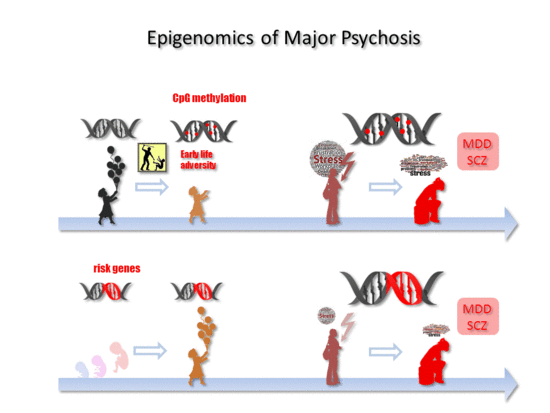
Epigenomics of Major Depressive Disorders and Schizophrenia: Early Life Decides
Abstract
:1. Introduction
2. The Genetic Architecture of MDD and SCZ
3. Early Life Events Preset Adult Behavior
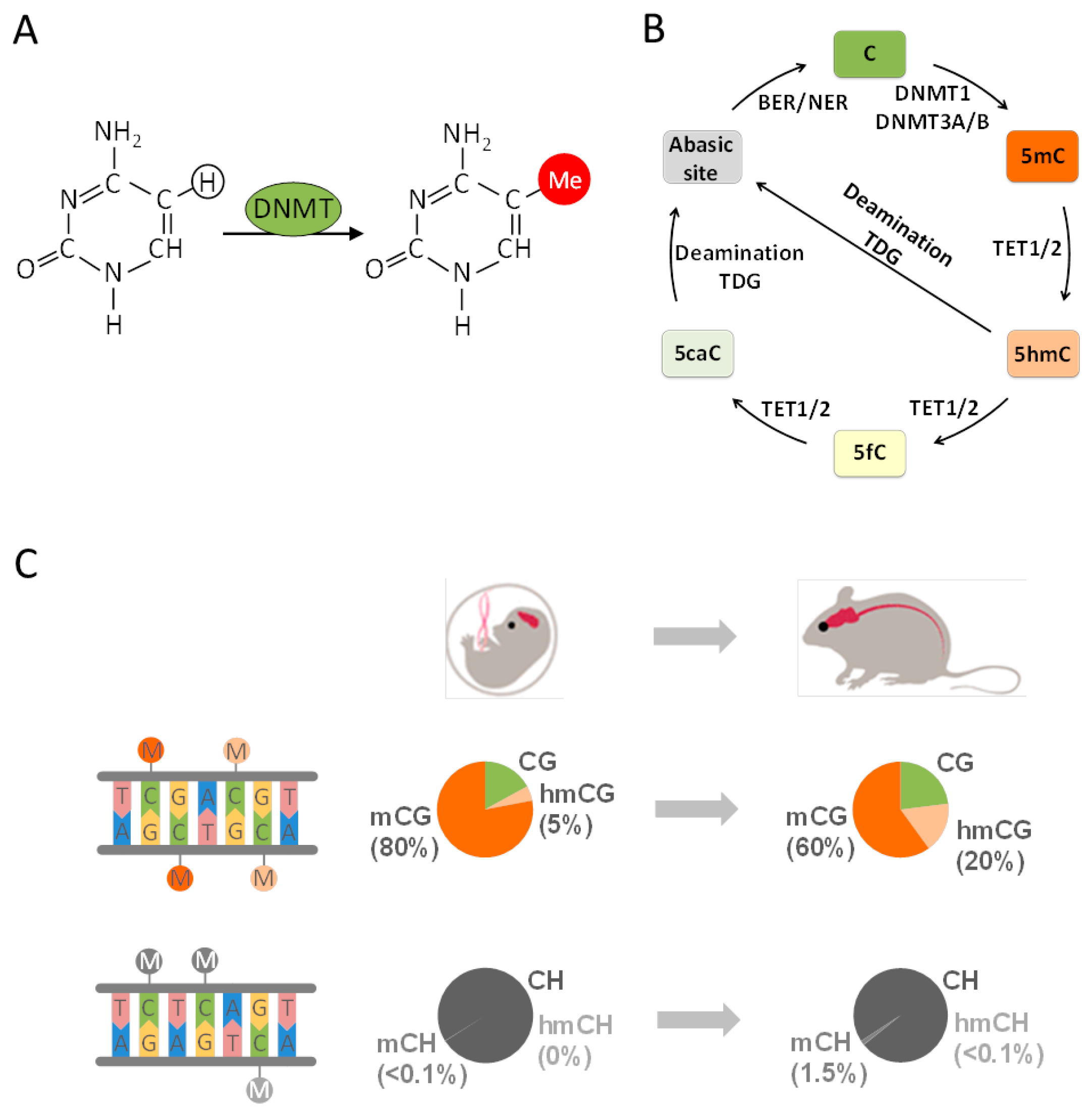
4. From Epigenetics to Epigenomics
