Development of a High-Density SNP-Based Linkage Map and Detection of QTL for β-Glucans, Protein Content, Grain Yield per Spike and Heading Time in Durum Wheat
Abstract
:1. Introduction
2. Results
2.1. Field Trait Analysis
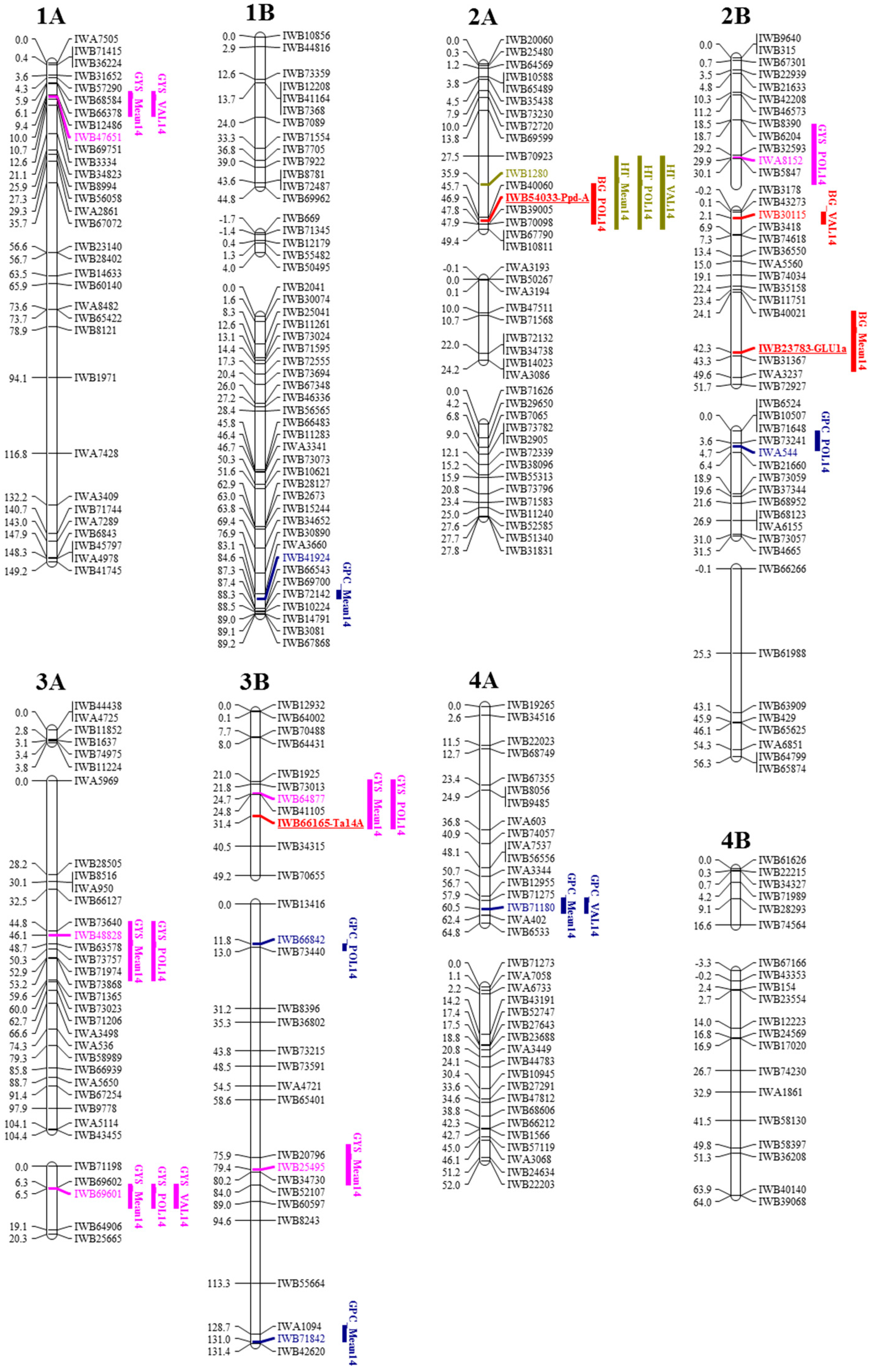
2.2. A Duilio × Avonlea Linkage Map
2.3. Detection of QTL for β-Glucan Contents, Protein Content, Grain Yield per Spike and Heading Time
2.4. Candidate Genes Related to All the Detected QTL
3. Discussion
4. Materials and Methods
4.1. Genetic Materials and Field Experiments
4.2. SNP Genotyping
4.3. Segregation Analysis and Map Construction
4.4. Statistical Analysis, and QTL and Genes Detections
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cobb, J.N.; DeClerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. TAG 2013, 126, 867–887. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.H.; Ph, H.Y.W.; Yang, M.H. Achievement and prospect of good quality breeding in winter cereals. Korean J. Crop Sci. 1998, 43, 11–15. [Google Scholar]
- Blanco, A.; Mangini, G.; Giancaspro, A.; Giove, S.; Colasuonno, P.; Simeone, R.; Signorile, A.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L.; et al. Relationships between grain protein content and grain yield components through quantitative trait locus analyses in a recombinant inbred line population derived from two elite durum wheat cultivars. Mol. Breed. 2012, 30, 79–92. [Google Scholar] [CrossRef]
- Collins, H.M.; Burton, R.A.; Topping, D.L.; Liao, M.-L.; Bacic, A.; Fincher, G.B. Variability in Fine structures of noncellulosic cell wall polysaccharides from cereal grains: Potential importance in human health and nutrition. Cereal Chem. 2010, 87, 272–282. [Google Scholar] [CrossRef]
- Guarda, A.; Rosell, C.M.; Benedito, C.; Galotto, M.J. Different hydrocolloids as bread improvers and antistaling agents. Food Hydrocoll. 2004, 18, 241–247. [Google Scholar] [CrossRef]
- Mohamed, A.; Rayas-Duarte, P.; Xu, J. Hard Red Spring wheat/C-TRIM 20 bread: Formulation, processing and texture analysis. Food Chem. 2008, 107, 516–524. [Google Scholar] [CrossRef]
- Kurek, M.A.; Wyrwisz, J.; Piwińska, M.; Wierzbicka, A. Influence of the wheat flour extraction degree in the quality of bread made with high proportions of β-glucan. Food Sci. Technol. 2015, 35, 273–278. [Google Scholar] [CrossRef]
- Wang, S.W.; Forrest, D.; Allen, K.; Chao, A.; Huang, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphismarray. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Colasuonno, P.; Lozito, M.L.; Marcotuli, I.; Nigro, D.; Giancaspro, A.; Mangini, G.; de Vita, P.; Mastrangelo, A.M.; Pecchioni, N.; Houston, K.; et al. The carotenoid biosynthetic and catabolic genes in wheat and their association with yellow pigments. BMC Genom. 2017, 18, 122–140. [Google Scholar] [CrossRef] [PubMed]
- Marcotuli, I.; Houston, K.; Schwerdt, J.G.; Waugh, R.; Fincher, G.B.; Burton, R.A.; Blanco, A.; Gataleda, A. Genetic diversity and genome wide association study of β-glucan content in tetraploid wheat grains. PLoS ONE 2016, 11, e0152590. [Google Scholar] [CrossRef] [PubMed]
