Whole-Cell Fluorescent Biosensors for Bioavailability and Biodegradation of Polychlorinated Biphenyls
Abstract
:1. Introduction
- Biosensors determine only the bioavailable fraction of compounds, thus giving a more accurate response on the toxicity of a sample. Bioavailability is also important in bioremediation. If substances are bioavailable, they are potentially biodegradable.
- Biosensors provide an inexpensive and simple way of determining contaminants.
- As they are living organisms, they provide information on toxicology of different compounds.
- Some stress-induced biosensors report the mutagenic effects of samples with great sensitivity.
- Biosensors are unsurpassed in studying gene expression and physiology of bacteria in complex environments.
1.1. Commonly Used Reporter Genes
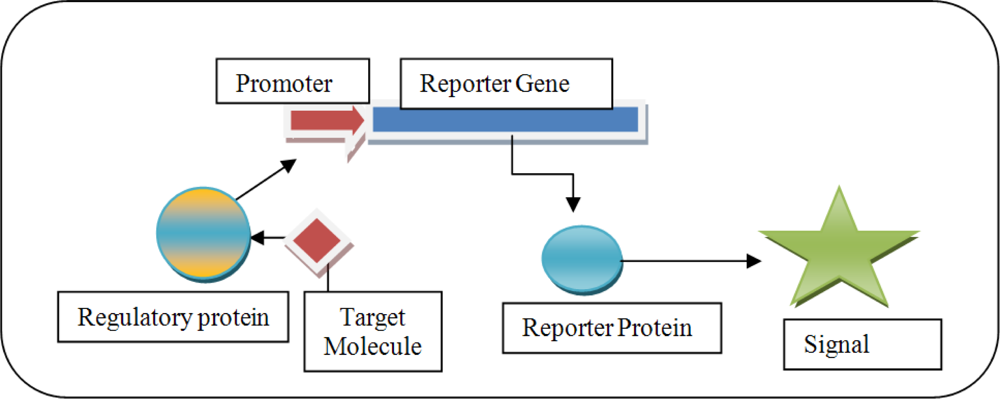
1.2. Promoters and Regulatory Elements for the Construction of Biosensors
2. Development of Biosensors to Detect PCB Biodegradation
2.1. Biosensors Based on Monitoring Chlorobenzoic Acids
3. Encapsulation of Biosensors for Environmental Use
- Agar/agarose: competent cells can be added to molten agar or agarose (1–5%). Gelation occurs as the agar or agarose cools to room temperature [125].
- Carrageenan: a 2% solution of carrageenan is warmed to 70–80 °C to initiate dissolution and then maintained at 35–50 °C. The cell culture is also warmed and added to the carrageenan solution. Gel formation occurs through the addition of cold 0.1 M potassium chloride [124].
- Polyurethane–polycarbomyl sulfonate (PCS): polyurethane or PCS at a polymer content of 30–50% is mixed with a 1% calcium-chloride solution, the pH is adjusted to approximately 6.5 and the cell mass is added. This mixture is sprayed into 0.75% calcium alginate, resulting in bead formation. After one hour, the beads are removed, washed and introduced into a 2% sodium-tripolyphosphate buffer, which dissolves the alginate layer leaving only a layer of polyurethane–PCS surrounding the cells [126].
