraClean™ Microbial DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA), following the manufacturer’s instructions.
2.3. Primers and Probes
The specific oligonucleotide probes (Table 2), each consisting of 30 nucleotides, were designed based on the polymorphic regions of 16S rRNA genes of the target pathogens using the unique probe selector program ( http://array.iis.sinica.edu.tw/ups/) [39]. The database was made by the retrieved sequences of the 26 reference strains (Table 1) from NCBI GenBank. Alignment of each probe to the 16S rDNA sequences of the species used in this study was then determined using the ClustalW alignment program (DS Gene version 1.5; Accelrys Inc., Tokyo, Japan). Discrimination by certain computer-derived probes was not satisfactory in practice, and therefore, we generated new probes by modifying one or two nucleotides from the original sequences. All oligonucleotides were normalized to a calculated annealing temperature of 65 ± 3 °C and commercially synthesized (Operon Biotechnologies, Inc., USA). Three positive control probes (U735, U1352, and EV71) were used in this study. U735 and U1352 were used to confirm the efficacy of PCR and were designed from the conserved regions of 16S rDNA for eubacteria [40]. EV71 was used to confirm the efficacy of hybridization and was designed from the capsid protein VP1 of the human enterovirus 71 gene (the biotin-labeled EV71 PCR amplicon was incorporated in the hybridization buffer supplied with the kit mentioned below). Thirty poly(A) oligonucleotides were used as the negative control probe. Each probe was chemically synthesized and 5′-amino-modified with the space linker of 15 poly(T) (Operon Biotechnologies, Inc.).
The 16S rDNA universal primers 16S-F and 16S-R were referred to as B27F and U1492R, respectively [41]. The primers were commercially synthesized (Operon Biotechnologies, Inc.) with biotin labeled on the 5′ end to generate biotinylated PCR amplicons that could react with streptavidin-conjugated alkaline phosphatase (Strep-AP) and NBT/BCIP for colormetric signaling on the chips.
2.4. Target DNA Amplification
The 16S rDNA of the 168 strains described above was amplified by PCR using universal 16S-F and 16S-R, as described previously [34]. In brief, PCR was performed in a 50 μL reaction mixture containing 0.5 μL of Taq DNA polymerase (5 U/μL; Promega), 5 μL of 10× NH4 buffer, 2 μL of 10 mM dNTP mix, 10 μL of 10 mM MgCl2, 2 μL of 10 μM forward primer 16S-F, 2 μL of 10 μM reverse primer 16S-R, 2 μL of bacterial genomic DNA (100 ng/μL), and 26.5 μL of sterile H2O. The cycling protocol was 1 cycle at 94 °C for 5 min, 30 cycles at 94 °C for 2 min, 48 °C for 1.5 min, and 72 °C for 2 min, followed by 1 cycle at 72 °C for 10 min. The resulting approximately 1.5 kbp PCR products were subsequently cloned into the pGEM-T Easy vector (Promega) and sequenced on an ABI 377 automated sequencer (Applied Biosystems, USA) using vector primers. Sequences were compared with GenBank databases using the BLAST program [42]. The sequencing-confirmed 16S rDNA PCR amplicons were then used to determine the positive probes (U735 and U1352) in DNA hybridization using the protocol described below.
2.5. Microarray Preparation and Hybridization
Spotting 10 μM of each probe to each specific position on the microarray organic polymer substrate (patent no. US-7109024, supplied with the DR. Chip DIY Kit™, DR. Chip Biotechnology, Inc., Miao-Li, Taiwan) was performed using a contact spotting machine (DR. Fast Spot™; DR. Chip Biotechnology, Inc.), and immobilization using a UV crosslinker (Spectroline XLE-1000; Spectronics Corp., New York, USA) with 0.8 J/cm2 for 10 min. A schematic diagram of the probe position on the microarray is illustrated in Figure 1(a). Hybridization and colorimetric development were performed using the DR. Chip DIY Kit™ (DR. Chip Biotechnology, Inc.), and all of the reagents including DR. Hyb™ Buffer, Strep-AP, wash buffer, NBT/BCIP and detection buffer were supplied with the Kit. In brief, 15 μL of PCR amplicons were mixed with 200 μL DR. Hyb™ Buffer (DR. Chip Biotechnology, Inc.; 6× SSC, 5× Denhardt’s reagent, 0.5% SDS, 100 μg/mL salmon sperm DNA), denatured in boiling water for 5 min, and immediately chilled on ice for 5 min. The hybridization mixture was transferred to the chip well, incubated at 55 °C with vibration for 60 min, and washed twice with wash buffer (DR. Chip Biotechnology, Inc.; 0.1 M maleic acid, 0.15 M NaCl, pH 7.5). The chip was then added to 0.2 μL Strep-AP (DR. Chip Biotechnology, Inc.; 0.5 μL/mL in blocking buffer) and 200 μL blocking reagent (Roche GmbH, cat. no. 11096176001; 1%). at room temperature (25–35 °C) for 30 min and washed twice again with wash buffer. The colorimetric reaction was implemented by adding 4 μL NBT/BCIP and 196 μL detection buffer (DR. Chip Biotechnology, Inc.; 0.1 M Tris-HCl, 0.1 M NaCl, pH 9.5) to the chip well, developing in the dark at room temperature for 5 min, and washing twice with distilled water. Hybridization results were indicated on the microarray as blue spots that could be read directly by the naked eye.
2.6. Specificity of Assay
The specificity of the fish pathogen microarray was evaluated using the genomic DNA extracted from all 168 strains, which was then used as a DNA template for 16S rDNA PCR. Hybridization to the microarray for each PCR amplicon was performed separately using the protocol described above.
2.7. Detection Limit of the Microarray
Two approaches for the limit of detection were analyzed. To assess the overall detection limit of the microarray with purified genomic DNA, a serial dilution (100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg) of genomic DNA extracted from the eight pathogen collection strains (ATCC7966, ATCC15947, NCIMB2248, MT2055, ATCC19264, ATCC33539, NCIMB2248, and ATCC29178; Table 1) was used as the template for 16S rDNA PCR followed by hybridization to the microarray. To assess the limit of detection of different pathogens in suspension, the eight target pathogens were cultured to the stationary phase in broth medium. Serial dilutions were produced in the range 101–109 CFU/mL in TE-buffer (1 mM Tris-HCl, and 0.5 mM EDTA, pH8). Total DNA of 2 mL of each dilution was extracted using the UltraClean™ Microbial DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA), following the manufacturer’s instructions. Two microliters of the extracted DNA was used as a DNA template for PCR and the following microarray assay.