5. Early Life Adversity-Dependent Epigenomic Responses as Risk Factor for MDD
6. Epigenomics in SCZ
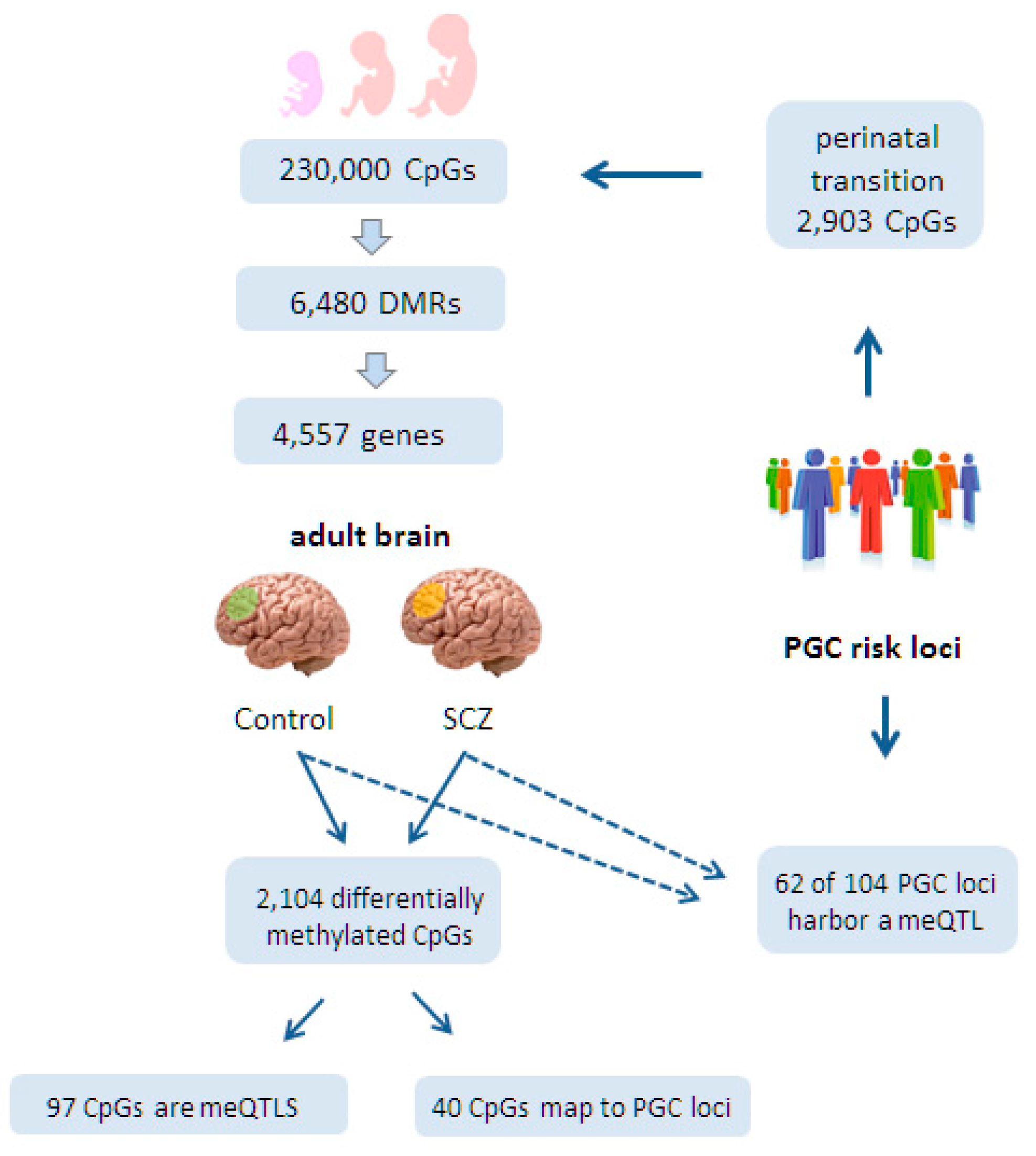
6.1. Epigenomic Changes in SCZ Are Enriched at Neurodevelopmental Loci
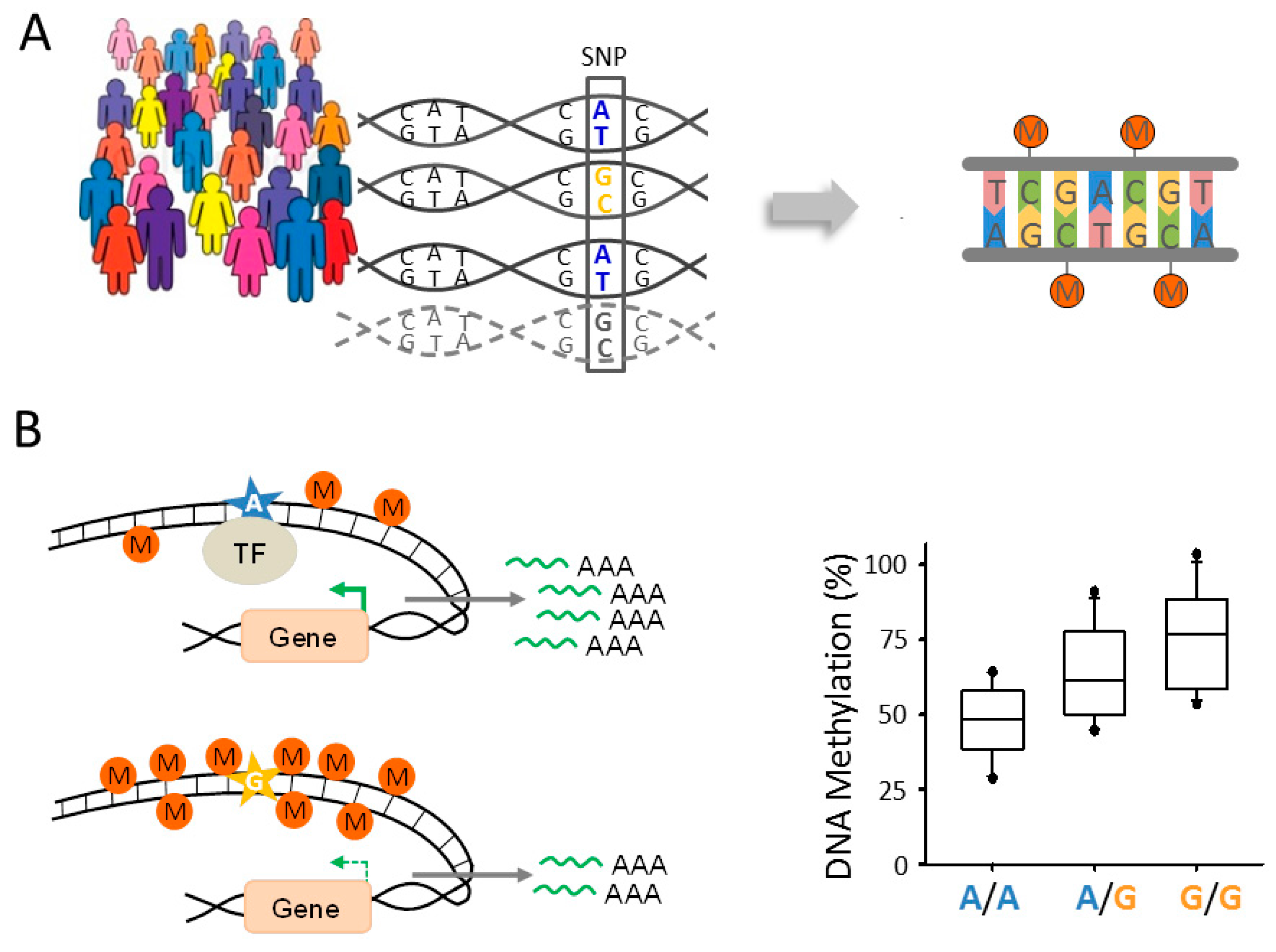
6.2. Epigenomic Changes in SCZ Are Genetically Controlled
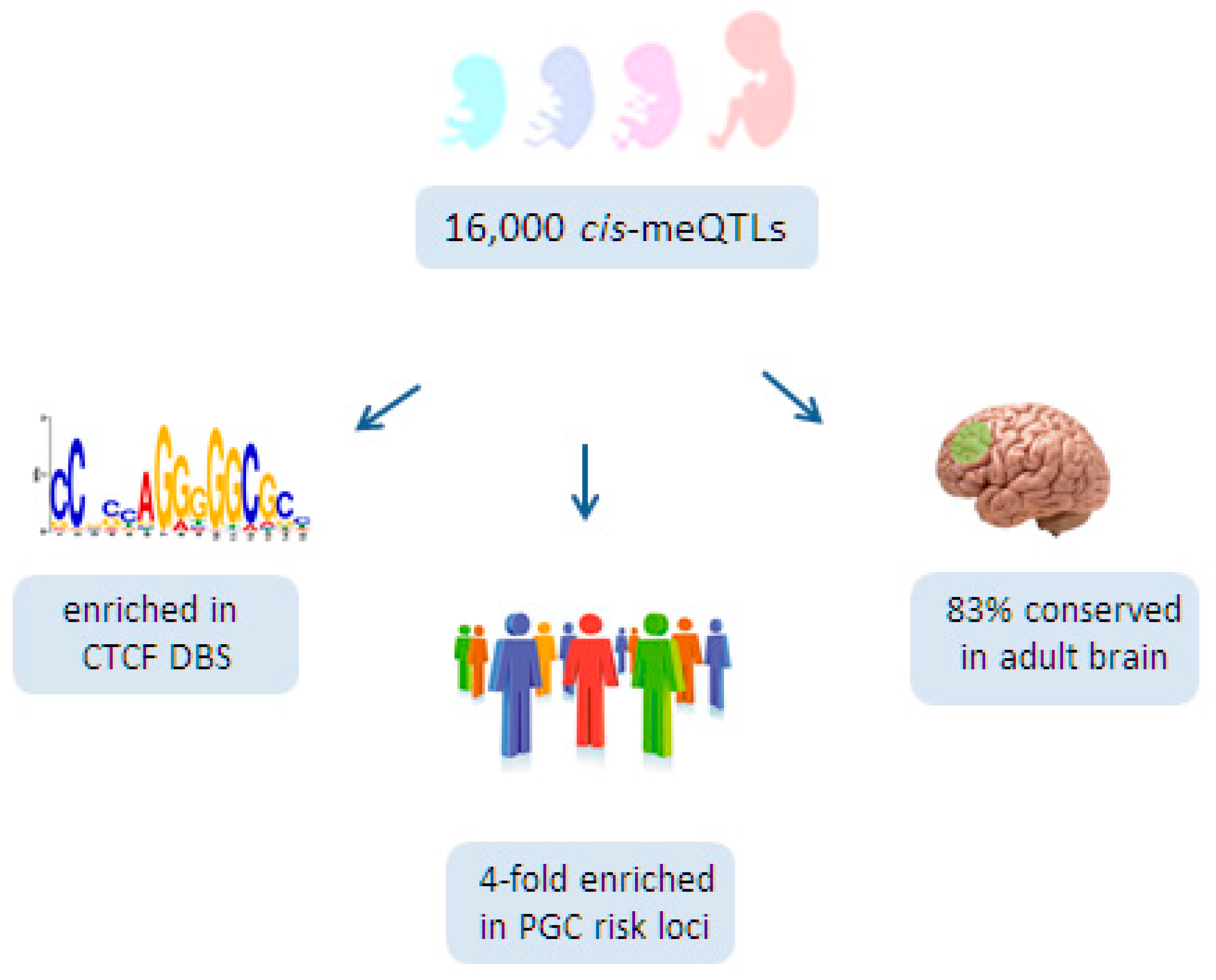
6.3. A Role for Fetal meQTLs in Mediating the Genetic Risk for SCZ
7. Conclusions and Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO Depression. Available online: http://www.who.int/mediacentre/factsheets/fs369/en/ (accessed on 24 April 2017).
- WHO Schizophrenia. Available online: http://www.who.int/mediacentre/factsheets/fs397/en/ (accessed on 24 April 2017).