- Marcotuli, I.; Houston, K.; Waugh, R.; Fincher, G.B.; Burton, R.A.; Blanco, A.; Gataleda, A. Genome Wide Association Mapping for Arabinoxylan Content in a Collection of Tetraploid Wheats. PLoS ONE 2015, 10, e0132787. [Google Scholar] [CrossRef] [PubMed]
- Laidò, G.; Mangini, G.; Taranto, F.; Gadaleta, A.; Blanco, A.; Cattivelli, L.; Marone, D.; Mastrangelo, A.M.; Papa, R.; de Vita, R. Genetic diversity and population structure of tetraploid wheats (L.) estimated by SSR, DArT and pedigree data. PLoS ONE 2013, 8, e67280. [Google Scholar] [CrossRef] [PubMed]
- Colasuonno, P.; Gadaleta, A.; Giancaspro, A.; Nigro, D.; Giove, S.; Incerti, O.; Mangini, G.; Signorile, A.; Simeone, R.; Blanco, A. Development of a high-density SNP-based linkage map and detection of yellow pigment content QTLs in durum wheat. Mol. Breed. 2014, 34, 1563–1578. [Google Scholar] [CrossRef]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2014, 110, 8057–8062. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Ricci, A.; Salvi, S.; Milner, S.G.; Noli, E.; Martelli, P.L.; Casadio, R.; Akhunov, E.; Scalabrin, S.; Vendramin, V.; et al. A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 2015, 13, 648–663. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Simeone, R.; Gadaleta, A. Detection of QTL for grain protein content in durum wheat. Theor. Appl. Genet. 2006, 112, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Oury, F.X.; Berard, P.; Brancourt-Hulmel, M.; Depatureaux, C.; Doussinault, G.; Galic, N.; Giraud, A.; Heumez, E.; Lecomte, C.; Pluchard, P.; et al. Yield and grain protein concentration in bread wheat: A review and a study of multi-annual data from a French breeding program. J. Genet. Breed. 2003, 57, 59–68. [Google Scholar]
- Zeng, Z. Precision mapping of quantitative trait loci. Genetics 1994, 136, 1457–1468. [Google Scholar] [PubMed]
- Tabbita, F.; Pearce, S.; Barneix, A.J. Breeding for increased grain protein and micronutrient content in wheat: Ten years of the GPC-B1 gene. J. Cereal Sci. 2017, 73, 183–191. [Google Scholar] [CrossRef]
- Kiseleva, A.A.; Shcherban, A.B.; Leonova, I.N.; Frenkel, Z.; Salina, E.A. Identification of new heading date determinants in wheat 5B chromosome. BMC Plant Biol. 2016, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, A.; Giancaspro, A.; Nigro, D.; Giove, S.L.; Incerti, O.; Simeone, R.; Piarulli, L.; Colasuonno, P.; Valè, G.; Cattivelli, L.; et al. A new genetic and deletion map of wheat chromosome 5A to detect candidate genes for quantitative traits. Mol. Breed. 2014, 34, 1599. [Google Scholar] [CrossRef]
- Russo, M.A.; Ficco, D.B.M.; Laidò, G.; Marone, D.; Papa, R.; Blanco, A.; Gadaleta, A.; De Vita, P.; Mastrangelo, A.M. A dense durum wheat × T. dicoccum linkage map based on SNP markers for the study of seed morphology. Mol. Breed. 2014, 34, 1579–1597. [Google Scholar] [CrossRef]