2.8. Field Test
2.8.1. Mixed Microbial Cultures and Fish Tissues
To evaluate the availability of the microarray to actual samples, the kidney of tilapia (Oreochris niloticus × Oreochris aureus) was used. The kidney samples were examined using the microbiological methods, to confirm that they were germfree. The eight target pathogens were suspended in TE-buffer and adjusted the density to approx. 3 × 106 CFU/mL each. Equal amounts of bacteria were mixed. The kidney tissues were homogenized with the bacterial mixture in a ratio of 1:10 (w/v). Total DNA of 2 mL of the homogenates was extracted using the UltraClean™ Microbial DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA), following the manufacturer’s instructions. Two microliters of the extracted DNA was used as a DNA template for PCR and the following microarray assay.
2.8.2. Fishpond Water Samples
The microarray was evaluated for used in surveys of fishpond water sampled from ten local fish farms (seawater, n = 5, freshwater, n = 5). For microarray assays, total DNA of 2 mL of each water sample was extracted using the UltraClean™ Microbial DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA), following the manufacturer’s instructions. Microarray assays were performed separately using the protocol described above with the extracted DNA as a template. In parallel, appropriate dilutions of the water samples were made in sterile 0.85% NaCl solution for bacteriological assays. One hundred microliters of each dilution was plated onto marine agar (BD, USA) for seawater samples or tryptic soy agar (BD, USA) for freshwater samples. The plates were then incubated at 28 °C for 48 h. Ten colonies from the agar plates were randomly picked, purified and identified by the homology of 16S rDNA sequence.
3. Results and Discussion
3.1. 16S rDNA Amplication and Microarray Hybridization
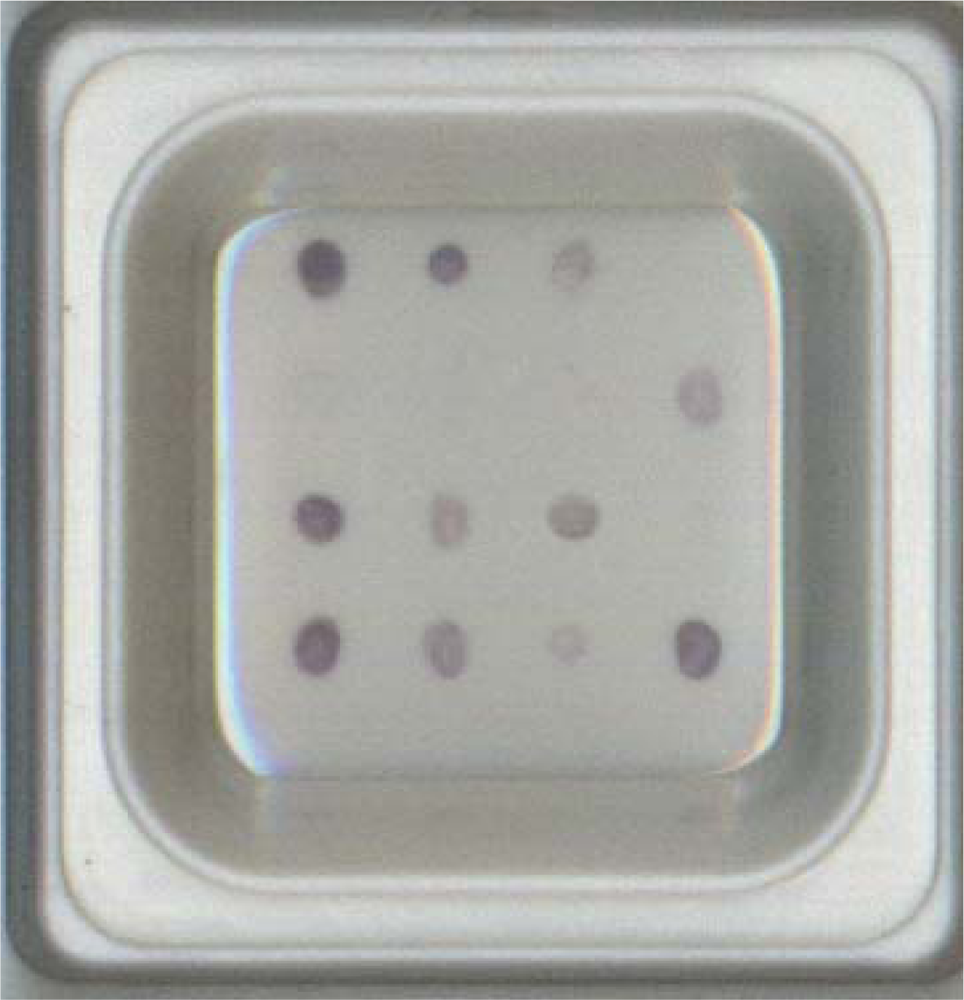
The 16S rDNA amplicon was obtained by PCR using 16S universal primers 16S-F and 16S-R for each genomic DNA of 168 strains. The resulting approximately 1.5 kbp PCR products were cloned and verified using the sequences retrieved from GenBank databases. For microarray hybridizations, the biotinylated PCR products obtained from pathogen-containing samples were incubated on an organic polymer substrate chip which served the traditional role of dot blot, except that the probe and target positions were reversed. After hybridization and a series of stringency washes, the bound PCR amplicons were reacted with streptavidin-conjugated alkaline phosphatase and NBT/BCIP, resulting in blue spots on the chip. These signals are readily visible to the naked eye, requiring no laser scanning or imaging systems. All PCR amplicons were confirmed by hybridization with either 16S-positive control probes U735 or U1352 on the microarray, which was observed as a blue spot visible to the naked eye (Figure 1).
3.2. Probe Specificity
3.2.1. Genomic DNA
A total of 168 strains were used to validate the probe specificity of the microarray. Eight species-specific probes, two PCR-positive control probes, one hybridization-positive control probe, and one negative control probe were finally selected and confirmed for the microarray (Table 2). The identities and gaps for each probe for the 16S rDNA of each representative strain were analyzed using the ClustalW alignment program and are listed in Table 3. In practice tests, only 30 bp DNA probes with fewer than three non-consecutive nucleotide differences and no gap in the 16S rDNA amplicons showed positive results for hybridization and colorization. Good discrimination was evidenced by the fact that all probes consistently distinguished their corresponding target strains with 100% specificity. The hybridization and colorization patterns obtained for the 8 fish pathogens are shown in Figure 1. None of the other 61 strains of bacterial species hybridized to specific probes on the microarray.