- Collins, P.Y.; Patel, V.; Joestl, S.S.; March, D.; Insel, T.R.; Daar, A.S.; Scientific Advisory Board and the Executive Committee of the Grand Challenges on Global Mental Health; Anderson, W.; Dhansay, M.A.; Phillips, A.; et al. Grand challenges in global mental health. Nature 2011, 475, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Wittchen, H.U.; Jacobi, F.; Rehm, J.; Gustavsson, A.; Svensson, M.; Jönsson, B.; Olesen, J.; Allgulander, C.; Alonso, J.; Faravelli, C.; et al. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur. Neuropsychopharmacol. 2011, 21, 655–679. [Google Scholar] [CrossRef] [PubMed]
- Sadock, B.J.; Ruiz, P.; Sadock, V.A. Kaplan & Sadock’s Synopsis of Psychiatry: Behavioral Sciences, Clinical Psychiatry, 11th ed.; Wolters Kluwer: Philadelphia, PA, USA, 2015. [Google Scholar]
- Sadock, B.J.; Sadock, V.A.; Ruiz, P. Kaplan & Sadock’s Concise Textbook of Clinical Psychiatry, 4th ed.; Wolters Kluwer: Philadelphia, PA, USA, 2017. [Google Scholar]
- Clouston, T.S. The Neuroses of Development: The Morison Lectures for 1980, 1st ed.; Oliver & Boyd: Edinburgh, UK, 1891. [Google Scholar]
- Kraepelin, E. Dementia Praecox. The Clinical Roots of the Schizophrenia Concept, 5th ed.; Cambridge University Press: Leipzig, Germany, 1896. [Google Scholar]
- Feinberg, I. Schizophrenia: Caused by a fault in programmed synaptic elimination during adolescence? J. Psychiatr. Res. 1982, 17, 319–334. [Google Scholar] [CrossRef]
- Murray, R.M.; Lewis, S.W. Is schizophrenia a neurodevelopmental disorder? Br. Med. J. 1988, 296, 63. [Google Scholar] [CrossRef]
- Schulsinger, F.; Parnas, J.; Petersen, E.T.; Schulsinger, H.; Teasdale, T.W.; Mednick, S.A.; Møller, L.; Silverton, L. Cerebral ventricular size in the offspring of schizophrenic mothers. A preliminary study. Arch. Gen. Psychiatry 1984, 41, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Freud, S.; Bernays, A. Gesammelte Werke: Chronologisch Geordnet; Imago Publishing: London, UK, 1940; ISBN 978-3-596-50300-1. [Google Scholar]
- Francis, D.D.; Meaney, M.J. Maternal care and the development of stress responses. Curr. Opin. Neurobiol. 1999, 9, 128–134. [Google Scholar] [CrossRef]
- Heim, C.; Nemeroff, C.B. The role of childhood trauma in the neurobiology of mood and anxiety disorders: Preclinical and clinical studies. Biol. Psychiatry 2001, 49, 1023–1039. [Google Scholar] [CrossRef]
- Ladd, C.O.; Huot, R.L.; Thrivikraman, K.V.; Nemeroff, C.B.; Meaney, M.J.; Plotsky, P.M. Long-term behavioral and neuroendocrine adaptations to adverse early experience. Prog. Brain Res. 2000, 122, 81–103. [Google Scholar] [PubMed]
- Olson, S. From Neurons to Neighborhoods: An Update: Workshop Summary; National Academies Press: Washington, DC, USA, 2012. [Google Scholar]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Murgatroyd, C.; Wu, Y.; Bockmühl, Y.; Spengler, D. Genes learn from stress: How infantile trauma programs us for depression. Epigenet. Off. J. DNA Methylation Soc. 2010, 5, 194–199. [Google Scholar] [CrossRef]
- Zhang, T.-Y.; Meaney, M.J. Epigenetics and the environmental regulation of the genome and its function. Annu. Rev. Psychol. 2010, 61, 439–466. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Spengler, D. The lasting legacy of social stress on the epigenome of the hypothalamic-pituitary-adrenal axis. Epigenomics 2012, 4, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Klengel, T.; Binder, E.B. Epigenetics of Stress-Related Psychiatric Disorders and Gene × Environment Interactions. Neuron 2015, 86, 1343–1357. [Google Scholar] [CrossRef] [PubMed]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichtenstein, P.; Björk, C.; Hultman, C.M.; Scolnick, E.; Sklar, P.; Sullivan, P.F. Recurrence risks for schizophrenia in a Swedish national cohort. Psychol. Med. 2006, 36, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Kendler, K.S.; Neale, M.C. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 2003, 60, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Daly, M.J.; O’Donovan, M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nat. Rev. Genet. 2012, 13, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium; Ripke, S.; Wray, N.R.; Lewis, C.M.; Hamilton, S.P.; Weissman, M.M.; Breen, G.; Byrne, E.M.; Blackwood, D.H.R.; Boomsma, D.I.; et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 2013, 18, 497–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casey, B.J.; Craddock, N.; Cuthbert, B.N.; Hyman, S.E.; Lee, F.S.; Ressler, K.J. DSM-5 and RDoC: Progress in psychiatry research? Nat. Rev. Neurosci. 2013, 14, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, B.N.; Insel, T.R. Toward the future of psychiatric diagnosis: The seven pillars of RDoC. BMC Med. 2013, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Converge Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 2015, 523, 588–591. [Google Scholar] [CrossRef]
- Haigis, M.C.; Guarente, L.P. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006, 20, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Hyde, C.L.; Nagle, M.W.; Tian, C.; Chen, X.; Paciga, S.A.; Wendland, J.R.; Tung, J.Y.; Hinds, D.A.; Perlis, R.H.; Winslow, A.R. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 2016, 48, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Olfson, M.; Blanco, C.; Marcus, S.C. Treatment of Adult Depression in the United States. JAMA Intern. Med. 2016, 176, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Cross-Disorder Group of the Psychiatric Genomics Consortium; Lee, S.H.; Ripke, S.; Neale, B.M.; Faraone, S.V.; Purcell, S.M.; Perlis, R.H.; Mowry, B.J.; Thapar, A.; Goddard, M.E.; et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 2013, 45, 984–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Power, R.A.; Tansey, K.E.; Buttenschøn, H.N.; Cohen-Woods, S.; Bigdeli, T.; Hall, L.S.; Kutalik, Z.; Lee, S.H.; Ripke, S.; Steinberg, S.; et al. Genome-wide Association for Major Depression Through Age at Onset Stratification: Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Biol. Psychiatry 2017, 81, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Kendler, K.S.; Gatz, M.; Gardner, C.O.; Pedersen, N.L. Age at onset and familial risk for major depression in a Swedish national twin sample. Psychol. Med. 2005, 35, 1573–1579. [Google Scholar] [CrossRef] [PubMed]
