Identification and Validation of Reference Genes for Seashore Paspalum Response to Abiotic Stresses
Abstract
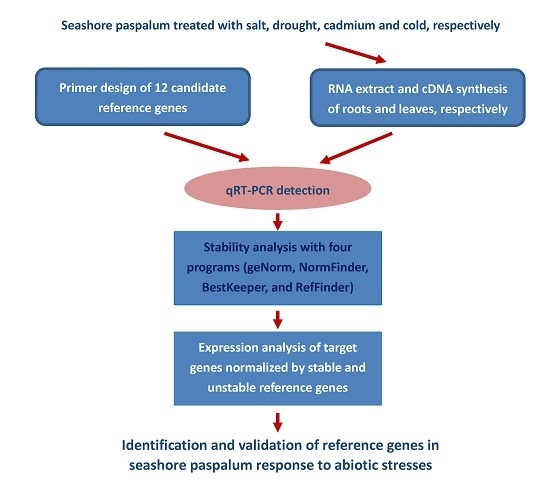
:1. Introduction
2. Results
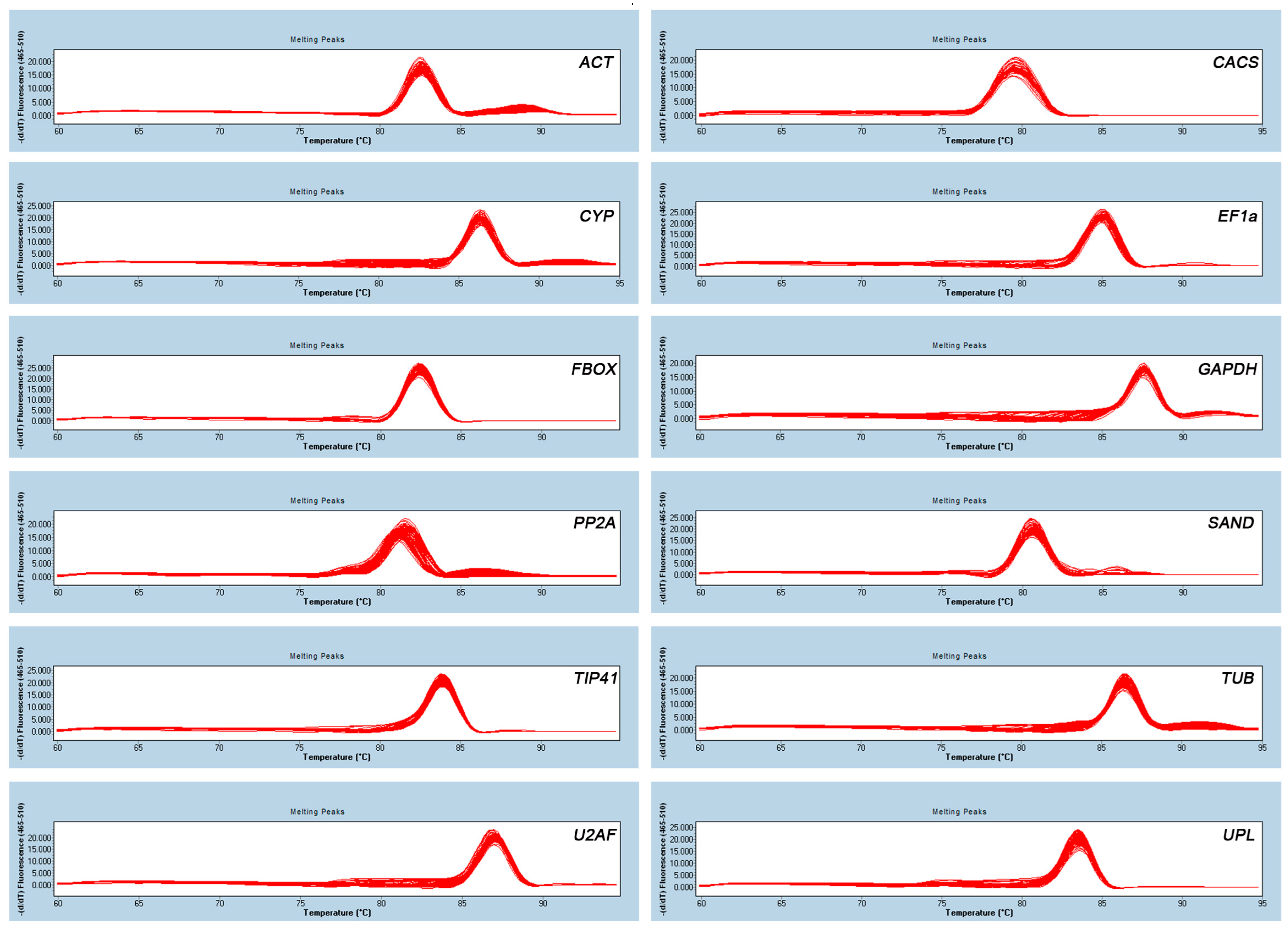
2.1. Identification of PCR Amplicons, Primer Specificity, and Amplification Efficiency of qRT-PCR
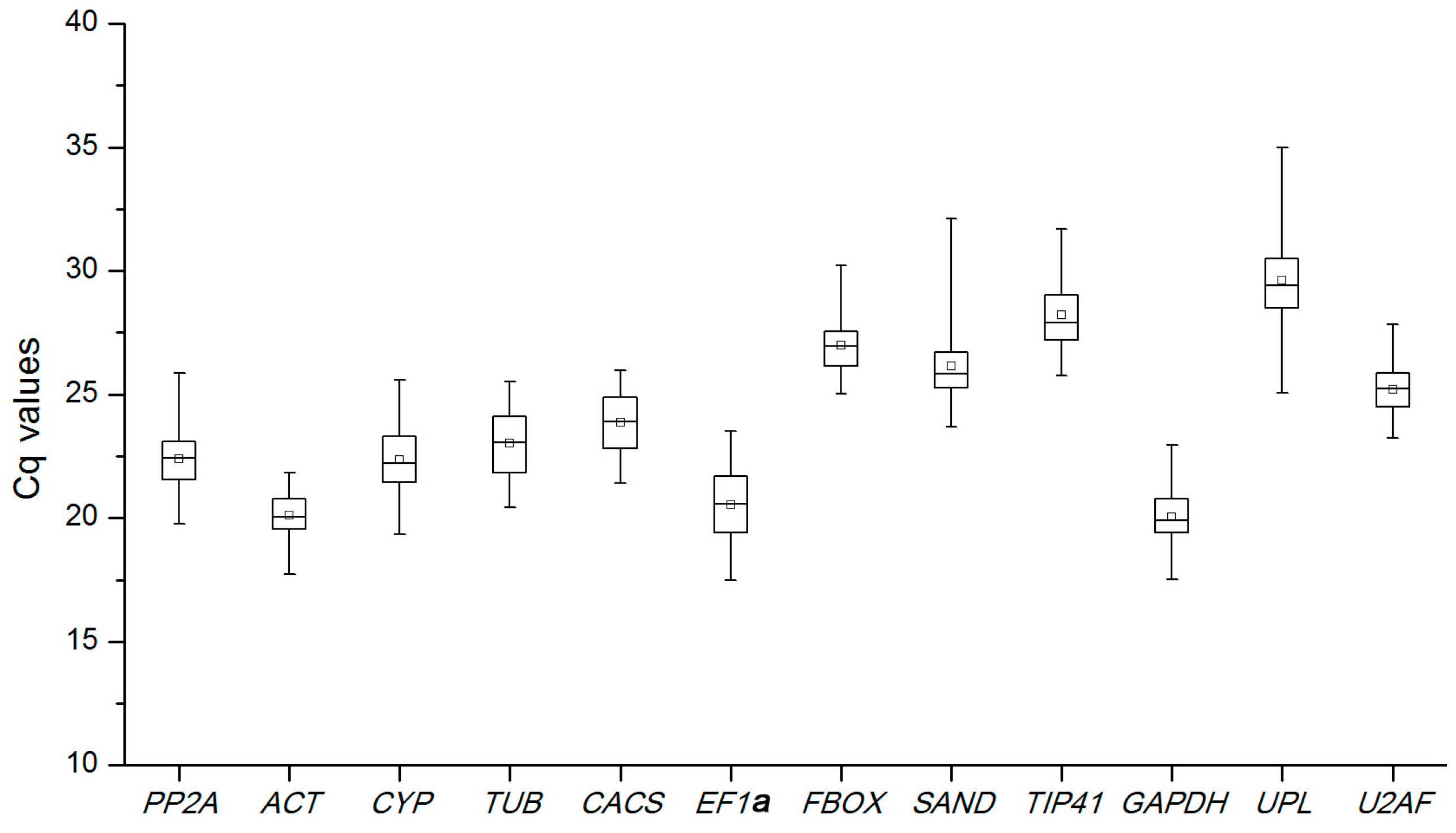
2.2. Expression Levels and Variations of Reference Genes
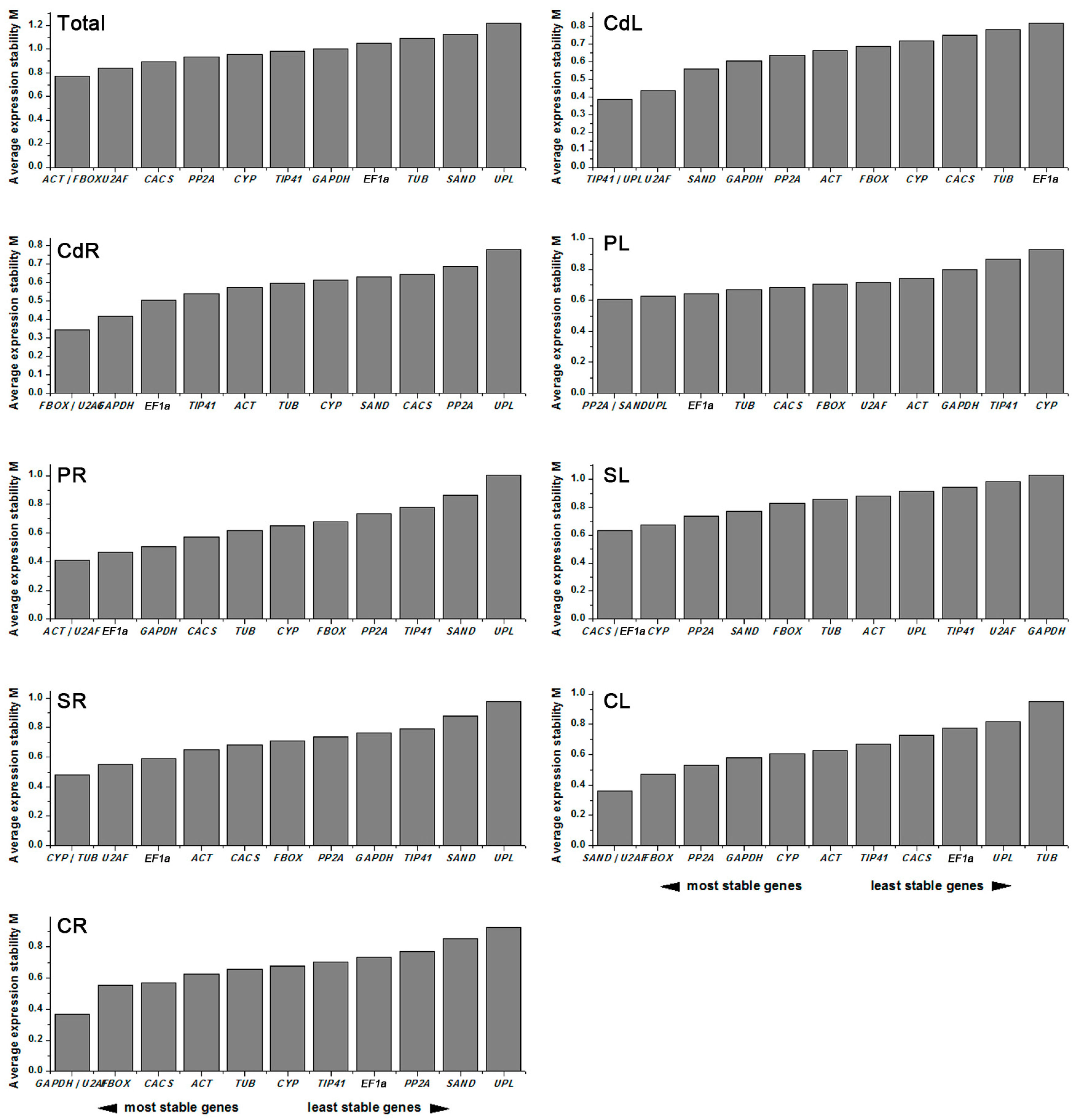
2.3. Stability of Candidate Reference Genes
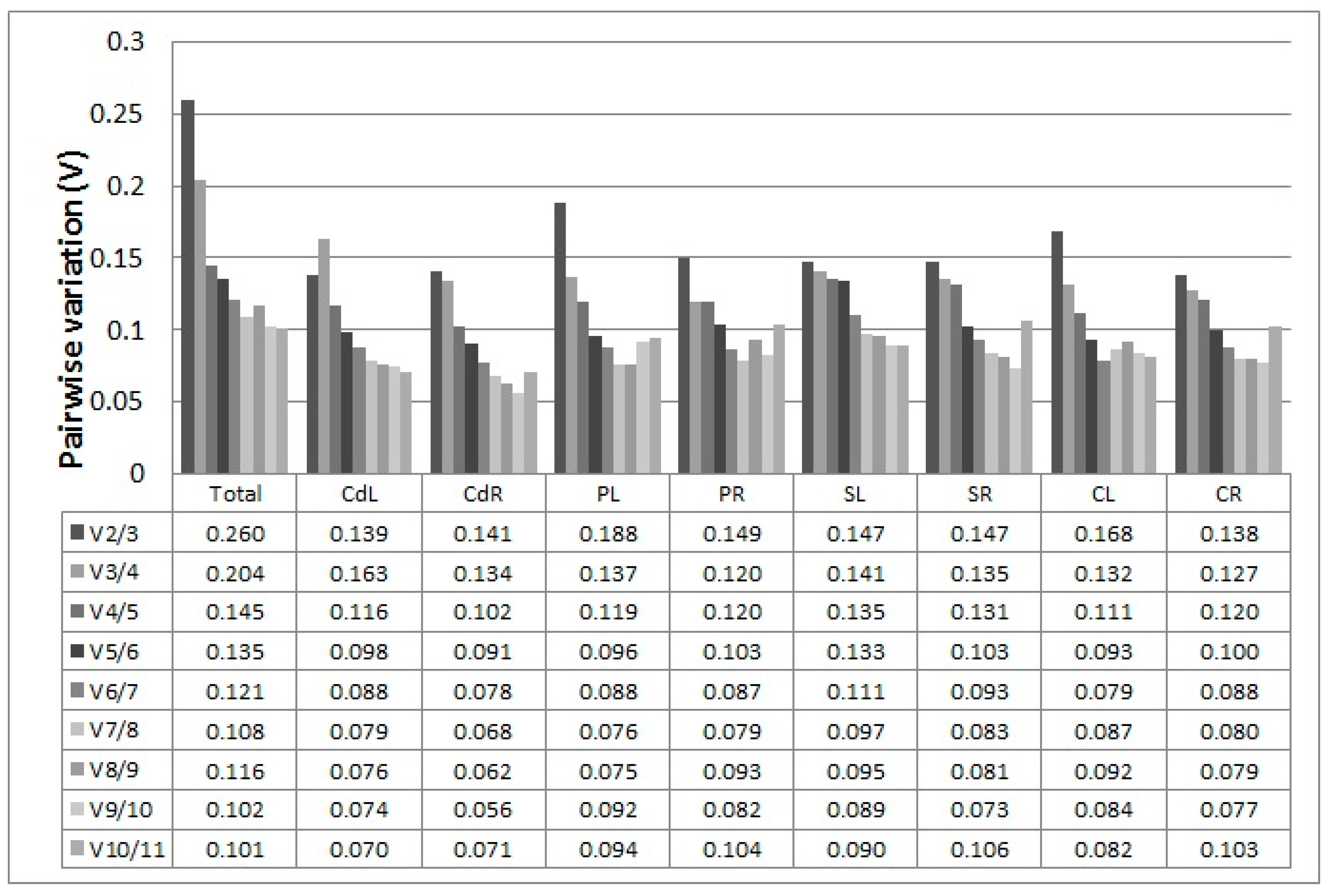
2.3.1. geNorm Analysis
2.3.2. NormFinder Analysis
2.3.3. BestKeeper Analysis
2.3.4. RefFinder Analysis
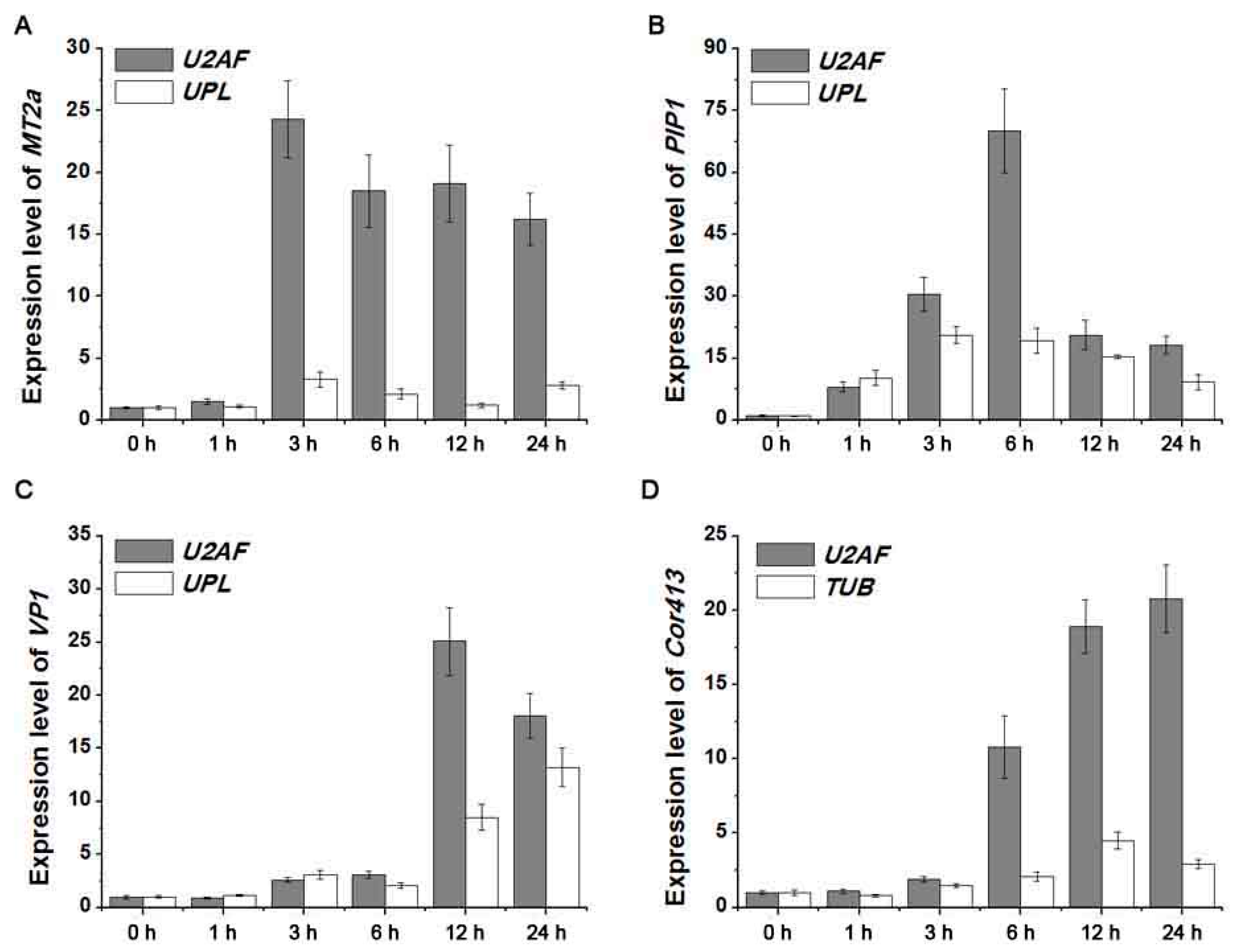
2.4. Detection of Four Target Gene Expression Levels Normalized by Screened Reference Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
4.2. Total RNA Isolation and cDNA Synthesis
4.3. Selection of Reference Genes and Primer Design
4.4. qRT-PCR Analysis
4.5. Stability Analysis
4.6. Validation of Reference Genes by Expression Analysis of Four Stress-Related Genes under Abiotic Stresses
4.7. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, K.C.; Fernandes de Melo, D.; Morais, V.; Vasconcelos, I.; Costa, J. Selection of suitable soybean EF1α genes as internal controls for real-time PCR analyses