- Polyacrylamide: cells are mixed in a solution of acrylamide and bisacrylamide. Ammonium persulfate and N,N,N′,N′-tetramethylethylenediamine (TEMED) are then added to initiate polymerization [128].
- Polyvinyl alcohol: the cell suspension is mixed with a 13% polyvinyl alcohol, 0.02% sodium-alginate mixture. Gel formation occurs on contact with a solution of saturated boric acid and 2% calcium chloride [129].
- Sol–gel: cells are combined with 0.1 M Tris-Cl and tetramethylorthosilicate, tetraethoxysilane, methyltrimethoxysilane, ethyltrimethoxysilane, propyltrimeth
- Oxysilane or polydimethylsiloxane. Solidification times vary depending on the concentrations used [130].
- Polyethylleneimine [131].
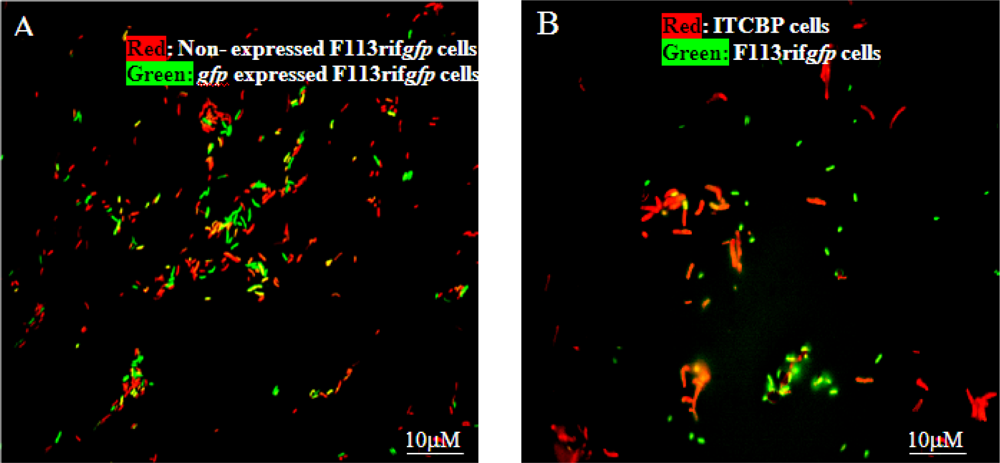
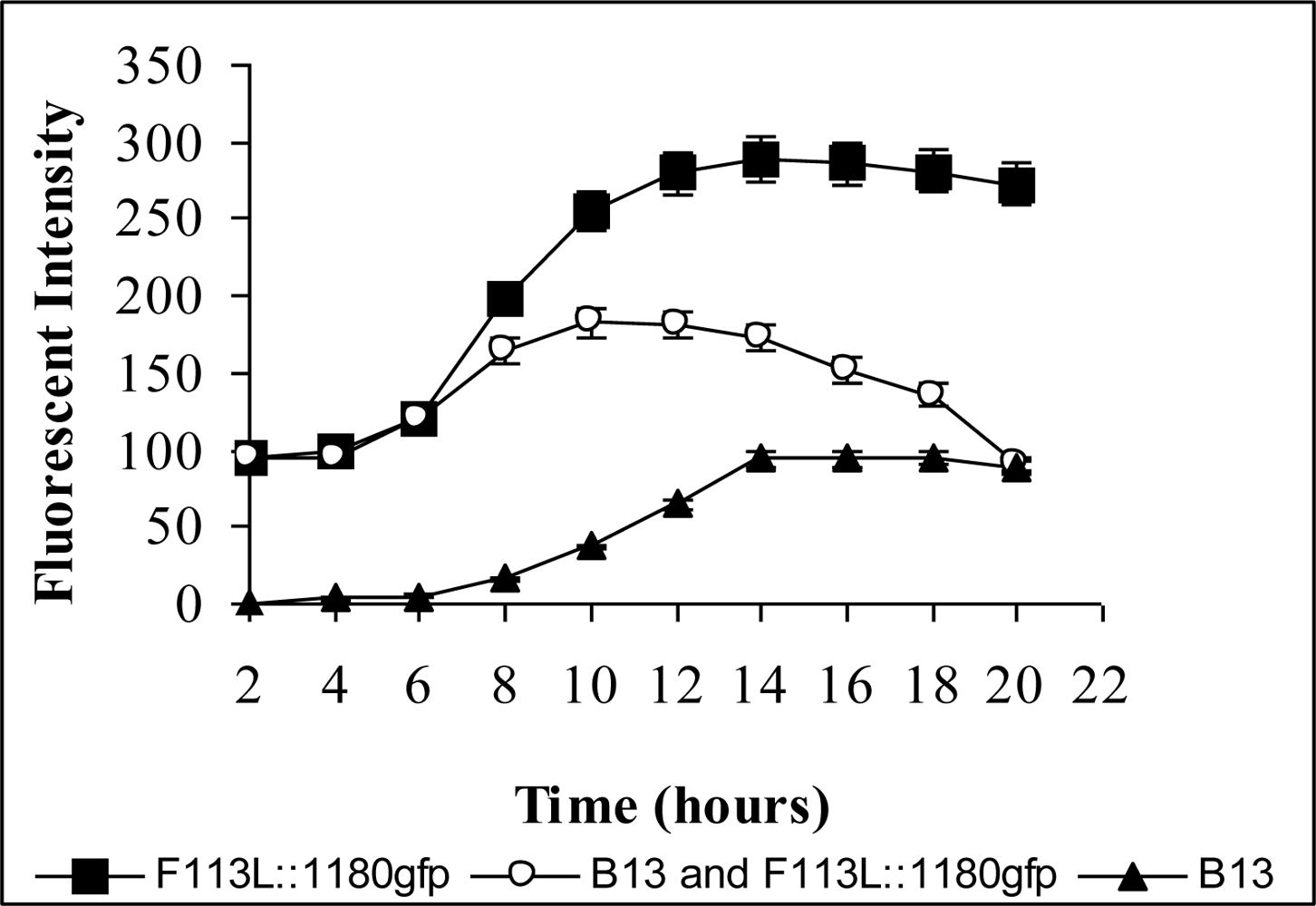
3.1. Monitoring PCB Degradation in Vitro and in Soil Using Encapsulated PCB Biosensors
3.2. Monitoring Chlorobenzoic Acids (CBA) Bioavailability and Biodegradation
4. Discussion and Conclusions
Acknowledgments
References and Notes
- Harmsen, J. Measuring bioavailability: From a scientific approach to standard methods. J. Environ. Qual 2007, 5, 1420–1428. [Google Scholar]
- Tecon, R.; van der Meer, J.R. Bacterial biosensors for measuring availability of environmental pollutants. Sensors 2008, 8, 4062–4080. [Google Scholar]
- Purohit, H.J. Biosensors as molecular tools for use in bioremediation. J. Cleaner. Prod 2003, 11, 293–301. [Google Scholar]
- Ron, E. Biosensening environmental pollution. Curr. Opin. Biotechnol 2007, 18, 252–256. [Google Scholar]
- Yagi, K. Applications of whole-cell bacterial biosensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol 2007, 73, 1251–1258. [Google Scholar]
- Mulchandani, A.; Rogers, K.R. (Eds.) Enzyme and Micriobial Biosensors: Techniques and Protocols; Humana Press: Totowa, NJ, USA, 1998.