- Giancaspro, A.; Giove, S.L.; Zito, D.; Blanco, A.; Gadaleta, A. Mapping QTLs for Fusarium head blight resistance in an interspecific wheat population. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sourdille, P.; Cadalen, T.; Guyomarc’h, H.; Snape, J.W.; Perretant, M.R.; Charmet, G.; Boeuf, C.; Bernard, S.; Bernard, M. An update of the Courtot × Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor. Appl. Genet. 2003, 106, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Hanocq, E.; Niarquin, M.; Heumez, E.; Rousset, M.; Le Gouis, J. Detection and mapping of QTL for earliness components in a bread wheat recombinant inbred lines population. Theor. Appl. Genet. 2004, 110, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Peterson, D.M.; Wesenberg, D.M.; Burrup, D.E. β-glucan content and its relationship to agronomic characteristics in elite oat germplasm. Crop Sci. 1995, 35, 965–970. [Google Scholar] [CrossRef]
- Peterson, D.M.; Wesenberg, D.M.; Burrup, D.E.; Erickson, C.A. Relationships among agronomic traits and grain composition in oat genotypes grown in different environments. Crop Sci. 2005, 45, 1249–1255. [Google Scholar] [CrossRef]
- Thomas, W.T.B.; Powell, W.; Swanston, J.S. The effects of major genes on quantitatively varying characters in barley. 4. The GPert and denso loci and quality characters. Heredity 1991, 66, 381–389. [Google Scholar] [CrossRef]
- Eticha, F.; Grausgruber, H.; Berghoffer, E. Multivariate analysis of agronomic and quality traits of hull-less spring barley (Hordeum vulgare L.). J. Plant Breed. Crop Sci. 2010, 2, 81–95. [Google Scholar]
- Prasad, M.; Kumar, N.; Kulwal, P.L.; Roder, M.S.; Balyan, H.S.; Dhaliwal, H.S.; Gupta, P.K. QTL analysis for grain protein content using SSR markers and validation studies using NILs in bread wheat. Theor. Appl. Genet. 2003, 106, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Kunert, A.; Naz, A.A.; Dedeck, O.; Pillen, K.; Leon, J. AB-QTL analysis in winter wheat: I. Synthetic hexaploid wheat (T. turgidum ssp. dicoccoides × T. tauschii) as a source of favourable alleles for milling and baking quality traits. Theor. Appl. Genet. 2007, 115, 683–695. [Google Scholar]
- Huang, X.Q.; Cloutier, S.; Lycar, L.; Radovanovic, N.; Humphreys, D.G.; Noll, J.S.; Somers, D.J.; Brown, P.D. Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor. Appl. Genet. 2006, 113, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Kulwal, P.; Kumar, N.; Kumar, A.; Gupta, R.K.; Balyan, H.S.; Gupta, P.K. Gene networks in hexaploid wheat: Interacting quantitative trait loci for grain protein content. Funct. Integr. Genom. 2005, 5, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Groos, C.; Robert, N.; Bervas, E.; Charmet, G. Genetic analysis of grain protein content, grain yield and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 2003, 106, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Bӧrner, A.; Schumann, E.; Furste, A.; Coster, H.; Leithold, B.; Rӧder, M.S.; Weber, W.E. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 105, 921–936. [Google Scholar]