- Weissman, M.M.; Wickramaratne, P.; Merikangas, K.R.; Leckman, J.F.; Prusoff, B.A.; Caruso, K.A.; Kidd, K.K.; Gammon, G.D. Onset of major depression in early adulthood. Increased familial loading and specificity. Arch. Gen. Psychiatry 1984, 41, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Ripke, S.; O’Dushlaine, C.; Chambert, K.; Moran, J.L.; Kähler, A.K.; Akterin, S.; Bergen, S.E.; Collins, A.L.; Crowley, J.J.; Fromer, M.; et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013, 45, 1150–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Schizophrenia Consortium; Purcell, S.M.; Wray, N.R.; Stone, J.L.; Visscher, P.M.; O’Donovan, M.C.; Sullivan, P.F.; Sklar, P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009, 460, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, H.; Ophoff, R.A.; Steinberg, S.; Andreassen, O.A.; Cichon, S.; Rujescu, D.; Werge, T.; Pietiläinen, O.P.H.; Mors, O.; Mortensen, P.B.; et al. Common variants conferring risk of schizophrenia. Nature 2009, 460, 744–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Gluckman, P.D.; Hanson, M.A.; Cooper, C.; Thornburg, K.L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008, 359, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Gluckman, P.D.; Hanson, M.A.; Bateson, P.; Beedle, A.S.; Law, C.M.; Bhutta, Z.A.; Anokhin, K.V.; Bougnères, P.; Chandak, G.R.; Dasgupta, P.; et al. Towards a new developmental synthesis: Adaptive developmental plasticity and human disease. Lancet 2009, 373, 1654–1657. [Google Scholar] [CrossRef]
- Hoffmann, A.; Spengler, D. DNA memories of early social life. Neuroscience 2014, 264, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Toga, A.W.; Thompson, P.M.; Sowell, E.R. Mapping brain maturation. Trends Neurosci. 2006, 29, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, J.-P. Brain synaptogenesis and epigenesis. Med. Sci. 2005, 21, 428–433. [Google Scholar] [CrossRef]
- Cowan, W.M. The development of the brain. Sci. Am. 1979, 241, 113–133. [Google Scholar] [CrossRef] [PubMed]
- Kessler, R.C.; McLaughlin, K.A.; Green, J.G.; Gruber, M.J.; Sampson, N.A.; Zaslavsky, A.M.; Aguilar-Gaxiola, S.; Alhamzawi, A.O.; Alonso, J.; Angermeyer, M.; et al. Childhood adversities and adult psychopathology in the WHO World Mental Health Surveys. Br. J. Psychiatry 2010, 197, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Green, J.G.; McLaughlin, K.A.; Berglund, P.A.; Gruber, M.J.; Sampson, N.A.; Zaslavsky, A.M.; Kessler, R.C. Childhood adversities and adult psychiatric disorders in the national comorbidity survey replication I: Associations with first onset of DSM-IV disorders. Arch. Gen. Psychiatry 2010, 67, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Dube, S.R.; Anda, R.F.; Felitti, V.J.; Chapman, D.P.; Williamson, D.F.; Giles, W.H. Childhood abuse, household dysfunction, and the risk of attempted suicide throughout the life span: Findings from the Adverse Childhood Experiences Study. JAMA J. Am. Med. Assoc. 2001, 286, 3089–3096. [Google Scholar] [CrossRef]
- Chapman, D.P.; Whitfield, C.L.; Felitti, V.J.; Dube, S.R.; Edwards, V.J.; Anda, R.F. Adverse childhood experiences and the risk of depressive disorders in adulthood. J. Affect. Disord. 2004, 82, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Akdeniz, C.; Tost, H.; Meyer-Lindenberg, A. The neurobiology of social environmental risk for schizophrenia: An evolving research field. Soc. Psychiatry Psychiatr. Epidemiol. 2014, 49, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Jablensky, A. Epidemiology of schizophrenia: The global burden of disease and disability. Eur. Arch. Psychiatry Clin. Neurosci. 2000, 250, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.; Malchow, B.; Hasan, A.; Falkai, P. The impact of environmental factors in severe psychiatric disorders. Front. Neurosci. 2014, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Bahari-Javan, S.; Varbanov, H.; Halder, R.; Benito, E.; Kaurani, L.; Burkhardt, S.; Anderson-Schmidt, H.; Anghelescu, I.; Budde, M.; Stilling, R.M.; et al. HDAC1 links early life stress to schizophrenia-like phenotypes. Proc. Natl. Acad. Sci. USA 2017, 114, E4686–E4694. [Google Scholar] [CrossRef] [PubMed]
- Gilman, S.E.; Hornig, M.; Ghassabian, A.; Hahn, J.; Cherkerzian, S.; Albert, P.S.; Buka, S.L.; Goldstein, J.M. Socioeconomic disadvantage, gestational immune activity, and neurodevelopment in early childhood. Proc. Natl. Acad. Sci. USA 2017, 114, 6728–6733. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.; Pearson, R.M.; Goodman, S.H.; Rapa, E.; Rahman, A.; McCallum, M.; Howard, L.M.; Pariante, C.M. Effects of perinatal mental disorders on the fetus and child. Lancet 2014, 384, 1800–1819. [Google Scholar] [CrossRef]
- Lupien, S.J.; McEwen, B.S.; Gunnar, M.R.; Heim, C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat. Rev. Neurosci. 2009, 10, 434–445. [Google Scholar] [CrossRef] [PubMed]
- McEwen, B.S. Physiology and Neurobiology of Stress and Adaptation: Central Role of the Brain. Physiol. Rev. 2007, 87, 873–904. [Google Scholar] [CrossRef] [PubMed]
- De Kloet, E.R.; Joëls, M.; Holsboer, F. Stress and the brain: From adaptation to disease. Nat. Rev. Neurosci. 2005, 6, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, L. Epigenetics; Garland Science: New York, NY, USA, 2014; ISBN 978-0-8153-6511-2. [Google Scholar]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Gu, H.; Müller, F.; Donaghey, J.; Tsai, L.T.-Y.; Kohlbacher, O.; de Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E.; et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Kriaucionis, S.; Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef] [PubMed]
- Mellén, M.; Ayata, P.; Dewell, S.; Kriaucionis, S.; Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012, 151, 1417–1430. [Google Scholar] [CrossRef] [PubMed]