- Tresse, O.; Errampalli, D.; Kostrzynska, M.; Leung, K.T.; Lee, H.; Trevors, J.T.; Van Elsas, J.D. Green fluorescent protein as a visual marker in a p-nitrophenol degrading Moraxella sp. FEMS Microbiol. Lett 1998, 164, 187–193. [Google Scholar]
- Errampalli, D.; Okamura, H.; Lee, H.; Trevors, J.T.; Van Elsas, J.D. Green fluorescent protein as a marker to monitor survival of phenanthrene-mineralising Pseudomonas sp. UG14Gr in creosote-contaminated soil. FEMS Microbiol. Ecol 1998, 26, 181–191. [Google Scholar]
- Errampalli, D.; Tress, O.; Lee, H.; Trevors, J.T. Bacterial survival and mineralization of p-nitrophenol in soil by green fluorescent protein-marked Moraxella sp. G21 encapsulated cells. FEMS Microbiol. Ecol 1999, 30, 229–236. [Google Scholar]
- Cassidy, M.B.; Leung, K.T.; Lee, H.; Trevors, J.T. A comparison of enumeration methods for culturable Pseudomonas fluorescens cells marked with green fluorescent protein. J. Microbiol. Meth 2000, 40, 135–145. [Google Scholar]
- Abby, A.M.; Beaudette, L.A.; Lee, H.; Trevors, J.T. Polychlorinated biphenyl (PCB) degradation and persistence of a gfp-marked Ralstonia eutropha H850 in PCB-contaminated soil. Appl. Microbiol. Biotechnol 2003, 63, 222–230. [Google Scholar]
- Moller, S.; Sternberg, C.; Andersen, J.B.; Christensen, B.B.; Ramos, J.L.; Givskow; Molin, S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol 1998, 64, 721–732. [Google Scholar]
- Boldt, T.S.; Sorensen, J.; Karlson, U.; Molin, S.; Ramos, C. Combined use of different Gfp reporters for monitoring single-cell activity of a genetically modified PCB degrader in the rhizosphere of Alfalfa. FEMS Microbiol. Ecol 2004, 48, 139–148. [Google Scholar]
- Germaine, K.; Keogh, E.; Garcia-Cabellos, G.; Borremans, B.; van der Lelie, D; Barac, T.; Oeyen, L.; Vangronsveld, J.; Porteous Moore, F.; Moore, E.R.B.; Campbell, C.D.; Ryan, D.; Dowling, D.N. Colonisation of poplar trees by gfp expressing bacterial endophytes. FEMS Microbiol. Ecol 2004, 48, 109–118. [Google Scholar]
- Schreiter, P.P.; Gillor, O.; Post, A.; Belkin, S.; Schmid, R.D.; Bachmann, T.T. Monitoring of phosphorus bioavailability in water by an immobilized luminescent cyanobacterial reporter strain Biosens. Bioelectron 2001, 16, 811–818. [Google Scholar]
- Taylor, C.J.; Bain, L.A.; Richardson, D.J.; Spiro, S.; Russell, D.A. Construction of whole-cell gene reporter for the fluorescent bioassay of nitrate. Anal. Biochem 2004, 328, 60–66. [Google Scholar]
- Brandt, K.K.; Holm, P.E.; Nybroe, O. Bioavailability and toxicity of soil particle-associated copper as determined by two bioluminescent Pseudomonas fluorescens biosensor strains. Environ. Toxicol. Chem 2006, 25, 1738–1741. [Google Scholar]
- Fujimoto, H.; Wakabayashi, M.; Yamashiro, H.; Maeda, I.; Isoda, K.; Kondoh, M.; Kawase, M.; Miyasaka, H.; Yagi, K. Whole-cell arsenite biosensor using photosynthetic bacterium Rhodovulum sulfidophilum: Rhodovulum sulfidophilum as an arsenite biosensor. Appl. Microbiol. Biotechnol 2006, 73, 332–338. [Google Scholar]
- Liu, X.; Germaine, K.; Ryan, D.; Dowling, D.N. Development of a GFP-based biosensor for detecting the bioavailability and biodegradation of polychlorinated biphenyls (PCBs). J. Environ. Eng. Landsc. Manag 2007, 15, 261–268. [Google Scholar]
- Lejon, D.; Martins, J.; Leveque, J.; Spanini, L.; Pascault, N.; Landry, M.; Milloux, M.; Nowak, V.; Chaussod, R.; Ranjard, L. Copper dynamics and impacts on microbial communities in soils of variable organic status. Environ. Sci. Technol 2008, 42, 2819–2825. [Google Scholar]
- Willardson, M.B.; Wilkins, F.J.; Rand, A.T.; Schupp, M.J.; Hill, K.K.; Keim, P.; Jackson, J.P. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl. Environ. Microbiol 1998, 64, 1006–1012. [Google Scholar]
- Diesel, E.; Schreiber, M.; van der Meer, J. Development of bactyeria-based bioassays for arsenic detection in natural waters. Anal. Bioanal. Chem 2009, 394, 687–693. [Google Scholar]
- Fu, Y.; Chen, W.; Huang, Q. Construction of two lux-tagged Hg2+ specific biosensors and their luminescence performance. Appl. Microbiol. Biotechnol 2008, 79, 363–370. [Google Scholar]
- Keane, A.; Lau, P.; Ghoshal, S. Use of a whole cell biosensor to assess the bioavailability enhancement of aromatic hydrocarbon compounds bu nonionic surfactants. Biotechnol. Bioeng 2008, 99, 86–98. [Google Scholar]
- Kohlmeier, S.; Mancuso, M.; Deepthike, U.; Tecon, R.; van der Meer, J.; Harms, H.; Wells, M. Comparison of naphthalene bioavailability determined by whole cell biosensing and availability determined by extraction with Tenax. Environ. Pollut 2008, 156, 803–808. [Google Scholar]
- Li, Y.; Li, F.; Ho, C.; Liao, V.H. Construction and comparison of fluorescence and bioluminescence bacterial biosensors for the detection of bioavailable toluene and related compounds. Environ. Pollut 2008, 152, 123–129. [Google Scholar]
- Fiorentino, G.; Ronca, R.; Bartolucci, S. A novel E. coli biosensor for detecting aromatic aldehydes based on a responsive inducible archaeal promoter fused to the green fluorescent protein. Appl. Microbiol. Biotechnol 2009, 82, 67–77. [Google Scholar]
- Applegate, M.B.; Kehrmeyer, R.S.; Sayler, S.G. A chromosomally based tod luxCDABE whole-cell reporter for benzene, toluene, ethylbenzene, and xylene (BTEX) sensing. Appl. Environ. Microbiol 1998, 64, 2730–2735. [Google Scholar]