- Huang, X.Q.; Cӧster, H.; Ganal, M.W.; Rӧder, M.S. Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor. Appl. Genet. 2003, 106, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Q.; Kempf, H.; Ganal, M.W.; Rӧder, M.S. Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Marza, F.; Bai, G.H.; Carver, B.F.; Zhou, W.C. Quantitative trait loci for yield and related traits in the wheat population Ning7840 × Clark. Theor. Appl. Genet. 2005, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- McCartney, C.A.; Somers, D.J.; Humphreys, D.G.; Lukow, O.; Ames, N.; Noll, J.; Cloutier, S.; McCallum, B.D. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × ‘AC Domain’. Genome 2005, 48, 870–883. [Google Scholar] [CrossRef] [PubMed]
- Quarrie, S.A.; Steed, A.; Calestani, C.; Semikhodskii, A.; Lebreton, C.; Chinoy, C.; Steele, N.; Pljevljakusić, D.; Waterman, E.; Weyen, J.; et al. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor. Appl. Genet. 2005, 110, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Kuchel, H.; Williams, K.J.; Langridge, P.; Eagles, H.A.; Jefferies, S.P. Genetic dissection of grain yield in bread wheat: I. QTL analysis. Theor. Appl. Genet. 2007, 115, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Kulwal, P.L.; Balyan, H.S.; Gupta, P.K. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol. Breed. 2007, 19, 163–177. [Google Scholar] [CrossRef]
- McIntyre, C.L.; Mathews, K.L.; Rattey, A.; Chapman, S.C.; Drenth, J.; Ghaderi, M.; Reynolds, M.; Shorter, R. Molecular detection of genomic regions associated with grain yield and yield-related components in an elite bread wheat cross evaluated under irrigated and rainfed conditions. Theor. Appl. Genet. 2010, 120, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.S.; Le Gouis, J.; Leflon, M.; Rong, W.Y.; Laperche, A.; Bran- court-Hulmel, M. Using probe genotypes to dissect QTL × environment interactions for grain yield components in winter wheat. Theor. Appl. Genet. 2010, 121, 1501–1517. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, J.D.; Greene, R.A.; Bermudez-Kandianis, C.E.; La Rota, C.M.; Edwards, H.; Sorrells, S.F.; Dake, T.; Benscher, D.; Kantety, R.; Linkiewicz, A.M.; et al. Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 2004, 168, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Dilbirligi, M.; Erayman, M.; Campbell, B.T.; Randhawa, H.S.; Baenziger, P.S.; Dweikat, I.; Gill, K.S. High-density mapping and comparative analysis of agronomically important traits on wheat chromosome 3A. Genomics 2006, 88, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.T.; Baezinger, P.S.; Gill, K.S.; Eskridge, K.M.; Budak, H.; Erayman, M.; Dweikat, I.; Yen, Y. Identification of QTLs and environmental interactions associated with agronomic traits on chromosome 3A of wheat. Crop Sci. 2003, 43, 1493–1505. [Google Scholar] [CrossRef]
- Basic Local Alignment Search Tool (BLAST). Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 20 June 2016).