3.2.2. Mixed Microbial Cultures and Fish Tissues
Results of the preliminary test for the applicability of the microarray is illustrated in Figure 2. The probes for all the 8 pathogens (A. hydrophila, E. tarda, F. columnare, L. garvieae, P. damselae, P. anguilliseptica, S. iniae and V. anguillarum) gave positive signals. This result demonstrated that the probes designed were specific to their corresponding species.
3.2.3. Fishpond Water Samples
A total of 10 rearing water samples (seawater, n = 5, freshwater, n = 5) from different local fish farms were analyzed and compared using the microarray and bacteriological methods parallelly. Hybridization results showed that three samples (pond Fw1, Fw2 & Fw4) contained A. hydrophila, one sample (pond Fw5) contained F. columnare, and one sample (pond Sw2) contained P. damselae (Table 4). This finding was consistently confirmed by both 16S rDNA sequencing and bacteriological methods. All samples tested generated positive signals for the control probes U735 and U1352, suggesting that bacteria besides the eight pathogens studied were present. The overall results yielded high accuracy, indicating that this microarray has the ability to detect and discriminate among different pathogens in aquaculture.
The genetic variation in 16S rRNA among species is a subject of debate. Fox et al. [43] considered that 16S rRNA sequencing might not be sufficient to guarantee species identity. González et al. [44] believed that a high degree of genetic similarity for 16S rRNA genes across species might compromise the specificity of PCR detection. It will detect different species by using this 16S rRNA, indicating more common instead of more specificity. The strategies of probe design and microarray technology we used in this study can overcome the obstacles mentioned above. Firstly, with assistance from the Unique Probe Selector program [39], we deliberately designed specific probes based on polymorphic regions of the 16S rRNA gene with as high a degree of variation as possible. Secondly, the length of each probe was designed to be as short as 30 nucleotides. According to a hybridization rule-of-thumb of 10–15% [38,45,46], a DNA duplex will form between targets and their complementary probes if genetic dissimilarity is <10–15%, i.e., three nucleotides of the 30-mer probe in our case. A duplex between targets (16S PCR amplicons, ∼1,500 bp) and probes (30 bp each) is less likely to form when dissimilarity exceeds three bases, particularly when base mismatches are distributed systematically [47]. Furthermore, hybridization stringency is positional- and context-sensitive. If mismatches occur at the terminal ends of the probe, the effects will be less compared with when they occur throughout the probe sequence [48]. These three general rules for DNA duplex formation were applied appropriately to probe design in this study. For example, the original sequence of the probe Phda (Table 2), as calculated from the Unique Probe Selector program, was 5′-cgggcctctcgcgtcaggattagcccaggt-3′, which is 100% identical to that of P. damselae. However, there were only two mismatches between the original Phda probe and the 16S PCR amplicons from the phylogenetically related Vibrio spp., e.g., V. marinus, V. proteolyticus, V. salmonicida, and V. vulnificus, which led to false-positive hybridization results. Therefore we revised two nucleotides from the probe Phda (G changed to T, T changed to A at positions 23 and 30, respectively, Table 2). The revised Phda (5′-cgggcctctcgcgtcaggattaTcccaggA-3′) showed two nucleotide mismatches to the 16S rDNA of P. damselae, but one mismatch occurred at the 3′ end of the probe with less effect on hybridization. In comparison with the phylogenetically related Vibrio spp., four nucleotides differed from their 16S rRNA genes (Table 3) and at least three mismatches were distributed throughout the probe sequences to interrupt duplex formation. Using a similar strategy, the microarray we constructed was demonstrated as discriminating the eight target pathogens from 26 ecologically and/or phylogenetically related bacteria (Table 1), some of which were not distinguishable by 16S PCR and electrophoresis.
3.3. Detection Limit of the Microarray
3.3.1. Genomic DNA
Under ideal conditions, genomic DNA was extracted from the eight purified pathogenic collection strains and serially diluted (100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg) as the template for 16S PCR to test the detection limit of the microarray. Positive signals were generated from DNA dilutions 1 pg (Table 5). The lowest detected concentration of genomic DNA in the microarray was 0.1 pg for A. hydrophila. Visible bands of 16S PCR amplicons were not observed at this concentration by standard gel electrophoresis. However, for other strains, DNA <1 pg generated either ambiguous or no signals. Therefore, we decided to use 1 pg as the DNA detection limit for this microarray. All 81 strains of the 8 target species were tested and were shown to have been detected successfully at this concentration.
3.3.2. Bacteria from Pure Culture
For the purpose of directly detecting pathogens in suspension samples, serially diluted cultures of the eight target strains were tested using the microarray. All target pathogens were detectable at concentrations in the range 103–104 CFU/mL (Table 6). For example, E. tarda was selected as representative and serial dilutions prepared in the range 4 × 101–4 × 109 CFU/mL. The detection limit was as low as 4 × 103 CFU/mL (Figure 3).
Several factors could result in the detection of false-negatives on microarray. Direct capturing of 16S rRNA with surface-immobilized oligonucleotides is strongly influenced by inaccessible secondary structures, which may encompass some of the most variable binding sites of target molecules [49]. The addition of “helper” oligonucleotides permitted greater accessibility of the corresponding target region for probe hybridization and had a clear impact on the signal intensity of particular probes [50]. However, the application of helper oligonucleotides resulted in a dramatic increase in overall signal intensity, including the mismatch controls [51]. Another parameter that has to be considered in the context of signal limitation is steric hindrance. When hybridizing on a solid support, the binding efficiency of target molecules may be reduced by unfavourable steric interactions mediated by the solid matrix [52]. Peplies et al. [51] indicated that the addition of 12-mer and 18-mer poly(A) spacers to the probe sequence can mitigate structural inhibition. These authors found a linear correlation between spacer length and measured signal intensity even without the addition of helper oligonucleotides. Since the molar concentration of PCR product is very high, the concentration of target molecules using our detection protocol was much greater than that by direct detection of 16S rRNA on microarray [50], we decided not to take any risk by increasing the non-specific signal. Therefore, we added 15-mer poly(T) spacer to the 5′-end of each probe but added no helper oligonucleotides to the hybridization buffer. With hybridization conditions being considered adequate in our study, false-negative signals in the testing of 81 strains of target species did not occur.