- Roberto, F.; Barnes, J.; Bruhn, D. Evaluation of a GFP reporter gene construct for environmental arsenic detection. Talanta 2002, 58, 181–188. [Google Scholar]
- Stiner, L.; Halverson, L.J. Development and characterization of a green fluorescent protein-based bacterial biosensor for bioavailable toluene and related compounds. Appl. Environ. Microbiol 2002, 68, 1962–1971. [Google Scholar]
- Ikeno, S.; Ogino, C.; Ito, T.; Shimizu, N. Detection of benzene derivatives by recombinant E. coli with Ps promoter and GFP as a reporter protein. J. Biochem. Eng 2003, 15, 193–197. [Google Scholar]
- Chang, S.T.; Lee, H.J.; Gu, M.B. Enhancement in the sensitivity of an immobilized cell-based soil biosensor for monitoring PAH toxicity. Sens. Actuat 2004, 97, 272–276. [Google Scholar]
- Peca, L.; Kos, P.; Mate, Z.; Farsang, A.; Vass, I. Construction of bioluminescent cyanobacterial reporter strains for detection of nickel, cobalt and zinc. FEMS Microbiol. Lett 2008, 289, 258–64. [Google Scholar]
- Layton, A.C.; Muccini, M.; Ghosh, M.M.; Sayler, G.S. Construction of a bioluminescent reporter strain to detect polychlorinated biphenyls. Appl. Environ. Microbiol 1998, 64, 5023–5026. [Google Scholar]
- Kohler, S.; Bachmann, T.T.; Schmitt, J.; Belkin, S.; Schmid, R.D. Detection of 4-chlorobenzoate using immobilized recombinant Escherichia coli reporter strains. Sens. Actuators 2000, 70, 139–144. [Google Scholar]
- Hansen, L.H.; Sorensen, S.J. The use of whole-cell biosensors to detect and quantify compounds or conditions affecting biological systems. Microb. Ecol 2001, 42, 483–494. [Google Scholar]
- Daunert, S.; Barrett, G.; Feliciano, J.S.; Shetty, R.S.; Shrestha, S.; Smith-Spencer, W. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem. Rev 2000, 100, 2705–2738. [Google Scholar]
- Turpeinen, R.; Virta, M.; Haggblom, M. Analysis of arsenic bioavailability in contaminated soils. Environ. Toxicol. Chem 2003, 22, 1–6. [Google Scholar]
- Tauriainen, S.; Virta, M.; Karp, M. Detecting bioavailable toxic metals and metalloids from natural water samples using luminescent sensor bacteria. Wat. Res 2000, 34, 2661–2666. [Google Scholar]
- Petanen, T.; Virta, M.; Karp, M.; Romantschuk, M. Construction and use broad host range mercury and arsenite sensor plasmids in the soil bacterium Pseudomonas fluorescens OS8. Microb. Ecol 2001, 41, 360–368. [Google Scholar]
- Heim, R.; Prasher, D.; Tsien, R. Wavelength mutations and post-translational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci USA 1994, 91, 12501–12504. [Google Scholar]
- Cubitt, A.B.; Heim, R.; Adams, S.R.; Boyd, A.E.; Gross, L.A.; Tsien, R.Y. Understanding, improving and using green fluorescent proteins. TIBS 1995, 20, 448–455. [Google Scholar]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.; Prasher, D. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar]
- Heim, R.; Cubitt, A.; Tsien, R. Improved green fluorescence. Nature 1995, 373, 663–664. [Google Scholar]
- Heim, R.; Tsien, R.Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol 1996, 6, 178–182. [Google Scholar]
- Cha, H.J.; Srivastava, R.; Vakharia, V.N.; Rao, G.; Bentley, W.E. Green fluorescent protein as a noninvasive stress probe in resting Escherichia coli cells. Appl. Environ. Microbiol 1999, 65, 409–414. [Google Scholar]
- Joyner, D.C.; Lindow, S.E. Heterogeneity of iron bioavailability on plants assessed with a whole-cell GFP-based bacterial biosensor. Microbiology 2000, 146, 2435–2445. [Google Scholar]
- Brandl, M.T.; Quinones, B.; Lindow, S.E. Heterogeneous transcription of an indoleacetic acid biosynthetic gene in Erwinia herbicola on plant surfaces. Proc. Natl. Acad. Sci. USA 2001, 98, 3454–3459. [Google Scholar]
- Leveau, J.H.; Lindow, S.E. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 2001, 98, 3446–3453. [Google Scholar]
- Miller, W.G.; Brandl, M.T.; Quinones, B.; Lindow, S.E. Biological sensor for sucrose availability: relative sensitivities of various reporter genes. Appl. Environ. Microbiol 2001, 67, 1308–1317. [Google Scholar]
- Meighen, E.A. Molecular biology of bacterial bioluminescence. Microbiol. Rev 1991, 55, 123–142. [Google Scholar]
- Heitzer, A.; Malachowsky, K.; Thonnard, J.E.; Bienkowski, P.R.; White, D.C.; Sayler, G.S. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol 1994, 60, 1487–1494. [Google Scholar]
- Simpson, M.L.; Sayler, G.S.; Applegate, B.M.; Ripp, S.; Nivens, D.E.; Paulus, M.J.; Jellison, G.E. Bioluminescent-bioreporter integrated circuits form novel whole-cell biosensors. Trends Biotechnol 1998, 16, 332–338. [Google Scholar]
- Girotti, S.; Ferri, E.N.; Fumo, M.G.; Maiolini, E. Monitoring of environmental pollutants by bioluminescent bacteria. Analytica Chimica. Acta 2008, 608, 2–29. [Google Scholar]
- Winther-Larsen, H.C.; Josefsen, K.D.; Brautaset, T.; Valla, S. Parameters Affecting Gene Expression from the Pm Promoter in Gram-Negative Bacteria. Metab. Eng 2000, 2, 79–91. [Google Scholar]
- Phoenix, P.; Keane, A.; Patel, A.; Bergeron, H.; Ghoshal, S.; Lau, P.C. Characterization of a new solvent-responsive gene locus in Pseudomonas putida F1 and its functionalization as a versatile biosensor. Environ. Microbiol 2003, 5, 1309–1327. [Google Scholar]
- Campbell, D.W.; Muller, C.; Reardon, K.F. Development of a fiber optic enzymatic biosensor for 1,2-dichloroethane. Biotechnol. Lett 2006, 28, 883–887. [Google Scholar]