- Lombard, V.; Golaconda, R.H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Beales, J.; Turner, A.; Griffiths, S.; Snape, J.W.; Laurie, D.A. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 115, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Scarth, R.; Law, C.N. The control of the day-length response in wheat by the group 2 chromosomes. Z. Pflanzenzüchtung 1984, 92, 140–150. [Google Scholar]
- Bernard, S.M.; BlomMøller, A.L.; Dionisio, G.; Kichey, T.; Jahn, T.P.; Dubois, F.; Baudo, M.; Lopes, M.S.; Tercé-Laforgue, T.; Foyer, C.H.; et al. Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008, 67, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, J.; Ravel, C.; Pageau, K.; Heumez, E.; Dubois, F.; Hirel, B.; Le Gouis, J. A quantitative genetic study for elucidating the contribution of glutamine synthetase; glutamate dehydrogenase and other nitrogen-related physiological traits to the agronomic performance of common wheat. Theor. Appl. Genet. 2009, 119, 645–662. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, A.; Nigro, D.; Giancaspro, A.; Blanco, A. The glutamine synthetase (GS2) genes in relation to grain protein content of durum wheat. Funct. Integr. Genom. 2011, 11, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Bordes, J.; Ravel, C.; Jaubertie, J.P.; Duperrier, B.; Gardet, O.; Balfourier, F. Use of a global wheat core collection for association analysis of flour and dough quality traits. J. Cereal Sci. 2011, 54, 137–147. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Seo, Y.-H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.-H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Lin, W.S.; Chen, R.K.; Chao, Y.Y.; Chin, S.W.; Chen, F.C.; Lee, C.Y. Establishment of a prediction model for the miRNA-based heading date characteristics of rice in the booting stage. Genet. Mol. Res. 2015, 14, 4381–4390. [Google Scholar] [CrossRef] [PubMed]
- Gil-Humanes, J.; Pistón, F.; Martín, A.; Barro, F. Comparative genomic analysis and expression of the APETALA2-like genes from barley, wheat, and barley-wheat amphiploids. BMC Plant Biol. 2009, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, X. Rhythmic expression of wheat TaGI genes introduced by photoperiod and ectopic expression of TaGI1 promoting flowering in transgenic Arabidopsis. Unpublished.
- Wang, C.; Ma, Q.H.; Lin, Z.B.; He, P.; Liu, J.Y. Cloning and characterization of a cDNA encoding 14–3-3 protein with leaf and stem-specific expression from wheat. DNA Seq. 2008, 19, 130–136. [Google Scholar] [CrossRef] [PubMed]
- McCleary, B.V.; Codd, R. Measurement of (1–3),(1–4)-β-d-glucan in barley and oats: A streamlined enzymatic procedure. J. Sci. Food Agric. 1991, 55, 303–312. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- TraitGenetics. Available online: http://www.traitgenetics.com/en/ (accessed on 20 June 2001).
- Akhunov, E.; Nicolet, C.; Dvorak, J. Single nucleotide polymorphism genotyping in polyploid wheat with the Illumina GoldenGate assay. Theor. Appl. Genet. 2009, 119, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. JoinMap, version 4.0; Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma B.V.: Wageningen, The Netherlands, 2006.
- Haldane, J.B.S. The combination of linkage values, and the calculation of distance between linked factors. J. Genet. 1919, 8, 299–309. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information (NCBI). Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 20 June 1988).
- URGI Website. Unité de Recherche Génomique Info. Available online: https://urgi.versailles.inra.fr/ (accessed on 21 September 2015).