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.-X.; Wu, H.; Dai, Q.; Irier, H.; Upadhyay, A.K.; Gearing, M.; Levey, A.I.; et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011, 14, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell 2014, 156, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Shao, N.; Szulwach, K.E.; Vialou, V.; Huynh, J.; Zhong, C.; Le, T.; Ferguson, D.; Cahill, M.E.; Li, Y.; et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat. Neurosci. 2015, 18, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Kaas, G.A.; Zhong, C.; Eason, D.E.; Ross, D.L.; Vachhani, R.V.; Ming, G.-L.; King, J.R.; Song, H.; Sweatt, J.D. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 2013, 79, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wei, W.; Zhao, Q.-Y.; Widagdo, J.; Baker-Andresen, D.; Flavell, C.R.; D’Alessio, A.; Zhang, Y.; Bredy, T.W. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, 7120–7125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Su, Y.; Shin, J.; Zhong, C.; Guo, J.U.; Weng, Y.-L.; Gao, F.; Geschwind, D.H.; Coppola, G.; Ming, G.; et al. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat. Neurosci. 2015, 18, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Ramsahoye, B.H.; Biniszkiewicz, D.; Lyko, F.; Clark, V.; Bird, A.P.; Jaenisch, R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 2000, 97, 5237–5242. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Barr, C.L.; Kim, A.; Yue, F.; Lee, A.Y.; Eubanks, J.; Dempster, E.L.; Ren, B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 2012, 148, 816–831. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.D.; He, Y.; Whitaker, J.W.; Hariharan, M.; Mukamel, E.A.; Leung, D.; Rajagopal, N.; Nery, J.R.; Urich, M.A.; Chen, H.; et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 2015, 523, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Müller, F.; Liao, J.; Zhang, Y.; Gu, H.; Bock, C.; Boyle, P.; Epstein, C.B.; Bernstein, B.E.; Lengauer, T.; et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011, 7, e1002389. [Google Scholar] [CrossRef] [PubMed]
- Beck, S.; Olek, A.; Walter, J. From genomics to epigenomics: A loftier view of life. Nat. Biotechnol. 1999, 17, 1144. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Hildmann, T.; Novik, K.L.; Lewin, J.; Tost, J.; Cox, A.V.; Andrews, T.D.; Howe, K.L.; Otto, T.; Olek, A.; et al. DNA methylation profiling of the human major histocompatibility complex: A pilot study for the human epigenome project. PLoS Biol. 2004, 2, e405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Champagne, F.A.; Francis, D.D.; Mar, A.; Meaney, M.J. Variations in maternal care in the rat as a mediating influence for the effects of environment on development. Physiol. Behav. 2003, 79, 359–371. [Google Scholar] [CrossRef]
- Heim, C.; Shugart, M.; Craighead, W.E.; Nemeroff, C.B. Neurobiological and psychiatric consequences of child abuse and neglect. Dev. Psychobiol. 2010, 52, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Francis, D.; Diorio, J.; Liu, D.; Meaney, M.J. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science 1999, 286, 1155–1158. [Google Scholar] [CrossRef] [PubMed]
- Weaver, I.C.G.; Meaney, M.J.; Szyf, M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc. Natl. Acad. Sci. USA 2006, 103, 3480–3485. [Google Scholar] [CrossRef] [PubMed]
- Weaver, I.C.G.; Champagne, F.A.; Brown, S.E.; Dymov, S.; Sharma, S.; Meaney, M.J.; Szyf, M. Reversal of Maternal Programming of Stress Responses in Adult Offspring through Methyl Supplementation: Altering Epigenetic Marking Later in Life. J. Neurosci. 2005, 25, 11045–11054. [Google Scholar] [CrossRef] [PubMed]
- Weaver, I.C.; Cervoni, N.; Champagne, F.A.; D’Alessio, A.C.; Sharma, S.; Seckl, J.R.; Dymov, S.; Szyf, M.; Meaney, M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004, 7, 847–854. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.O.; Suderman, M.; Sasaki, A.; Huang, T.C.T.; Hallett, M.; Meaney, M.J.; Szyf, M. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS ONE 2011, 6, e14739. [Google Scholar] [CrossRef] [PubMed]
- Junghans, D.; Haas, I.G.; Kemler, R. Mammalian cadherins and protocadherins: About cell death, synapses and processing. Curr. Opin. Cell Biol. 2005, 17, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Moisiadis, V.G.; Matthews, S.G. Glucocorticoids and fetal programming part 1: Outcomes. Nat. Rev. Endocrinol. 2014, 10, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Moisiadis, V.G.; Matthews, S.G. Glucocorticoids and fetal programming part 2: Mechanisms. Nat. Rev. Endocrinol. 2014, 10, 403–411. [Google Scholar] [CrossRef] [PubMed]
- French, N.P.; Hagan, R.; Evans, S.F.; Mullan, A.; Newnham, J.P. Repeated antenatal corticosteroids: Effects on cerebral palsy and childhood behavior. Am. J. Obstet. Gynecol. 2004, 190, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Glover, V. Annual Research Review: Prenatal stress and the origins of psychopathology: An evolutionary perspective. J. Child Psychol. Psychiatry 2011, 52, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Crudo, A.; Suderman, M.; Moisiadis, V.G.; Petropoulos, S.; Kostaki, A.; Hallett, M.; Szyf, M.; Matthews, S.G. Glucocorticoid programming of the fetal male hippocampal epigenome. Endocrinology 2013, 154, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Dobbing, J.; Sands, J. Growth and development of the brain and spinal cord of the guinea pig. Brain Res. 1970, 17, 115–123. [Google Scholar] [CrossRef]
- Crudo, A.; Petropoulos, S.; Suderman, M.; Moisiadis, V.G.; Kostaki, A.; Hallett, M.; Szyf, M.; Matthews, S.G. Effects of antenatal synthetic glucocorticoid on glucocorticoid receptor binding, DNA methylation, and genome-wide mRNA levels in the fetal male hippocampus. Endocrinology 2013, 154, 4170–4181. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.O.; Sasaki, A.; D’Alessio, A.C.; Dymov, S.; Labonté, B.; Szyf, M.; Turecki, G.; Meaney, M.J. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009, 12, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Suderman, M.; McGowan, P.O.; Sasaki, A.; Huang, T.C.T.; Hallett, M.T.; Meaney, M.J.; Turecki, G.; Szyf, M. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc. Natl. Acad. Sci. USA 2012, 109 (Suppl. S2), 17266–17272. [Google Scholar] [CrossRef] [PubMed]
- Labonté, B.; Suderman, M.; Maussion, G.; Navaro, L.; Yerko, V.; Mahar, I.; Bureau, A.; Mechawar, N.; Szyf, M.; Meaney, M.J.; et al. Genome-wide epigenetic regulation by early-life trauma. Arch. Gen. Psychiatry 2012, 69, 722–731. [Google Scholar] [CrossRef] [PubMed]