The microarray system described herein could detect as little as 1 pg of purified genomic DNA, which is equivalent to 200–250 cells. This is not as sensitive as that previously reported in the literature, where lower detection limits were observed, e.g., 675 fg of Yersinia ruckeri DNA that was amplified with 16S universal primers [36] and 10 fg of Bacillus anthracis DNA that was amplified with species-specific primers [53]. Not surprisingly, regular microarray systems detect fluorescent signals of hybridization by laser reader, which is 10- to100-fold more sensitive than our naked-eye reading system, especially for small amounts of target molecules in samples. However, with our system we demonstrated the limit of detection with 103–104 CFU/mL of target pathogen in serially diluted suspensions (Figure 3). These results are comparable to the detection limit of the fluorescent-labeled microarray system. Zhou et al. [54] reported that in 102–105 CFU/mL serial dilutions of S. aureus, the optimal positive signal was obtained with 104 CFU/mL. In studies by Maynard et al. [55], using a combination of PCR followed by microarray hybridization, the detection limit for Salmonella enterica was estimated to be on the order of 104 CFU/mL. Agreement between these data indicates that when the amount of nucleic acid is not limiting, the economic naked-eye reading microarray system may prove very valuable as a tool for discriminating multiple pathogens in aquaculture.
4. Conclusions
The need to instantaneously monitor pathogen threats in aquaculture has led to the development of simultaneous detection systems. Oligonucleotide microarray, combining PCR technology with hybridization of the resulting amplification products, and post hybridization image processing have produced extremely powerful tools for pathogen detection, differentiation, and identification. In this report, we used this technology to design a DNA microarray containing specific oligonucleotide probes for the 16S rDNA polymorphic regions of eight aquacultural candidate pathogens. It was demonstrated to discriminate the 8 target pathogens from 26 ecologically or phylogenetically related bacteria, some of them were not distinguishable by 16S PCR and electrophoresis. Furthermore, we chose a naked-eye reading microarray system. The resulting signals are readily visible to the naked eye, requiring no laser scanning or imaging systems. The entire microarray manipulation time was less than 2 h, equivalent to the time needed for gel electrophoresis. This DNA microarray is well suited for detection of multiple fish pathogens in aquaculture.
Acknowledgments
This work was supported by project 95AS-6.1.6-AI-A8 from the Council of Agriculture, Executive Yuan, Taiwan.
References
- Lu, C.P. Pathogenic Aeromonas hydrophila and the fish diseases caused by it. J. Fish. China 1992, 16, 282–288. [Google Scholar]
- Egusa, S. Some bacterial diseases of freshwater fishes in Japan. Fish Pathol 1976, 10, 103–114. [Google Scholar]
- Liu, C.I.; Tsai, S.S. Edwardsielosis in pond-cultured eel in Taiwan. CAPD Fish. Ser. 3 Rep. Fish Dis. Res 1980, 3, 108–115. [Google Scholar]
- Nakatsugawa, T. Edwardsiella tarda isolated from cultured young flouders. Fish Pathol 1983, 18, 99–101. [Google Scholar]
- Aoki, T.; Kitao, T. Drug resistance and transferable R plasmids in Edwardsiella tarda from fish culture ponds. Fish Pathol 1981, 15, 277–281. [Google Scholar]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens, 4th ed; Praxis Publishing Ltd: Chichester, UK, 2007; pp. 272–275. [Google Scholar]
- Decostere, A.; Haesebrouck, F.; Devriese, L.A. Characterization of four Flavobacterium columnare (Flexibacter columnaris) strains isolated from tropical fish. Vet. Microbiol 1998, 62, 35–45. [Google Scholar]
- Janssen, W.A.; Surgalla, M.J. Morphology, physiology, and serology of a Pasteurella species pathogenic for white perch (Roccus americanus). J. Bacteriol 1968, 96, 1606–1610. [Google Scholar]
- Kubota, S.S.; Kimura, M.; Egusa, S. Studies of a bacterial tuberculoidosis of yellowtail. I. Symptomatology and histopathology. Fish Pathol 1970, 4, 111–118. [Google Scholar]
- Toranzo, A.E.; Barreiro, S.; Casal, J.F.; Figueras, A.; Magariños, B.; Barja, J.L. Pasteurellosis in cultured gilthead seabream (Sparus aurata): First report in Spain. Aquaculture 1991, 99, 1–15. [Google Scholar]
- Balebona, M.C.; Moriñigo, M.A.; Sedano, J.; Martinez-Manzanares, E.; Vidaurreta, A.; Borrego, J.J.; Toranzo, A.E. Isolation of Pasteurella piscicida from sea bass in Southwestern Spain. Bull. Eur. Ass. Fish Pathol 1992, 12, 168–170. [Google Scholar]
- Nakai, T.; Fujiie, N.; Muroga, K.; Arimoto, M.; Mizuta, Y.; Matsuoka, S. Pasteurella piscicida infection in hatchery-reared juvenile striped jack. Fish Pathol 1992, 27, 103–108. [Google Scholar]
- Fukuda, Y.; Matsuoka, S.; Mizuno, Y.; Narita, K. Pasteurella piscicida infection in cultured juvenile Japanese flounder. Fish Pathol 1996, 31, 33–38. [Google Scholar]
- Lopez, C.; Rajam, P.R.; Lin, J.H.-Y.; Yang, H.-L. Disease outbreak in sea farmed cobia, Rachycentron canadum associated with Vibrio spp., Photobacterium damselae ssp. piscicida, monogenean and myxosporean parasites. Bull. Eur. Ass. Fish Pathol 2002, 23, 206–211. [Google Scholar]