- Leedjarv, A.; Ivask, A.; Virta, M.; Kahru, A. Analysis of bioavailable phenols from natural samples by recombinant luminescent bacterial sensors. Chemosphere 2006, 64, 1910–1919. [Google Scholar]
- Tizzard, A.C.; Bergsma, J.H.; Lloyd-Jones, G. A resazurin-based biosensor for organic pollutants. Biosens. Bioelectron 2006, 22, 759–763. [Google Scholar]
- Dawson, J.J.C.; Iroegbu, C.O.; Maciel, H.; Paton, G.I. Application of luminescent biosensors for monitoring the degradation and toxicity of BTEX compounds in soils. J. Appl. Microbiol 2008, 1, 141–151. [Google Scholar]
- Bontidean, I.; Mortari, A.; Leth, S.; Brown, N.L.; Karlson, U.; Larsen, M.M.; Vangvonsveld, J.; Corbisier, P.; Csoregi, E. Biosensors for detection of mercury in contaminated soils. Environ. Pollut 2004, 131, 255–262. [Google Scholar]
- Shetty, R.S.; Deo, S.K.; Liu, Y.; Daunert, S. Fluorescence-based sensing system for copper using genetically engineered living yeast cells. Biotechnol. Bioeng 2004, 88, 664–670. [Google Scholar]
- Magrisso, S.; Erel, Y.; Belkin, S. Microbial reporters of metal bioavailability. Microb. Biotechnol 2008, 4, 320–330. [Google Scholar]
- Farre, M.; Goncalves, C.; Lacorte, S.; Barcelo, D.; Alpendurada, M.F. Pesticide toxicity assessment using an electrochemical biosensor with Pseudomonas putida and a bioluminescence inhibition assay with Vibrio fischeri. Anal. Bioanal. Chem 2002, 373, 696–703. [Google Scholar]
- Chinalia, F.A.; Paton, G.I.; Killham. Physiological and toxicological characterisation of an engineered whole-cell biosensor. Bioresour. Technol 2008, 99, 714–721. [Google Scholar]
- Huang, W.E.; Wang, H.; Zheng, H.; Huang, L.; Singer, A.C.; Thompson, I.; Whiteley, A.S. Chromosomally located gene fusions constructed in Acinetobacter sp. ADP1 for the detection of salicylate. Environ. Microbiol 2005, 7, 1339–1348. [Google Scholar]
- Lei, Y.; Mulchandani, P.; Wang, J.; Chen, W.; Mulchandani, A. Highly sensitive and selective amperometric microbial biosensor for direct determination of p-nitrophenyl-substituted organophosphate nerve agents. Environ. Sci. Technol 2005, 39, 8853–8857. [Google Scholar]
- Mulchandani, P.; Chen, W.; Mulchandani, A. Direct determination of p-nitrophenyl substituent organophosphorus nerve agents using a recombinant Pseudomonas putida js444-modified clark oxygen electrode. J. Agric. Food. Chem 2005, 53, 524–527. [Google Scholar]
- Norman, A.; Hansen, L.H.; Sorensen, S.J. A flow cytometry-optimized assay using an SOS-green fluorescent protein (SOS-GFP) whole-cell biosensor for the detection of genotoxins in complex environments. Mutat. Res 2006, 603, 164–172. [Google Scholar]
- Matsui, N.; Kaya, T.; Nagamine, K.; Yasukawa, T.; Shiku, H.; Matsue, T. Electrochemical mutagen screening using microbial chip. Biosens. Bioelectron 2006, 21, 1202–1209. [Google Scholar]
- Belkin, S.; Smulski, D.R.; Vollmer, A.C.; Van Dyk, T.K.; LaRossa, R.A. Oxidative stress detection with Escherichia coli harboring a katG’:lux fusion. Appl. Environ. Microbiol 1996, 62, 2252–2256. [Google Scholar]
- Choi, J.W.; Park, K.W.; Lee, D.B.; Lee, W.; Lee, W.H. Cell immobilization using self-assembled synthetic oligopeptide and its application to biological toxicity detection using surface plasmon resonance. Biosens. Bioelectron 2005, 20, 2300–2305. [Google Scholar]
- Neufeld, T.; Biran, D.; Popovtzer, R.; Erez, T.; Ron, E.Z.; Rishpon, J. Genetically engineered pfabA pfabR bacteria: an electrochemical whole cell biosensor for detection of water toxicity. Anal. Chem 2006, 78, 4952–4956. [Google Scholar]
- Sorensen, S.J.; Burmolle, M.; Hansen, L.H. Making bio-sense of toxicity: new developments in whole-cell biosensors. Curr. Opin. Biotechnol 2006, 17, 11–16. [Google Scholar]
- Cases, I.; de Lorenzo, V. Promoters in the environment: transcriptional regulation in its natural context. Nat. Rev. Microbiol 2005, 3, 105–118. [Google Scholar]
- Jensen, S. Report of a new chemical hazard. New. Sci 1966, 32, 612. [Google Scholar]
- Risebrough, R.W.; Walker, W.; Schmidt, T.T.; deLappe, B.W.; Connors, C.W. Transfer of chlorinated biphenyls to Antarctica. Nature 1976, 264, 738–739. [Google Scholar]
- Triska, J.; Kuncova, G.; Mackova, M.; Novakova, H.; Paasivirta, J.; Lahtipera, M.; Vrchotova, N. Isolation and identification of intermediates from biodegradation of low chlorinated biphenyls (Delor 103). Chemosphere 2004, 54, 725–733. [Google Scholar]
- Weigel, W.; Wu, Q. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol. Ecol 2000, 32, 1–15. [Google Scholar]
- Arnett, C.M.; Parales, J.V.; Haddock, J.D. Influence of chlorine substituents on rates of oxidation of chlorinated biphenyls by the biphenyl dioxygenase of Burkholderia sp. strain LB400. Appl. Environ. Microbiol 2000, 66, 2928–2933. [Google Scholar]
- Abraham, W.R.; Nogales, B.; Golyshin, P.N.; Pieper, D.H.; Timmis, K.N. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr. Opin. Microbiol 2002, 5, 246–253. [Google Scholar]
- Johnson, B.L.; Hicks, H.E.; Cibulas, W.; Faroon, O.; Ashizawa, A.E.; De Rosa, C.T.; Cogliano, V.J.; Clark, M. Public Health implications of exposure to polychlorinated biphenyls (PCBs). Agency for Toxic Substances and Disease Registry: Atlanta, GA, 1998; Available online: www.atsdr.cdc.gov/DT/pcb007.html/ (accessed on 24 January 2010).