- Labonté, B.; Suderman, M.; Maussion, G.; Lopez, J.P.; Navarro-Sánchez, L.; Yerko, V.; Mechawar, N.; Szyf, M.; Meaney, M.J.; Turecki, G. Genome-wide methylation changes in the brains of suicide completers. Am. J. Psychiatry 2013, 170, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Yen, S.; Shea, M.T.; Sanislow, C.A.; Skodol, A.E.; Grilo, C.M.; Edelen, M.O.; Stout, R.L.; Morey, L.C.; Zanarini, M.C.; Markowitz, J.C.; et al. Personality traits as prospective predictors of suicide attempts. Acta Psychiatr. Scand. 2009, 120, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.; Mechawar, N.; Turecki, G. Suicide neurobiology. Prog. Neurobiol. 2009, 89, 315–333. [Google Scholar] [CrossRef] [PubMed]
- Jollant, F.; Lawrence, N.L.; Olié, E.; Guillaume, S.; Courtet, P. The suicidal mind and brain: A review of neuropsychological and neuroimaging studies. World J. Biol. Psychiatry 2011, 12, 319–339. [Google Scholar] [CrossRef] [PubMed]
- Bockmühl, Y.; Patchev, A.V.; Madejska, A.; Hoffmann, A.; Sousa, J.C.; Sousa, N.; Holsboer, F.; Almeida, O.F.X.; Spengler, D. Methylation at the CpG island shore region upregulates Nr3c1 promoter activity after early-life stress. Epigenetics 2015, 10, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellani, C.A.; Melka, M.G.; Diehl, E.J.; Laufer, B.I.; O’Reilly, R.L.; Singh, S.M. DNA methylation in psychosis: Insights into etiology and treatment. Epigenomics 2015, 7, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Pishva, E.; Kenis, G.; van den Hove, D.; Lesch, K.-P.; Boks, M.P.M.; van Os, J.; Rutten, B.P.F. The epigenome and postnatal environmental influences in psychotic disorders. Soc. Psychiatry Psychiatr. Epidemiol. 2014, 49, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Swathy, B.; Banerjee, M. Understanding epigenetics of schizophrenia in the backdrop of its antipsychotic drug therapy. Epigenomics 2017. [Google Scholar] [CrossRef] [PubMed]
- Mill, J.; Tang, T.; Kaminsky, Z.; Khare, T.; Yazdanpanah, S.; Bouchard, L.; Jia, P.; Assadzadeh, A.; Flanagan, J.; Schumacher, A.; et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008, 82, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Wockner, L.F.; Noble, E.P.; Lawford, B.R.; Young, R.M.; Morris, C.P.; Whitehall, V.L.J.; Voisey, J. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl. Psychiatry 2014, 4, e339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pidsley, R.; Viana, J.; Hannon, E.; Spiers, H.; Troakes, C.; Al-Saraj, S.; Mechawar, N.; Turecki, G.; Schalkwyk, L.C.; Bray, N.J.; et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014, 15, 483. [Google Scholar] [CrossRef] [PubMed]
- Ball, M.P.; Li, J.B.; Gao, Y.; Lee, J.-H.; LeProust, E.M.; Park, I.-H.; Xie, B.; Daley, G.Q.; Church, G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009, 27, 361–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckhardt, F.; Lewin, J.; Cortese, R.; Rakyan, V.K.; Attwood, J.; Burger, M.; Burton, J.; Cox, T.V.; Davies, R.; Down, T.A.; et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006, 38, 1378–1385. [Google Scholar] [CrossRef] [PubMed]
- Naeve, G.S.; Ramakrishnan, M.; Kramer, R.; Hevroni, D.; Citri, Y.; Theill, L.E. Neuritin: A gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc. Natl. Acad. Sci. USA 1997, 94, 2648–2653. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Banasr, M.; Choi, M.; Chae, S.Y.; Licznerski, P.; Lee, B.; Voleti, B.; Li, N.; Lepack, A.; Fournier, N.M.; et al. Neuritin produces antidepressant actions and blocks the neuronal and behavioral deficits caused by chronic stress. Proc. Natl. Acad. Sci. USA 2012, 109, 11378–11383. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.C.; Nap, J. P. Genetical genomics: The added value from segregation. Trends Genet. 2001, 17, 388–391. [Google Scholar] [CrossRef]
- Rockman, M.V.; Kruglyak, L. Genetics of global gene expression. Nat. Rev. Genet. 2006, 7, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.T.; Pai, A.A.; Pickrell, J.K.; Gaffney, D.J.; Pique-Regi, R.; Degner, J.F.; Gilad, Y.; Pritchard, J.K. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011, 12, R10. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.T.; Tsai, P.-C.; Yang, T.-P.; Pidsley, R.; Nisbet, J.; Glass, D.; Mangino, M.; Zhai, G.; Zhang, F.; Valdes, A.; et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012, 8, e1002629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drong, A.W.; Nicholson, G.; Hedman, A.K.; Meduri, E.; Grundberg, E.; Small, K.S.; Shin, S.-Y.; Bell, J.T.; Karpe, F.; Soranzo, N.; et al. The presence of methylation quantitative trait loci indicates a direct genetic influence on the level of DNA methylation in adipose tissue. PLoS ONE 2013, 8, e55923. [Google Scholar] [CrossRef] [PubMed]
- Gertz, J.; Varley, K.E.; Reddy, T.E.; Bowling, K.M.; Pauli, F.; Parker, S.L.; Kucera, K.S.; Willard, H.F.; Myers, R.M. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet. 2011, 7, e1002228. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.R.; van der Brug, M.P.; Hernandez, D.G.; Traynor, B.J.; Nalls, M.A.; Lai, S.-L.; Arepalli, S.; Dillman, A.; Rafferty, I.P.; Troncoso, J.; et al. Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain. PLoS Genet. 2010, 6, e1000952. [Google Scholar] [CrossRef] [PubMed]