- Nakai, T.; Muroga, K. Studies on red spot disease of pond cultured eels—V. Immune response of the Japanese eel to the causative bacterium Pseudomonas anguilliseptica. Bull. Eur. Ass. Fish Pathol 1979, 45, 817–821. [Google Scholar]
- Kuo, S.-C.; Kou, G.-H. Pseudomonas anguilliseptica isolated from red spot disease of pond-cultured eel, Anguilla japonica. Rep. Inst. Fish. Biol. Min. Econ. Aff. Nat. Taiwan Univ 1978, 3, 19–23, (in Chinese with English abstract).. [Google Scholar]
- Ellis, A.E.; Dear, G.; Stewart, D.J. Histopathology of ‘sekiten-byo’ caused by Pseudomonas anguilliseptica in the European eel, Anguilla anguilla L, in scotland. J. Fish Dis 1983, 6, 77–79. [Google Scholar]
- Michel, C.; Bernardet, J.F.; Dinand, D. Phenotypic and genotypic studies of Pseudomonas anguilliseptica strains isolated from farmed European eels (Anguilla anguilla) in France. Fish Pathol 1992, 27, 229–232. [Google Scholar]
- Nakajima, K.; Muroga, K.; Hancock, R.E.W. Comparison of fatty acid, protein and serological properties distinguishing outer membranes of Pseudomonas anguilliseptica strains from those of fish pathogens and other pseudomonads. Int. J. Syst. Bacteriol 1983, 33, 1–8. [Google Scholar]
- Wiklund, T.; Bylund, G. Pseudomonas anguilliseptica as a pathogen of salmonid fish in Finland. Dis. Aquat. Org 1990, 8, 13–19. [Google Scholar]
- Nakai, T.; Hanada, H.; Muroga, K. First records of Pseudomonas anguilliseptica infection in cultured ayu, Plecoglossus altivelis. Fish Pathol 1985, 20, 481–484. [Google Scholar]
- Tajima, K.; Ezura, Y.; Kimura, T. Studies on the taxonomy and serology of causative organisms of fish vibriosis. Fish Pathol 1985, 20, 179–183. [Google Scholar]
- Toranzo, A.E.; Santos, Y.; Bandin, I.; Ramalde, J.L.; Ledo, A.; Fouz, B. Five year survey of bacterial fish infection in continental and marine aquaculture in noethwest Spain. World Aquacult 1990, 21, 91–94. [Google Scholar]
- Fryer, J.L.; Nelson, J.S.; Garrison, R.L. Vibriosis in fish. Prog. Fish Food Sci 1972, 5, 129–133. [Google Scholar]
- Levin, M.A.; Wolke, R.E.; Cabelli, V.J. Vibrio anguillarum as a cause of disease in winter flounder (Pseudopleuronectes americanus). Can. J. Microbiol 1972, 18, 1585–1592. [Google Scholar]
- Larsen, J.L.; Rasmuseen, H.B.; Dalsgaard, I. Study of Vibrio anguillarum strains from different sources with emphasis on ecological and psthological properties. Appl. Environ. Microbiol 1988, 54, 2264–2267. [Google Scholar]
- Rodgers, C.J.; Furones, M.D. Disease problems in cultured marine fish in the Mediterranean. Fish Pathol 1998, 33, 157–164. [Google Scholar]
- Vendrell, D.; Balcázar, J.L.; Ruiz-Zarzuela, I.; Gironés, O.; Múzquiz, J.L. Lactococcus garvieae in fish: A review. Comp. Immunol. Microbiol. Infect. Dis 2006, 29, 177–198. [Google Scholar]
- Perera, R.P.; Johnson, S.K. Streptococcus iniae associated with mortality of Tilapia nilotica × T. aurea hybrids. J. Aquat. Anim. Health 1994, 6, 335–340. [Google Scholar]
- Bekker, A.; Hugo, C.; Albertyn, J.; Boucher, C.E.; Bragg, R.R. Pathogenic Gram-positive cocci in South African rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis 2011, 34, 483–487. [Google Scholar]
- Kusuda, R.; Salati, F. Fish Diseases and Disorders; Woo, P.T.K., Bruno, D., Eds.; CAB International: Wallingford, UK, 1999; Volume 3, pp. 303–317. [Google Scholar]
- Suzuki, K.; Sakai, D.K. Real-time PCR for quantification of viable Renibacterium salmoninarum in chum salmon Oncorhynchus keta. Dis. Aquat. Org 2007, 74, 209–223. [Google Scholar]
- Balcázar, J.L.; Vendrell, D.; de Blas, I.; Ruiz-Zarzuela, I.; Gironés, O.; Múzquiz, J.L. Quantitative detection of Aeromonas salmonicida in fish tissue by real-time PCR using self-quenched, fluorogenic primers. J. Med. Microbiol 2007, 56, 323–328. [Google Scholar]
- Chang, C.-I.; Wu, C.-C.; Cheng, T.C.; Tsai, J.-M.; Lin, K.-J. Multiplex nested-polymerase chain reaction for the simultaneous detection of Aeromonas hydrophila, Edwardsiells tarda, Photobacterium damselae and Streptococcus iniae, four important fish pathogens in subtropical Asia. Aquac. Res 2009, 40, 1182–1190. [Google Scholar]
- Hussein, M.M.A.; Hatai, K. Multiplex PCR for detection of Lactococcus garvieae, Streptococcus iniae and S. dysgalactiae in cultured yellowtail. Aquac. Sci 2006, 54, 269–274. [Google Scholar]
- Warsen, A.E.; Krug, M.J.; LaFrentz, S.; Stanek, D.R.; Loge, F.J.; Call, D.R. Simultaneous discrimination between 15 fish pathogens by using 16S ribosomal DNA PCR and DNA microarrays. Appl. Environ. Microbiol 2004, 70, 4216–4221. [Google Scholar]
- Lipshutz, R.J.; Fodor, S.P.A.; Gingeras, T.R.; Lockhart, D.J. High density synthetic oligonucleotide arrays. Nat. Genet 1999, 21, 20–24. [Google Scholar]
- Call, D.R.; Borucki, M.; Loge, F. Detection of bacterial pathogens in environmental samples using DNA microarrays. J. Microbiol. Methods 2003, 53, 235–243. [Google Scholar]
- Chen, S.-H.; Lo, C.-Z.; Su, S.-Y.; Kuo, B.-H.; Hsiung, C.A.; Lin, C.-Y. UPS 2.0: Unique probe selector for probe design and oligonucleotide microarrays at the pangenomic/genomic level. BMC Genomics 2010. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol 1991, 173, 697–703. [Google Scholar]
- James, G. PCR for Clinical Microbiology; Schuller, M., Sloots, T.P., James, G.S., Halliday, C.L., Carter, I.W.J., Eds.; Springer: London, UK, 2010; pp. 209–214. [Google Scholar]