of tissues during plant development and under stress conditions. Plant Cell. Rep. 2014, 33, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Shi, Y.; Ma, H. Selection of Candidate Reference Genes for Gene Expression Analysis in Kentucky Bluegrass (Poa pratensis L.) under Abiotic Stress. Front. Plant Sci. 2017, 8, 193. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yin, J.; Li, G.; Qi, L.; Yang, F.; Wang, R. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol. Biol. Rep. 2014, 41, 2325–2334. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.L.; Qu, Y.L.; Shan, X.Y.; Wan, Y.L. Screening and Validation of Housekeeping Genes of the Root and Cotyledon of Cunninghamia lanceolata under Abiotic Stresses by Using Quantitative Real-Time PCR. Int. J. Mol. Sci. 2016, 17, 1198. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.J.; Tao, Y.B.; Chen, M.S.; Fu, Q.T.; Li, C.Q.; Dong, Y.L.; Wang, X.L.; He, H.Y.; Xu, Z.F. Selection of Reliable Reference Genes for Gene Expression Studies of a Promising Oilseed Crop, Plukenetia volubilis, by Real-Time Quantitative PCR. Int. J. Mol. Sci. 2015, 16, 12513–12530. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tan, Z.; Hu, B.; Yang, Z.; Xu, B.; Zhuang, L.; Huang, B. Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses. Physiol. Plant 2015, 155, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, X.; Chen, S.; Zheng, L.; He, X.; Liu, M.; Qiao, G.; Wang, Y.; Zhuo, R. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci. Rep. 2017, 7, 40290. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, N.V.; Logacheva, M.D.; Penin, A.A. Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS ONE 2011, 6, e19434. [Google Scholar] [CrossRef] [PubMed]
- Marum, L.; Miguel, A.; Ricardo, C.P.; Miguel, C. Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS ONE 2012, 7, e35113. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Li, W.; Han, S.; Yang, W.; Qi, L. Reference Gene Selection for Quantitative Real-time PCR Normalization in Caragana intermedia under Different Abiotic Stress Conditions. PLoS ONE 2013, 8, e53196. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, J.; Eattock, N.; Van Deynze, A.; Blumwald, E. Selection and Validation of Reference Genes for Gene Expression Analysis in Switchgrass (Panicum virgatum) Using Quantitative Real-Time RT-PCR. PLoS ONE 2014, 9, e91474. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, C.; Tan, Z.; Liu, J.; Zhuang, L.; Yang, Z.; Huang, B. Functional Identification and Characterization of Genes Cloned from Halophyte Seashore Paspalum Conferring Salinity and Cadmium Tolerance. Front. Plant Sci. 2016, 7, 102. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yan, H.; Jiang, X.; Yin, G.; Zhang, X.; Qi, X.; Zhang, Y.; Yan, Y.; Ma, X.; Peng, Y. Identification of candidate reference genes in perennial ryegrass for quantitative RT-PCR under various abiotic stress conditions. PLoS ONE 2014, 9, e93724. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, B.; Tan, Z.; Liu, J.; Yang, Z.; Li, Z.; Huang, B. Selection of reference genes for quantitative real-time PCR normalization in creeping bentgrass involved in four abiotic stresses. Plant Cell Rep. 2015, 34, 1825–1834. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, Y.; Hu, B.; Tan, Z.; Huang, B. Identification and Validation of Reference Genes for Quantification of Target Gene Expression with Quantitative Real-time PCR for Tall Fescue under Four Abiotic Stresses. PLoS ONE 2015, 10, e0119569. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.C.; Liu, Q.H.; Dong, H.Y.; Zhou, Z.C.; Hao, Y.P.; Chen, X.L.; Xu, L.Y. Selection of Reference Genes for Real-Time Quantitative PCR in Pinus massoniana Post Nematode Inoculation. PLoS ONE 2016, 11, e0147224. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Han, X.; Chen, Y.; Wu, Q.; Wang, Y. Identification of appropriate reference genes for normalizing transcript expression by quantitative real-time PCR in Litsea cubeba. Mol. Genet. Genom. 2013, 288, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, Y.; Fang, H.; Shi, H.; Chen, K.; Zhang, Z.; Tan, X. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol. Genet. Genom. 2014, 289, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.M.; Liu, Y.P.; Ma, X.; Shuai, Q.; Gai, J.Y.; Li, Y. Evaluation of Reference Genes for Normalization of Gene Expression Using Quantitative RT-PCR under Aluminum, Cadmium, and Heat Stresses in Soybean. PLoS ONE 2017, 12, e0168965. [Google Scholar] [CrossRef] [PubMed]
- He, Y.H.; Yan, H.L.; Hua, W.P.; Huang, Y.Y.; Wang, Z.Z. Selection and Validation of Reference Genes for Quantitative Real-time PCR in Gentiana macrophylla. Front. Plant Sci. 2016, 7, 945. [Google Scholar] [CrossRef] [PubMed]
- Kundu, A.; Patel, A.; Pal, A. Defining reference genes for qPCR normalization to study biotic and abiotic stress responses in Vigna mungo. Plant Cell Rep. 2013, 32, 1647–1658. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.H.; Niu, H.W.; Liu, C.J.; Zhang, J.; Hou, C.Y.; Wang, D.M. Expression Stabilities of Candidate Reference Genes for RT-qPCR under Different Stress Conditions in Soybean. PLoS ONE 2013, 8, e75271. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Chen, J.; Tian, Q.; Wang, S.; Xia, X.; Yin, W. Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol. Plant 2014, 152, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qian, Y.Q.; Han, L.; Liu, J.X.; Sun, Z.Y. Identification of suitable reference genes in buffalo grass for accurate transcript normalization under various abiotic stress conditions. Gene 2014, 547, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper−Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, J.; Chang, Q.; Gu, C.; Song, A.; Chen, S.; Dong, B.; Chen, F. Cold acclimation induces freezing tolerance via antioxidative enzymes, proline metabolism and gene expression changes in two chrysanthemum species. Mol. Biol. Rep. 2014, 41, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, L.; Zong, J.; Chen, J.; Guo, H.; Guo, A.; Liu, J. Heterologous expression of the halophyte Zoysia matrella H+-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 91, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Hu, W.; Deng, X.; Ma, Z.; Chen, L.; Huang, C.; Wang, C.; Wang, J.; He, Y.; Yang, G.; He, G. Overexpression of the wheat aquaporin gene, TaAQP7, enhances drought tolerance in transgenic tobacco. PLoS ONE 2012, 7, e52439. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.S.; Liu, L.Q.; Zhao, Y.H.; Deng, Y.M.; Zhu, X.D.; Huang, S.Z. Overexpression of Iris lactea var. chinensis metallothionein llMT2a enhances cadmium tolerance in Arabidopsis thaliana. Ecotoxicol. Environ. Safe 2014, 105, 22–28. [Google Scholar] [CrossRef] [PubMed]
| Gene Symbol | Gene Name | GenBank Accession | Arabidopsis Homolog Locus | 5′-Primer Sequences (Forward/Reverse)-3′ | Amplicon Length (bp) |
|---|---|---|---|---|---|
| EF1a | Elongation factor 1a | KU049721 | AT5G60390 | GCGGACTGTGCTGTGCTTATC/AGTGGTGGCATCCATCTTGTT | 153 |
| CACS | Clathrin adaptor complex subunit | KX268090 | AT5G46630 | CACTGTCGAGTGGGTTCGCTAC/GCCGATGAATTTTACTTGTTGC | 109 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | KX268091 | AT1G13440 | GTCGCATGGTACGACAACGAGT/ACGGAAAACAAAAGGCAACTCA | 221 |
| TIP41 | TIP41-like family protein | KX268092 | AT4G34270 | TGATGAGATTGAGGGATACTCG/TACAGACGGTGGTCACCTTTGG | 244 |
| SAND | SAND family protein | KX268093 | AT2G28390 | CGGGGATTATGTTCTATTTTGC/TTATGGTACTGCCTGTGTCGGT | 266 |
| ACT | Actin 7 | KX268094 | AT5G09810 | CTTCTCTCAGCACTTTCCAACA/AAACATAACCTGCAATCTCTCC | 162 |
| TUB | Alpha Tubulin | KX268095 | AT5G19780 | GTCGGTGAGGGTATGGAGGAAG/ATGGAAACACACAGCAGCAGTT | 237 |