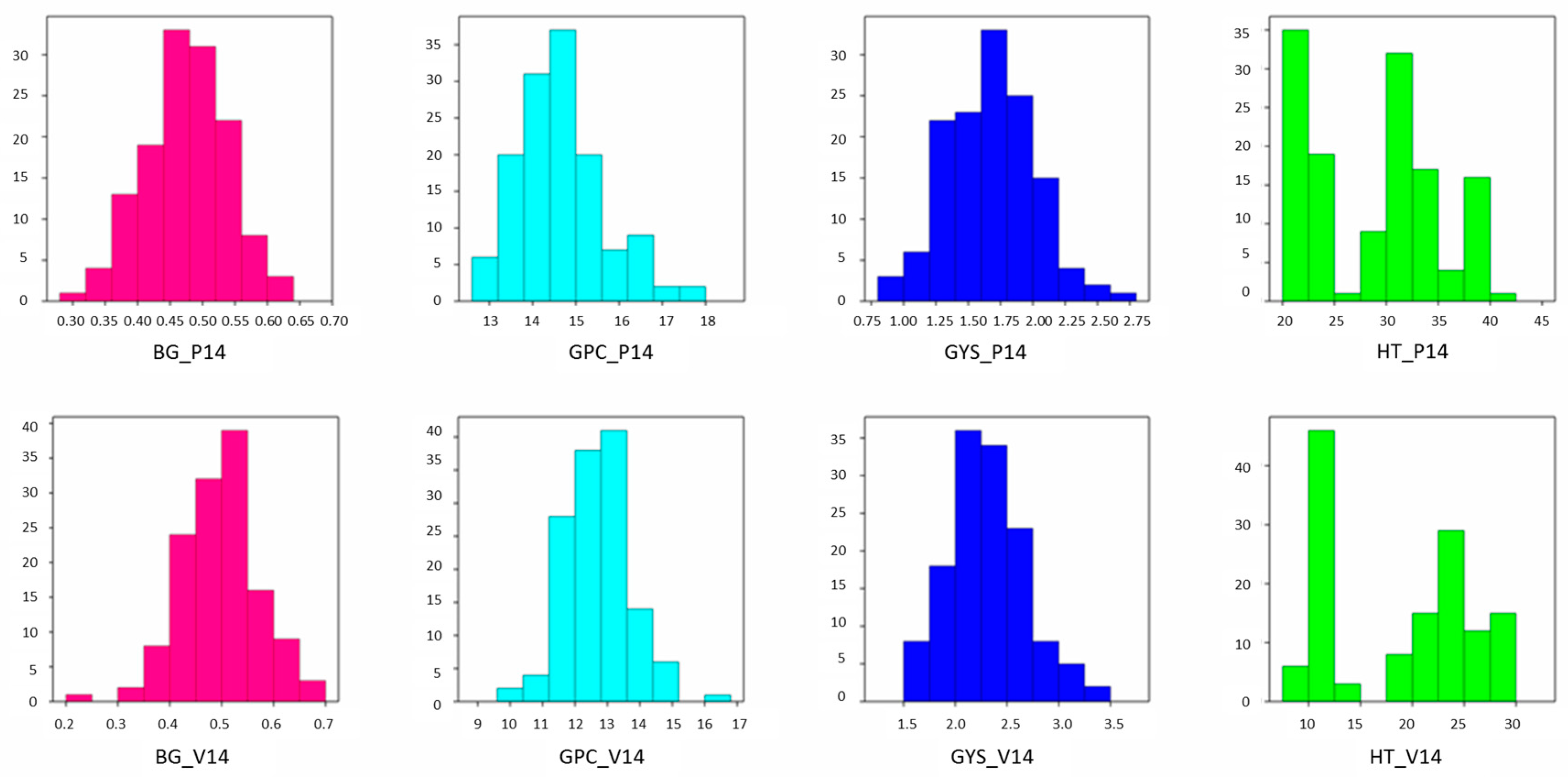

| Trait | Environment | |
|---|---|---|
| Policoro 2014 | Valenzano 2014 | |
| β-glucan (%) | ||
| Duilio | 0.48 | 0.48 |
| Avonlea | 0.56 | 0.59 |
| RIL mean | 0.47 | 0.49 |
| Range | 0.30–0.63 | 0.22–0.68 |
| s2G | 0.004 | 0.004 |
| h2 | 0.80 | 0.82 |
| Grain protein content (%) | ||
| Duilio | 14.3 | 12.0 |
| Avonlea | 13.9 | 12.1 |
| RIL mean | 14.63 | 12.7 |
| Range | 12.6–17.6 | 10.2–16.5 |
| s2G | 0.611 | 0.550 |
| h2 | 0.52 | 0.44 |
| Grain yield per spike (g) | ||
| Duilio | 1.71 | 2.37 |
| Avonlea | 1.93 | 2.55 |
| RIL mean | 1.67 | 2.31 |
| Range | 0.94–2.64 | 1.58–3.47 |
| s2G | 0.068 | 0.091 |
| h2 | 0.47 | 0.53 |
| Heading time (days) | ||
| Duilio | 20 | 12 |
| Avonlea | 38 | 26 |
| RIL mean | 29 | 19 |
| Range | 20–41 | 9–30 |
| s2G | 35.163 | 44.044 |
| h2 | 0.99 | 0.98 |
| Trait | Environment | GPC | GYS | HT |
|---|---|---|---|---|
| BG | Policoro | −0.04 | 0.12 | −0.10 |
| Valenzano | −0.24 ** | −0.06 | −0.02 | |
| GPC | Policoro | −0.36 *** | −0.11 | |
| Valenzano | −0.38 *** | 0.20 ** | ||
| GYS | Policoro | 0.16 | ||
| Valenzano | 0.05 |
| Chromosome | Number of Linkage Groups | Number of SNPs | Chromosome Length (cM) | Number of SNPs/cM |
|---|---|---|---|---|
| 1A | 1 | 302 | 149.2 | 2.0 |
| 1B | 3 | 522 | 137.9 | 3.8 |
| 2A | 3 | 422 | 101.4 | 4.2 |
| 2B | 4 | 492 | 169.6 | 2.9 |
| 3A | 3 | 283 | 128.5 | 2.2 |
| 3B | 2 | 294 | 180.6 | 1.6 |
| 4A | 2 | 273 | 116.8 | 2.3 |
| 4B | 3 | 230 | 80.6 | 2.8 |
| 5A | 2 | 473 | 214.7 | 2.2 |
| 5B | 4 | 454 | 151.2 | 3.0 |
| 6A | 2 | 287 | 143.7 | 2.0 |
| 6B | 2 | 376 | 123.5 | 3.0 |
| 7A | 4 | 500 | 106.9 | 4.6 |
| 7B | 2 | 536 | 157.9 | 3.4 |
| Total genome A | 17 | 2540 | 961.2 | 2.6 |
| Total genome B | 19 | 2904 | 1001.3 | 2.9 |
| Total genomes AB | 36 | 5444 | 1962.5 | 2.7 |
| QTL | Closest Marker | Chrom | QTL Interval | Mean Across Environments | Policoro | Valenzano | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cM | Effect | LOD | R2 | Effect | LOD | R2 | Effect | LOD | R2 | |||