- Pearson, W.R.; Lipman, D.I. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar]
- Fox, G.E.; Wisotzkey, J.D.; Jurtshuk, P., Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol 1992, 42, 166–170. [Google Scholar]
- González, S.F.; Krug, M.J.; Nielsen, M.E.; Santos, Y.; Call, D.R. Simultaneous detection of marine fish pathogens by using multiplex PCR and a DNA microarrar. J. Clin. Microbiol 2004, 42, 1414–1419. [Google Scholar]
- Wu, L.; Thompson, D.K.; Li, G.; Hurt, R.A.; Tiedge, J.M.; Zhou, J. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol 2001, 67, 5780–5790. [Google Scholar]
- Lemarchand, K.; Masson, L.; Brousseau, R. Molecular biology and DNA microarray technology for microbial quality monitoring of water. Crit. Rev. Microbiol 2004, 30, 145–172. [Google Scholar]
- Letowski, J.; Brousseau, R.; Masson, L. Designing better probes: Effect of probe size, mismatch position and number on hybridization in DNA oligonucleotide microarrays. J. Microbiol. Meth 2004, 57, 269–278. [Google Scholar]
- Call, D.R. Challenges and opportunities for pathogen detection using DNA microarrays. Crit. Rev. Microbiol 2005, 31, 91–99. [Google Scholar]
- Fuchs, B.M.; Glöckner, F.O.; Wulf, J.; Amann, R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol 2000, 66, 3603–3607. [Google Scholar]
- Chandler, D.P.; Newton, G.J.; Small, J.A.; Daly, D.S. Sequence versus structure for the direct detection of 16S rRNA on planar oligonucleotide microarrays. Appl. Environ. Microbiol 2003, 69, 2950–2958. [Google Scholar]
- Peplies, J.; Glockner, F.O.; Amann, R. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol 2003, 69, 1397–1407. [Google Scholar]
- Shchepinov, M.S.; Case-Green, S.C.; Southern, E.M. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res 1997, 25, 1155–1161. [Google Scholar]
- Wilson, W.J.; Strout, C.L.; DeSantis, T.Z.; Stilwell, J.L.; Carrano, A.V.; Andersen, G.L. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes 2002, 16, 119–127. [Google Scholar]
- Zhou, G.; Wen, S.; Liu, Y.; Li, R.; Zhong, X.; Feng, L.; Wang, L.; Cao, B. Development a DNA microarray for detection and identification of Legionella pneumophila and ten other pathogens in drinking water. Int. J. Food Microbiol 2011, 145, 293–300. [Google Scholar]
- Maynard, C.; Berthiaume, F.; Lemarchand, K.; Harel, J.; Payment, P.; Bayardelle, P.; Masson, L.; Brousseau, R. Waterborne pathogen detection by use of oligonucleotide-based microarrays. Appl. Environ. Microbiol 2005, 71, 8548–8557. [Google Scholar]


| Species | Number of strains from different sources | Collection strain a | Total number |
|---|---|---|---|
| Aeromonas hydrophila | 1 a, 18 b | ATCC7966 | 19 |
| A. sobria | 1 a | ATCC43979 | 1 |
| A. salmonicida | 1 c | MT423 | 1 |
| Edwardsiella tarda | 1 a, 30 b | ATCC15947 | 31 |
| Enterococcus faecalis | 1 a | ATCC19433 | 1 |
| E. faecium | 1 a, 1 b | ATCC19434 | 2 |
| Flavobacterium columnare | 1 d | NCIMB2248 | 1 |
| Lactococcus garvieae | 1 c, 14 b | MT2055 | 2 |
| L. pelagia | 1 a | ATCC25916 | 1 |
| Mycobacterium fortuitum | 1 a | ATCC19709 | 1 |
| M. marinum | 1 a | ATCC927 | 1 |
| Photobacterium damselae subsp. damselae | 1 a, 3 b | ATCC33539 | 4 |
| P. damselae subsp. piscicida | 1 a, 13 b | ATCC51736 | 14 |
| Pseudomonas aeruginosa | 1 a, 1 b | ATCC10145 | 2 |
| P. anguilliseptica | 1 d | NCIMB2248 | 1 |
| Staphylococcus epidermidis | 1 a, 1 e | ATCC12228 | 2 |
| Streptococcus iniae | 1 a, 1 b | ATCC29178 | 2 |
| Vibrio aestuarianus | 1 a | ATCC35048 | 1 |
| V. alginolyticus | 1 a, 26 b | ATCC17749 | 27 |
| V. anguillarum | 1 a, 4 b | ATCC19264 | 5 |
| V. harveyi | 1 a, 22 b | ATCC14126 | 23 |
| V. marinus | 1 a | ATCC15382 | 1 |
| V. parahaemolyticus | 1 a, 4 b | ATCC27969 | 5 |
| V. proteolyticus | 1 a | ATCC15338 | 1 |
| V. salmonicida | 1 a | ATCC43839 | 1 |
| V. vulnificus | 1 a, 4 b | ATCC27562 | 5 |
aAmerican Type Culture Collection (ATCC);bFisheries Research Institute, Taiwan (FRI),cFRS Marine Laboratory, UK (MT);dNCIMB, National Collection of Industrial, Marine and Food Bacteria, UK (NCIMB);eBioresource Collection and Research Center, Taiwan (BCRC).
| Oligo nucleotide | Sequence * | Tm (°C) | Target organism | Accession no. |
|---|---|---|---|---|
| Probe | ||||
| Aehy | ggttAatgcctaatacgtatcaactgtgac | 62.21 | A. hydrophila | DQ207728 |
| Edta | ctcatgccatcaTatgaacccagatgggat | 62.62 | E. tarda | DQ233654 |
| Flco | ccctgttgctagttgccagcgagtcatgtc | 65.01 | F. columnare | AY095342 |
| Laga | tcgccaacccgcgagggtgcgctaatctct | 67.68 | L. garvieae | AY699289 |
| Phda | cgggcctctcgcgtcaggattaTcccaggA | 65.50 | P. damselae | AY147861 |
| Psan | ccgttggaatccttgagattttagtggcgc | 66.29 | P. anguilliseptica | HM103328 |
| Stin | ggtgttaggccctttccggggcttagtgcc | 66.99 | S. iniae | AF335572 |
| Vian | tgacatctacagaatcctgcggagacgcgg | 68.45 | V. anguillarum | X16895 |
| U735 | actgaggtgcgaaagcgtggggagcaaaca | 65.28 | Eubacteria | AF233451 |
| U1352 | tgaatacgttcccgggccttgtacacaccg | 65.83 | Eubacteria | AF233451 |
| EV71 | atgaagcatgtcagggcttggatacctcg | 63.17 | Human enterovirus 71 | HQ283840 |
| poly(A) | aaaaaaaaaaaaaaaaaaaaaaaaaaaaaa | |||
| Primer | ||||
| 16S-F | agagtttgatcatggctcag | 49.73 | Eubacteria | AF233451 |
| 16S-R | ggttaccttgttacgactt | 46.77 | Eubacteria | AF233451 |
*Nucleotides designed differently from the original sequences are shown in uppercase.