- Hannon, E.; Spiers, H.; Viana, J.; Pidsley, R.; Burrage, J.; Murphy, T.M.; Troakes, C.; Turecki, G.; O’Donovan, M.C.; Schalkwyk, L.C.; et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 2016, 19, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.E.; Gao, Y.; Deep-Soboslay, A.; Tao, R.; Hyde, T.M.; Weinberger, D.R.; Kleinman, J.E. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2016, 19, 40–47. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium Human Genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Kilpinen, H.; Waszak, S.M.; Gschwind, A.R.; Raghav, S.K.; Witwicki, R.M.; Orioli, A.; Migliavacca, E.; Wiederkehr, M.; Gutierrez-Arcelus, M.; Panousis, N.I.; et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science 2013, 342, 744–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Rosario, R.C.-H.; Poschmann, J.; Rouam, S.L.; Png, E.; Khor, C.C.; Hibberd, M.L.; Prabhakar, S. Sensitive detection of chromatin-altering polymorphisms reveals autoimmune disease mechanisms. Nat. Methods 2015, 12, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Westra, H.-J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, A.D.; Hu, M.; Ren, B. Genome-wide mapping and analysis of chromosome architecture. Nat. Rev. Mol. Cell Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Badner, J.A.; Cheng, L.; Zhang, C.; Zhang, D.; Cox, N.J.; Gershon, E.S.; Kelsoe, J.R.; Greenwood, T.A.; Nievergelt, C.M.; et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol. Psychiatry 2013, 18, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Cheng, L.; Badner, J.A.; Chen, C.; Chen, Q.; Luo, W.; Craig, D.W.; Redman, M.; Gershon, E.S.; Liu, C. Genetic control of individual differences in gene-specific methylation in human brain. Am. J. Hum. Genet. 2010, 86, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Banovich, N.E.; Lan, X.; McVicker, G.; van de Geijn, B.; Degner, J.F.; Blischak, J.D.; Roux, J.; Pritchard, J.K.; Gilad, Y. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014, 10, e1004663. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Arcelus, M.; Lappalainen, T.; Montgomery, S.B.; Buil, A.; Ongen, H.; Yurovsky, A.; Bryois, J.; Giger, T.; Romano, L.; Planchon, A.; et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2013, 2, e00523. [Google Scholar] [CrossRef] [PubMed]
- Elkabetz, Y.; Studer, L. Human ESC-derived neural rosettes and neural stem cell progression. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Numata, S.; Ye, T.; Herman, M.; Lipska, B.K. DNA methylation changes in the postmortem dorsolateral prefrontal cortex of patients with schizophrenia. Front. Genet. 2014, 5, 280. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Arcelus, M.; Ongen, H.; Lappalainen, T.; Montgomery, S.B.; Buil, A.; Yurovsky, A.; Bryois, J.; Padioleau, I.; Romano, L.; Planchon, A.; et al. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet. 2015, 11, e1004958. [Google Scholar] [CrossRef] [PubMed]
- Flavahan, W.A.; Drier, Y.; Liau, B.B.; Gillespie, S.M.; Venteicher, A.S.; Stemmer-Rachamimov, A.O.; Suvà, M.L.; Bernstein, B.E. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016, 529, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Juraeva, D.; Haenisch, B.; Zapatka, M.; Frank, J.; GROUP Investigators; PSYCH-GEMS SCZ Working Group; Witt, S.H.; Mühleisen, T.W.; Treutlein, J.; Strohmaier, J.; et al. Integrated pathway-based approach identifies association between genomic regions at CTCF and CACNB2 and schizophrenia. PLoS Genet. 2014, 10, e1004345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laird, P.W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Stamenova, E.K.; Gu, H.; Gnirke, A.; Meissner, A. Targeted bisulfite sequencing of the dynamic DNA methylome. Epigenet. Chromatin 2016, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Vialou, V.; Feng, J.; Robison, A.J.; Nestler, E.J. Epigenetic mechanisms of depression and antidepressant action. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 59–87. [Google Scholar] [CrossRef] [PubMed]
- Holsboer, F. How can we realize the promise of personalized antidepressant medicines? Nat. Rev. Neurosci. 2008, 9, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Hayashi-Takagi, A.; Vawter, M.P.; Iwamoto, K. Peripheral biomarkers revisited: Integrative profiling of peripheral samples for psychiatric research. Biol. Psychiatry 2014, 75, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Bakulski, K.M.; Halladay, A.; Hu, V.W.; Mill, J.; Fallin, M.D. Epigenetic Research in Neuropsychiatric Disorders: The ‘Tissue Issue’. Curr. Behav. Neurosci. Rep. 2016, 3, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Guintivano, J.; Aryee, M.J.; Kaminsky, Z.A. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 2013, 8, 290–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Farlik, M.; Sheffield, N.C.; Nuzzo, A.; Datlinger, P.; Schönegger, A.; Klughammer, J.; Bock, C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015, 10, 1386–1397. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Baker-Andresen, D.; Ratnu, V.S.; Bredy, T.W. Dynamic DNA methylation: A prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 2013, 36, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeltner, N.; Studer, L. Pluripotent stem cell-based disease modeling: Current hurdles and future promise. Curr. Opin. Cell Biol. 2015, 37, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Brennand, K.; Savas, J.N.; Kim, Y.; Tran, N.; Simone, A.; Hashimoto-Torii, K.; Beaumont, K.G.; Kim, H.J.; Topol, A.; Ladran, I.; et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psychiatry 2015, 20, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.G.; Badsha, F.; Florio, M.; Kanton, S.; Gerber, T.; Wilsch-Bräuninger, M.; Lewitus, E.; Sykes, A.; Hevers, W.; Lancaster, M.; et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 2015, 112, 15672–15677. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Lancaster, M.A.; Castanon, R.; Nery, J.R.; Knoblich, J.A.; Ecker, J.R. Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Rep. 2016, 17, 3369–3384. [Google Scholar] [CrossRef] [PubMed]
- Quadrato, G.; Nguyen, T.; Macosko, E.Z.; Sherwood, J.L.; Yang, S.M.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 2017, 545, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Umezawa, A. DNA methylation dynamics in human induced pluripotent stem cells. Hum. Cell 2016, 29, 97–100. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffmann, A.; Sportelli, V.; Ziller, M.; Spengler, D. Epigenomics of Major Depressive Disorders and Schizophrenia: Early Life Decides. Int. J. Mol. Sci. 2017, 18, 1711. https://doi.org/10.3390/ijms18081711
Hoffmann A, Sportelli V, Ziller M, Spengler D. Epigenomics of Major Depressive Disorders and Schizophrenia: Early Life Decides. International Journal of Molecular Sciences. 2017; 18(8):1711. https://doi.org/10.3390/ijms18081711
Chicago/Turabian StyleHoffmann, Anke, Vincenza Sportelli, Michael Ziller, and Dietmar Spengler. 2017. "Epigenomics of Major Depressive Disorders and Schizophrenia: Early Life Decides" International Journal of Molecular Sciences 18, no. 8: 1711. https://doi.org/10.3390/ijms18081711