- EWG. PCBs in Farmed Salmon. Internet Commun. 2005. Available online: http://www.ewg.org/reports/farmedPCBs/ (accessed on 1 February 2010).
- ATSDR. Proceedings of the expert panel workshop to evaluate the public health implications for the treatment and disposal of polychlorinated biphenyls contaminated waste. Internet Commun. 1993. Available online: http://www.atsdr.cdc.gov/HAC/PCB/b_pcb_c1.html/ (accessed on 1 February 2010).
- Kolar, A.B.; Hrsak, D.; Fingler, S.; Cetkovic, H.; Petric, I.; Kolic, N.U. PCB degrading potential of aerobic bacteria enriched from marine sediments International. Int. Biodeterior. Biodegrad 2007, 60, 16–24. [Google Scholar]
- Catelani, D.; Mosselmans, G.; Nienhaus, J.; Sorlini, C.; Treccani, V. Microbial degradation of aromatic hydrocarbons used as reator coolants. Experientia 1970, 26, 922–923. [Google Scholar]
- Lunt, D.; Evans, W.C. The microbial metabolism of biphenyl. J. Biochem 1970, 118, 54–55. [Google Scholar]
- Ahmed, M.; Focht, D.D. Degradation of polychlorinated biphenyls by two species of Achromobacter. Can. J. Microbiol 1973, 19, 47–52. [Google Scholar]
- Furukawa, K.; Matsumura, F. Microbial metabolism of polychlorinated biphenyls-studies on relative degradability of polychlorinated biphenyl components by Alcaligenes sp. J. Agric. Food Chem 1976, 24, 251–256. [Google Scholar]
- Sylvestre, M.; Fauteux, J. A new facultative anaerobe capable of growth on 4-chlorobiphenyl. J. Gen. Appl. Microbiol 1982, 28, 61–72. [Google Scholar]
- Bedard, D.L.; Unterman, R.; Ropp, L.H.; Brennan, M.I.; Haber, M.L.; Johnson, C. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol 1986, 51, 761–768. [Google Scholar]
- Asturias, J.A.; Timmis, K.N. Three different 2,3-dihydroxybiphenyl- 1,2 dioxygenase genes in the grampositive polychlorobiphenyl degrading bacterium Rhodococcus globerulus P6. J. Bacteriol 1993, 175, 4631–4640. [Google Scholar]
- Seto, M.; Kimbara, K.; Shimura, M.; Hatta, T.; Fukuda, M. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Applied Appl. Environ. Microbiol 1995, 61, 3353–3358. [Google Scholar]
- Sakai, M.; Masai, E.; Asami, H.; Sugiyama, K.; Kimbara, K.; Fukuda, M. Diversity of 2,3-dihydroxybiphenyl dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1. J. Biosci. Bioeng 2002, 93, 421–427. [Google Scholar]
- Sierra, A.; Valera, J.L.; Marina, M.L.; Laborda, F. Study of the biodegradation process of polychlorinated biphenyls in liquid medium and soil by a new isolated aerobic bacterium (Janibacter sp). Chemosphere 2003, 53, 609–618. [Google Scholar]
- Abramowicz, D.A. Aerobic and anaerobic biodegradation of PCBs: a review. Crit. Rev. Biotechnol 1990, 10, 241–251. [Google Scholar]
- Furukawa, K. Engineering dioxygenases for efficient degradation of environmental pollutants. Curr. Opin. Biotechnol 2000, 11, 244–249. [Google Scholar]
- Borja, J.; Taleon, D.M.; Auresenia, K.; Gallardo, S. Polychlorinated biphenyls and their biodegradation. Process Biochem 2005, 40, 1999–2013. [Google Scholar]
- Pieper, D.H. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol 2005, 67, 170–191. [Google Scholar]
- Sylvestre, M. Genetically modified organisms to remediate polychlorinated biphenyls. Where Do We Stand? Int. Biodeterior. Biodegrad 2004, 54, 153–162. [Google Scholar]
- Bedard, D.L. Polychlorinated biphenyls in aquatic sediments: environmental fate and outlook for biological treatment. In Dehalogenation: Microbial Processes and Environmental Applications; Haggblom, M.M., Bossert, I.D., Eds.; Kluwer Academic Publishers: Boston, MA, USA, 2003; pp. 443–465. [Google Scholar]
- Turner, K.; Xu, S.; Pasini, P.; Deo, S.; Bachas, L.; Daunert, S. Hydroxylated polychlorinated biphenyls detection based on a genetically engineering bioluminescent whole-cell sensing system. Anal. Chem 2007, 79, 5740–5745. [Google Scholar]
- Liu, X. Development and application of biosensor technologies for the biodegradation of environmental pollutants. PhD. Thesis. Institute of Technology, Carlow, Ireland. 2008. [Google Scholar]
- Franklin, F.C.; Bagdasarian, M.; Bagdasarian, M.M.; Timmis, K.N. Molecular andfunctional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 1981, 78, 7458–7462. [Google Scholar]
- Ramos, J.L.; Gonzalez-Carrero, M.; Timmis, K.N. Broad-host range expression vector containing manipulated meta-cleavagepathway regulatory elements of the TOL plasmid. FEBS Lett 1988, 226, 241–246. [Google Scholar]
- de Lorenzo, V.; Fernandez, M.H.; Jakubzik, U.; Timmis, K.N. Engineering of alkyl-and haloaromatic-responsive gene expression mini-transposon containing regulated promoters of biodegradative pathways of Pseudomonas. Gene 1993, 130, 41–46. [Google Scholar]
- Herrero, M.; de Lorenzo, V.; Ensley, B.; Timmis, K.N. A T7 RNA polymerase-based system for the construction of Pseudomonas strain with phenotypes dependent on TOL-meta pathway effectors. Gene 1993, 134, 103–106. [Google Scholar]