| Probe | Identity (upper row)/gap (lower row) | |||||||
|---|---|---|---|---|---|---|---|---|
| Aehy | Edta | Flco | Laga | Phda | Psan | Stin | Vian | |
| Organism | ||||||||
| A. hydrophila | 29/30 * | 19/30 | 21/30 | 18/30 | 21/30 | 20/30 | 21/30 | 24/30 |
| 0/30 | 0/30 | 0/30 | 0/30 | 0/30 | 2/30 | 9/30 | 0/30 | |
| A. sobria | 21/30 | 22/30 | 21/30 | 18/30 | 23/30 | 20/30 | 19/30 | 23/30 |
| 0/30 | 0/30 | 0/30 | 0/30 | 0/30 | 2/30 | 1/30 | 0/30 | |
| A. salmonicida | 26/30 | 22/30 | 21/30 | 18/30 | 22/30 | 20/30 | 21/30 | 23/30 |
| 0/30 | 0/30 | 0/30 | 0/30 | 0/30 | 2/30 | 09/30 | 0/30 | |
| E. tarda | 19/30 | 29/30 | 22/30 | 18/30 | 23/30 | 23/30 | 19/30 | 23/30 |
| 1/30 | 0/30 | 2/30 | 0/30 | 2/30 | 11/30 | 3/30 | 0/30 | |
| E. faecalis | 22/30 | 18/30 | 21/30 | 22/30 | 16/30 | 19/30 | 18/30 | 21/30 |
| 7/30 | 0/30 | 1/30 | 0/30 | 0/30 | 0/30 | 0/30 | 2/30 | |
| E. faecium | 22/30 | 18/30 | 22/30 | 23/30 | 16/30 | 19/30 | 21/30 | 20/30 |
| 7/30 | 0/30 | 1/30 | 0/30 | 0/30 | 0/30 | 3/30 | 4/30 | |
| F. columnare | 22/30 | 23/30 | 30/30 | 19/30 | 23/30 | 20/30 | 22/30 | 22/30 |
| 3/30 | 5/30 | 0/30 | 1/30 | 5/30 | 4/30 | 7/30 | 2/30 | |
| L. garvieae | 20/30 | 18/30 | 21/30 | 30/30 | 20/30 | 19/30 | 19/30 | 21/30 |
| 10/30 | 1/30 | 1/30 | 0/30 | 6/30 | 0/30 | 1/30 | 2/30 | |
| L. pelagia | 18/30 | 23/30 | 25/30 | 20/30 | 25/30 | 22/30 | 19/30 | 21/30 |
| 1/30 | 2/30 | 1/30 | 0/30 | 0/30 | 11/30 | 3/30 | 3/30 | |
| M. fortuitum | 20/30 | 18/30 | 20/30 | 20/30 | 19/30 | 20/30 | 20/30 | 22/30 |
| 2/30 | 2/30 | 0/30 | 3/30 | 1/30 | 2/30 | 1/30 | 1/30 | |
| M. marinum | 20/30 | 19/30 | 20/30 | 21/30 | 20/30 | 23/30 | 20/30 | 18/30 |
| 4/30 | 2/30 | 0/30 | 3/30 | 1/30 | 8/30 | 1/30 | 0/30 | |
| P. damselae | 20/30 | 23/30 | 20/30 | 19/30 | 28/30 | 22/30 | 21/30 | 21/30 |
| 1/30 | 2/30 | 1/30 | 0/30 | 0/30 | 11/30 | 7/30 | 0/30 | |
| P. aeruginosa | 22/30 | 22/30 | 20/30 | 23/30 | 20/30 | 24/30 | 22/30 | 22/30 |
| 1/30 | 0/30 | 0/30 | 0/30 | 0/30 | 0/30 | 3/30 | 2/30 | |
| P. anguilliseptica | 23/30 | 22/30 | 20/30 | 23/30 | 19/30 | 30/30 | 20/30 | 22/30 |
| 1/30 | 0/30 | 0/30 | 0/30 | 0/30 | 0/30 | 4/30 | 2/30 | |
| S. epidermidis | 20/30 | 19/30 | 21/30 | 19/30 | 22/30 | 17/30 | 22/30 | 22/30 |
| 2/30 | 0/30 | 2/30 | 0/30 | 6/30 | 0/30 | 0/30 | 2/30 | |
| S. iniae | 22/30 | 23/30 | 22/30 | 22/30 | 21/30 | 17/30 | 30/30 | 21/30 |
| 10/30 | 6/30 | 1/30 | 0/30 | 4/30 | 0/30 | 0/30 | 2/30 | |
| V. aestuarianus | 18/30 | 23/30 | 25/30 | 20/30 | 25/30 | 23/30 | 19/30 | 26/30 |
| 1/30 | 2/30 | 1/30 | 0/30 | 0/30 | 1/30 | 3/30 | 0/30 | |
| V. alginolyticus | 22/30 | 23/30 | 25/30 | 21/30 | 25/30 | 22/30 | 19/30 | 21/30 |
| 5/30 | 2/30 | 1/30 | 0/30 | 0/30 | 11/30 | 3/30 | 0/30 | |
| V. anguillarum | 18/30 | 23/30 | 25/30 | 18/30 | 25/30 | 22/30 | 20/30 | 30/30 |
| 1/30 | 2/30 | 1/30 | 0/30 | 0/30 | 11/30 | 4/30 | 0/30 | |
| V. harveyi | 24/30 | 23/30 | 20/30 | 21/30 | 25/30 | 21/30 | 19/30 | 21/30 |
| 10/30 | 2/30 | 1/30 | 0/30 | 0/30 | 10/30 | 3/30 | 3/30 | |
| V. marinus | 20/30 | 19/30 | 21/30 | 18/30 | 26/30 | 23/30 | 21/30 | 22/30 |
| 6/30 | 0/30 | 1/30 | 0/30 | 0/30 | 1/30 | 4/30 | 1/30 | |
| V. parahaemolyticus | 18/30 | 23/30 | 25/30 | 20/30 | 25/30 | 22/30 | 19/30 | 21/30 |
| 1/30 | 2/30 | 1/30 | 0/30 | 0/30 | 11/30 | 3/30 | 0/30 | |
| V. proteolyticus | 23/30 | 24/30 | 25/30 | 20/30 | 26/30 | 22/30 | 20/30 | 22/30 |
| 10/30 | 2/30 | 1/30 | 0/30 | 0/30 | 11/30 | 4/30 | 0/30 | |
| V. salmonicida | 23/30 | 19/30 | 21/30 | 18/30 | 26/30 | 22/30 | 19/30 | 22/30 |
| 7/30 | 0/30 | 1/30 | 0/30 | 0/30 | 11/30 | 2/30 | 1/30 | |
| V. vulnificus | 22/30 | 24/30 | 25/30 | 20/30 | 26/30 | 22/30 | 19/30 | 25/30 |
| 7/30 | 2/30 | 1/30 | 0/30 | 0/30 | 11/30 | 3/30 | 0/30 | |
*Features in boldface represent the positive results for hybridization and color development.