| β-glucan content | ||||||||||||
| QGbg.mgb-2A.1 | IWB1280 | 2A | (35.8–48.0) | −0.03 | 4.5 | 0.14 | −0.02 | 6.1 | 0.19 | - | - | - |
| QGbg.mgb-2B.1 | IWB30115 | 2B | (0.1–3.9) | 0.02 | 4.7 | 0.15 | - | - | - | 0.03 | 6.1 | 0.19 |
| QGbg.mgb-2B.2 | IWB23783 | 2B | (29.9–47.9) | 0.02 | 3.8 | 0.12 | 0.03 | 3.1 | 0.10 | - | - | - |
| Protein content | ||||||||||||
| QGpc.mgb-1B.1 | IWB41924 | 1B | (81.9–84.5) | 0.64 | 3.0 | 0.10 | - | - | - | - | - | - |
| QGpc.mgb-2B.1 | IWA544 | 2B | (0.1–5.9) | - | - | - | −0.53 | 4.1 | 0.13 | - | - | - |
| QGpc.mgb-3B.1 | IWB66842 | 3B | (11.7–14.0) | - | - | - | 0.22 | 4.8 | 0.15 | - | - | - |
| QGpc.mgb-3B.2 | IWB71842 | 3B | (126.0–130.9) | 0.18 | 3.6 | 0.12 | - | - | - | - | - | - |
| QGpc.mgb-4A.1 | IWB71180 | 4A | (57.1–62.0) | −0.19 | 3.7 | 0.12 | - | - | - | - | - | - |
| QGpc.mgb-5A.1 | IWB28350 | 5A | (63.4–80.5) | −0.43 | 4.3 | 0.14 | −0.38 | 5.4 | 0.17 | - | - | - |
| QGpc.mgb-7A.1 | IWB20381 | 7A | (1.9–15.9) | - | - | - | −0.31 | 5.6 | 0.17 | - | - | - |
| QGpc.mgb-7B.1 | IWB71499 | 7B | (53.9–61.9) | 0.27 | 3.6 | 0.12 | 0.56 | 6.3 | 0.29 | 0.36 | 3.3 | 0.12 |
| Grain yield per spike | ||||||||||||
| QGys.mgb-1A.1 | IWB47651 | 1A | (8.3–16.0) | 0.15 | 7.1 | 0.22 | - | - | - | 0.24 | 9.5 | 0.28 |
| QGys.mgb-2B.1 | IWA8152 | 2B | (19.9–37.9) | - | - | - | 0.07 | 3.0 | 0.10 | - | - | - |
| QGys.mgb-3A.1 | IWB69601 | 3A | (5.5–12.5) | 0.09 | 9.3 | 0.28 | 0.10 | 5.3 | 0.17 | 0.09 | 4.7 | 0.15 |
| QGys.mgb-3A.2 | IWB48828 | 3A | (42.0–50.0) | −0.06 | 4.5 | 0.14 | −0.12 | 8.1 | 0.24 | - | - | - |
| QGys.mgb-3B.1 | IWB64877 | 3B | (20.6–28.5) | 0.08 | 3.6 | 0.12 | 0.13 | 4.1 | 0.13 | - | - | - |
| QGys.mgb-3B.2 | IWB25495 | 3B | (72.0–84.0) | −0.06 | 4.1 | 0.13 | - | - | - | - | - | - |
| QGys.mgb-6B.1 | IWB23659 | 6B | (26.0–32.1) | −0.08 | 7.0 | 0.21 | - | - | - | −0.09 | 5.5 | 0.17 |
| Heading time | ||||||||||||
| QHt.mgb-2A.1 | IWB54033 | 2A | (27.4–49.3) | −9.96 | 25.5 | 0.58 | −10.48 | 26.0 | 0.59 | −9.31 | 23.8 | 0.56 |