- Cebolla, A.; Guzman, C.; de Lorenzo, V. Nondisruptive detection of activity of catabolic promoters of Pseudomonas putida with an antigenic surface reporter system. Appl. Environ. Microbiol 1996, 62, 214–220. [Google Scholar]
- Mermod, N.; Ramos, J.L.; Lehrbach, P.R.; Timmis, K.N. Vectors for regulated expression of cloned genes in a wide range of gram-negative bacteria. J. Bacteriol 1986, 167, 447–454. [Google Scholar]
- Blatny, J.M.; Brautaset, T.; Winther-Larsen, H.C.; Haugan, K.; Valla, S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol 1997, 63, 370–379. [Google Scholar]
- Blatny, J.M.; Brautaset, T.; Winther-Larsen, H.C.; Karunakaran, P.; Valla, S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 1997, 38, 35–51. [Google Scholar]
- Cormack, B.P.; Valdivia, R.H.; Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996, 173, 33–38. [Google Scholar]
- Andersen, J.B.; Sternberg, C.; Poulsen, L.K.; Bjorn, S.P.; Givskov, M.; Molin, S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol 1998, 64, 2240–2246. [Google Scholar]
- Shanahan, P.; O’Sullivan, D.J.; Simpson, P.; Glennon, J.D.; O’Gara, F. Isolation of 2, 4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol 1992, 58, 353–358. [Google Scholar]
- Goris, J.; Vos, P.D.; Caballero-Mellado, J.; Park, J.; Falsen, E.; Quensen, J.F., III; Tiedje, J.M.; Vandamme, P. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp nov. Int. J. Syst. Evol. Microbiol 2004, 54, 1677–168. [Google Scholar]
- Brazil, G.M.; Kenefick, L.; Callanan, M.; Haro, A.; De Lorenzo, V.; Dowling, D.N.; O'Gara, F. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl. Environ. Microbiol 1995, 61, 1946–1952. [Google Scholar]
- Villacieros, M.; Whelan, C.; Mackova, M.; Molgaard, J.; Sánchez-Contreras, M.; Lloret, J.; Aguirre de Cárcer, D.; Oruezábal, R.I.; Bolanos, L.; Macek, T.; Karlson, U.; Dowling, D.N.; Martín, M.; Rivilla, R. Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Appl. Environ. Microbiol 2005, 71, 2687–2694. [Google Scholar]
- Bjerketorp, J.; Hakansson, S.; Belkin, S.; Jansson, J.K. Advances in preservation methods: keeping biosensor microorganisms alive and active. Curr. Opin. Biotechnol 2006, 17, 43–49. [Google Scholar]
- Marques, S.; Aranda-Olmedo, I.; Ramos, J.L. Controlling bacterial physiology for optimal expression of gene reporter constructs. Curr. Opin. Biotechnol 2006, 17, 50–56. [Google Scholar]
- Sticher, P.; Jaspers, M.C.; Stemmler, K.; Harms, H.; Zehnder, A.J.; van der Meer, J.R. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl. Environ. Microbiol 1997, 63, 4053–4060. [Google Scholar]
- Premkumar, J.R.; Lev, O.; Rosen, R.; Belkin, S. Encapsulation of Luminous Recombinant E. coli in Sol-Gel Silicate Films. Adv. Mater 2001, 13, 1773–1775. [Google Scholar]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol 2003, 6, 206–212. [Google Scholar]
- Trogl, J.; Ripp, S.; Kuncova, G.; Sayler, G.S.; Churava, A.; Parik, P.; Demnerova, K.; Halova, J.; Kubicova, L. Selectivity of whole cell optical biosensor with immobilized bioreporter Pseudomonas fluorescens HK44. Sen. Actuat 2005, 107, 98–103. [Google Scholar]
- Cassidy, M.B.; Lee, H.; Trevors, J.T. Environmental applications of immobilized microbial cells: A review. J. Ind. Microbiol 1996, 16, 79–101. [Google Scholar]
- Mamo, G.; Gessesse, A. Immobilization of alkaliphilic Bacillus sp. cells for xylanase production using batch and continuous culture. Appl. Biochem. Biotechnol 2000, 87, 95–101. [Google Scholar]
- Kanasawud, P.; Hjorleifsdottir, S.; Holst, O.; Mattiasson, B. Studies on immobilization of the thermophilic bacterium Thermus aquaticus YT-1 by entrapment in various matrices. Appl. Microbiol. Biotechnol 1989, 31, 228–233. [Google Scholar]
- Heitzer, A.; Webb, O.F.; Thonnard, J.E.; Sayler, G.S. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl. Envion. Microbiol 1992, 58, 1839–1846. [Google Scholar]
- Vorlop, K.; Muscat, A.; Beyersdorf, J. Entrapment of microbial cells withinpolyurethane hydrogel beads with the advantage of low toxicity. Biotechnol. Tech 1992, 6, 483–488. [Google Scholar]
- Wu, K.Y.A.; Wisecarver, K.D. Cell immobilization using PVA crosslinked with boric acid. Biotechnol. Bioeng 1992, 39, 447–449. [Google Scholar]
- Rietti-Shati, M.; Ronen, D.; Mandelbaum, R.T.J. Atrazine degradation by Pseudomonas strain ADP entrapped in sol-gel glass. J. Sol-Gel Sci. Technol 1996, 7, 77–79. [Google Scholar]
- Chu, Y.F.; Hsu, C.; Soma, P.; Lo, Y. Immobilization of bioluminescent Escherishia coli cells using natural and artificial fibers treated with polyethyleneimine. Bioresour. Technol 2009, 100, 3167–3174. [Google Scholar]
- Russo, A.; Moënne-Loccoz, Y.; Fedi, S.; Higgins, P.; Fenton, A.; Dowling, D.N.; O'Regan, M.; O'Gara, F. Improved delivery of biocontrol Pseudomonas and their antifungal metabolites using alginate polymers. Appl. Microbiol. Biotechnol. 1996, 44, 740–745. [Google Scholar]
- Power, B.; Dowling, D.N. Alginate studies on Pseudomonas fluorescens F113rifPCB. (Unpublished).