| Probe Sample | Hybridization signal | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aehy | Edta | Flco | Laga | Phda | Psan | Stin | Vian | U735 | U1352 | EV71 | poly(A) | |
| Seawater pond | ||||||||||||
| Sw1 | − | − | − | − | − | − | − | − | + | + | + | − |
| Sw2 | − | − | − | − | + | − | − | − | + | + | + | − |
| Sw3 | − | − | − | − | − | − | − | − | + | + | + | − |
| Sw4 | − | − | − | − | − | − | − | − | + | + | + | − |
| Sw5 | − | − | − | − | − | − | − | − | + | + | + | − |
| Freshwater pond | ||||||||||||
| Fw1 | + | − | − | − | − | − | − | − | + | + | + | − |
| Fw2 | + | − | − | − | − | − | − | − | + | + | + | − |
| Fw3 | − | − | − | − | − | − | − | − | + | + | + | − |
| Fw4 | + | − | − | − | − | − | − | − | + | + | + | − |
| Fw5 | − | − | + | − | − | − | − | − | + | + | + | − |
| Bacterial composition (number of species identified by 16S rDNA homology) | ||||||||||||
| Sw1 | Alteromonas sp. (2), Pseudoalteromonas sp. (3), Rhodobacteraceae bacterium (3), Vibrio alginolyticus (2) | |||||||||||
| Sw2 | Photobacterium damselae (2), Pseudoalteromonas sp. (2), Vibrio fortis (1), Vibrio harveyi (1), Vibrio sp. (4) | |||||||||||
| Sw3 | Gamma proteobacterium (3), Maribacter dokdonensis (1), Pseudoalteromonas sp. (2), Pseudomonas sp. (1), Roseobacter gallaeciensis (1), Tenacibaculum sp. (2) | |||||||||||
| Sw4 | Vibrio alginolyticus (1), Vibrio harveyi (6), Vibrio sp. (3) | |||||||||||
| Sw5 | Alteromonas sp. (2), Gamma proteobacterium (3), Pseudoalteromonas sp. (2), Rhodobacteraceae bacterium (1), Sulfitobacter sp. (2) | |||||||||||
| Fw1 | Aeromonas hydrophila (3), Aeromonas sp. (3), Citrobacter freundii (2), Plesiomonas shigelloides (1), Pseudomonas sp. (1) | |||||||||||
| Fw2 | Aeromonas hydrophila (3), Aeromonas sobria (1), Bacillus sp. (3), Citrobacter freundii (1), Citrobacter sp. (2) | |||||||||||
| Fw3 | Aeromonas sobria (2), Aeromonas sp. (1), Citrobacter sp. (3), Plesiomonas shigelloides (3), Pseudomonas sp. (1) | |||||||||||
| Fw4 | Aeromonas hydrophila (1), Bacillus cereus (3), Bacillus subtilis (3), Plesiomonas shigelloides (3) | |||||||||||
| Fw5 | Aeromonas sobria (1), Bacillus subtilis (1), Citrobacter freundii (1), Citrobacter sp. (2), Flavobacterium columnare (1), Plesiomonas shigelloides (2), Pseudomonas sp. (2) | |||||||||||
| Species | Genomic DNA (pg) | |||||
|---|---|---|---|---|---|---|
| 100 | 10 | 1 | 0.1 | 0.01 | 0.001 | |
| Aeromonas hydrophila | + | + | + | + | − | − |
| Edwardsiella tarda | + | + | + | ± | − | − |
| Flavobacterium columnare | + | + | ± | − | − | − |
| Lactococcus garvieae | + | + | ± | − | − | − |
| Photobacterium damselae | + | + | + | − | − | − |
| Pseudomonas anguilliseptica | + | + | + | ± | − | − |
| Streptococcus iniae | + | + | ± | − | − | − |
| Vibrio anguillarum | + | + | + | ± | − | − |
+ positive signal; − negative signal; ± weak or ambiguous signal.
| Species | Bacterial counts (CFU/mL) | |||||
|---|---|---|---|---|---|---|
| >106 | 105 | 104 | 103 | 102 | 101 | |
| Aeromonas hydrophila | + | + | + | + | ± | − |
| Edwardsiella tarda | + | + | + | + | ± | − |
| Flavobacterium columnare | + | + | + | ± | − | − |
| Lactococcus garvieae | + | + | + | + | ± | − |
| Photobacterium damselae | + | + | + | ± | − | − |
| Pseudomonas anguilliseptica | + | + | + | + | ± | − |
| Streptococcus iniae | + | + | + | ± | − | − |
| Vibrio anguillarum | + | + | + | + | − | − |
+ positive signal; − negative signal; ± weak or ambiguous signal.
© 2012 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chang, C.-I.; Hung, P.-H.; Wu, C.-C.; Cheng, T.C.; Tsai, J.-M.; Lin, K.-J.; Lin, C.-Y. Simultaneous Detection of Multiple Fish Pathogens Using a Naked-Eye Readable DNA Microarray. Sensors 2012, 12, 2710-2728. https://doi.org/10.3390/s120302710
Chang C-I, Hung P-H, Wu C-C, Cheng TC, Tsai J-M, Lin K-J, Lin C-Y. Simultaneous Detection of Multiple Fish Pathogens Using a Naked-Eye Readable DNA Microarray. Sensors. 2012; 12(3):2710-2728. https://doi.org/10.3390/s120302710
Chicago/Turabian StyleChang, Chin-I, Pei-Hsin Hung, Chia-Che Wu, Ta Chih Cheng, Jyh-Ming Tsai, King-Jung Lin, and Chung-Yen Lin. 2012. "Simultaneous Detection of Multiple Fish Pathogens Using a Naked-Eye Readable DNA Microarray" Sensors 12, no. 3: 2710-2728. https://doi.org/10.3390/s120302710
APA StyleChang, C.-I., Hung, P.-H., Wu, C.-C., Cheng, T. C., Tsai, J.-M., Lin, K.-J., & Lin, C.-Y. (2012). Simultaneous Detection of Multiple Fish Pathogens Using a Naked-Eye Readable DNA Microarray. Sensors, 12(3), 2710-2728. https://doi.org/10.3390/s120302710