| Trait | Gene | Enzyme | Contig | Marker ID | SNP | Wheat Map Position | ||
|---|---|---|---|---|---|---|---|---|
| Chrom | D × A Map | Durum Map | ||||||
| β-glucan content | GLU1a | β-glucosidase 1a | contig_2BL_8006334 | IWB23783 | T/C | 2B | 42.3 | 45.9 |
| Grain yield per spike | TaAP2 | APETALA2 | contig_2BS_5227696 | wmc213-2B | - | 2B | - | 31 |
| contig_2BS_5208541 | wmc243-2B | - | 2B | - | 31.1 | |||
| TaGI3 | gigantea 3 | contitg_3AS_200_3324175 | IWB65703 | T/C | 3A | 50.6 | 51.5 | |
| Ta14A | 14-3-3 protein | contig_3B_10411870 | IWB3714 | A/G | 3B | 31.5 | 33.2 | |
| contig_3B_10411871 | IWB11529 | A/G | 3B | 31.6 | 33.5 | |||
| Heading time | Ppd-A1 | Photoperiod sensitivity | contig_2AS_5262553 | IWB54033 | A/C | 2A | 47.8 | 46.2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcotuli, I.; Gadaleta, A.; Mangini, G.; Signorile, A.M.; Zacheo, S.A.; Blanco, A.; Simeone, R.; Colasuonno, P. Development of a High-Density SNP-Based Linkage Map and Detection of QTL for β-Glucans, Protein Content, Grain Yield per Spike and Heading Time in Durum Wheat. Int. J. Mol. Sci. 2017, 18, 1329. https://doi.org/10.3390/ijms18061329
Marcotuli I, Gadaleta A, Mangini G, Signorile AM, Zacheo SA, Blanco A, Simeone R, Colasuonno P. Development of a High-Density SNP-Based Linkage Map and Detection of QTL for β-Glucans, Protein Content, Grain Yield per Spike and Heading Time in Durum Wheat. International Journal of Molecular Sciences. 2017; 18(6):1329. https://doi.org/10.3390/ijms18061329
Chicago/Turabian StyleMarcotuli, Ilaria, Agata Gadaleta, Giacomo Mangini, Antonio Massimo Signorile, Silvana Addolorata Zacheo, Antonio Blanco, Rosanna Simeone, and Pasqualina Colasuonno. 2017. "Development of a High-Density SNP-Based Linkage Map and Detection of QTL for β-Glucans, Protein Content, Grain Yield per Spike and Heading Time in Durum Wheat" International Journal of Molecular Sciences 18, no. 6: 1329. https://doi.org/10.3390/ijms18061329