- Gilmartin, N. Biodegradation of PCBs: Characterisation of a Rhodococcus strain and possible function of a glutathione S-transferase (BphK) from Burkholderia LB400. PhD thesis. Institute of Technology Carlow, Carlow, Ireland. 2004. [Google Scholar]
- Dorn, E.; Hellwig, M.; Reineke, W.; Knackmuss, H.J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol 1974, 99, 61–70. [Google Scholar]
- Leff, L.G.; Leff, A.A. Use of green fluorescent protein to monitor survival of genetically engineered bacteria in aquatic environments. Appl. Environ. Microbiol 1996, 62, 3486–3488. [Google Scholar]
- Wang, Y.; Rawlings, M.; Gibson, D.T.; Labbe, D.; Bergeron, H.; Brousseau, R.; Lau, P.C. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol. Gen. Genet 1995, 246, 570–579. [Google Scholar]
- Fuchslin, H.P.; Ruegg, I.; van der Meer, J.R.; Egli, T. Effect of integration of a GFP reporter gene on fitness of Ralstonia eutropha during growth with 2,4-dichlorophenoxyacetic acid. Environ. Microbiol 2003, 5, 878–887. [Google Scholar]
- Marks, T.S.; Smith, A.R.; Quirk, A.V. Degradation of 4-Chlorobenzoic Acid by Arthrobacter sp. Appl._Environ. Microbiol 1984, 48, 1020–1025. [Google Scholar]
- Nawaz, M.S.; Chapatawala, K.D. Simultaneous degradation of acetonitrile and biphenyl by Pseudomonas aenrginosa. Can. J. Microbiol 1991, 37, 411–418. [Google Scholar]
- Sondossi, M.; Sylvestre, M.; Ahmad, D. Effects of chlorobenzoate on the Pseudomonas testosterone biphenyl and chlorobiphenyl degradation pathway. Appl. Environ. Microbiol 1992, 58, 485–495. [Google Scholar]
- Harms, H.; Rime, J.; Leupin, O.; Hug, S.J.; van der Meer, J. Effect of groundwater composition on arsenic detection by bacterial biosensors. Microchimica Acta 2005, 151, 217–222. [Google Scholar]
- Pettigrew, C.A.; Breen, A.; Corcoran, C.; Sayler, G.S. Chlorinated biphenyl mineralization by individual populations and consortia of freshwater bacteria. Appl. Environ. Microbiol 1990, 56, 2036–2045. [Google Scholar]
- Hickey, W.J.; Searles, D.B.; Focht, D.D. Enhanced mineralization of polychlorinated biphenyls in soil inoculated with chlorobenzoate-degrading bacteria. Appl. Environ. Microbiol 1993, 59, 1194–1200. [Google Scholar]
- Di Gioia, D.; Bertin, L.; Zanaroli, G.; Marchetti, L.; Fava, F. Polychlorinated biphenyl degradation in aqueous wastes by employing continuous fixed-bed bioreactors. Process Biochemistry 2006, 41, 935–940. [Google Scholar]
- Ikariyama, Y.; Nishiguchi, S.; Koyama, T.; Kobatake, E.; Aizawa, M. Fiber-optic-based biomonitoring of benzene derivatives by recombinant E. coli bearing luciferase gene-fused TOL-plasmid immobilized on the fiber-optic end. Anal. Chem 1997, 69, 2600–2605. [Google Scholar]
- Gil, G.C.; Mitchell, R.J.; Chang, S.T.; Gu, M.B. A biosensor for the detection of gas toxicity using a recombinant bioluminescent bacterium. Biosens. Bioelectron 2000, 15, 23–30. [Google Scholar]
- Feliciano, J.; Liu, Y.; Daunert, S. Novel reporter gene in a fluorescent-based whole cell sensing system. Biotechnol. Bioeng 2006, 93, 989–997. [Google Scholar]




© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, X.; Germaine, K.J.; Ryan, D.; Dowling, D.N. Whole-Cell Fluorescent Biosensors for Bioavailability and Biodegradation of Polychlorinated Biphenyls. Sensors 2010, 10, 1377-1398. https://doi.org/10.3390/s100201377
Liu X, Germaine KJ, Ryan D, Dowling DN. Whole-Cell Fluorescent Biosensors for Bioavailability and Biodegradation of Polychlorinated Biphenyls. Sensors. 2010; 10(2):1377-1398. https://doi.org/10.3390/s100201377
Chicago/Turabian StyleLiu, Xuemei, Kieran J. Germaine, David Ryan, and David N. Dowling. 2010. "Whole-Cell Fluorescent Biosensors for Bioavailability and Biodegradation of Polychlorinated Biphenyls" Sensors 10, no. 2: 1377-1398. https://doi.org/10.3390/s100201377