| PP2A | Protein phosphatase 2A | KX268096 | AT1G13320 | TAAGGTACTACGCAAACCAAGC/CAACACAATACATACACAGCACACA | 289 |
| FBOX | F-box/kelch-repeat protein | KX268097 | AT5G15710 | GTGCTAGCCAGCTCTGCAATAG/ACACATCCGACATCAACGATTC | 184 |
| UPL | E3 ubiquitin protein ligase | KX268098 | AT3G53090 | TACTTGGATTCAAATACCTACAGCC/TTAGAACCCCAGAAACACCGCT | 250 |
| CYP | Cyclophilin | KX268099 | AT2G29960 | CTGGAAGAGATACAAACGGATC/GCCACTAATGACAGTTATAGAACG | 275 |
| U2AF | Splicing factor U2af | KX268100 | AT5G42820 | AGGAGCCCAGTCAGGGAAA/CACGCAGAATAGCAACTCAAAT | 190 |
| MT2a | Metallothionein2a | KX268101 | AT3G09390 | CAGACTCTCGTCATGGGCGT/TCTCATCGGATCAGGTAGCA | 247 |
| VP1 | vacuolar H+-pyrophosphatase 1 | KX268102 | AT1G15690 | GTCCCTCAACATCCTCATCAAG/TAAGTCTAAGGTAACGCCTCCA | 281 |
| PIP1 | plasma membrane intrinsic protein 1 | KX268103 | AT4G00430 | AGGGCCATCCCGTTCAAGAG/ATAACAGCGGCGGCATATTA | 239 |
| Cor413 | cold-regulated 413 plasma membrane protein | KX268104 | AT3G50830 | TCAGGAACGCCTTCAGGAAG/GGATGGCAGAGGAGCACACT | 134 |
| Gene | CdL | CdR | PL | PR | SL | SR | CL | CR |
|---|---|---|---|---|---|---|---|---|
| ACT | 1.93 ± 0.01 | 1.96 ± 0.03 | 1.95 ± 0.02 | 1.93 ± 0.02 | 1.97 ± 0.02 | 1.92 ± 0.02 | 1.95 ± 0.03 | 1.94 ± 0.01 |
| CACS | 1.94 ± 0.02 | 1.97 ± 0.02 | 1.93 ± 0.02 | 1.92 ± 0.02 | 1.96 ± 0.02 | 1.96 ± 0.01 | 1.92 ± 0.02 | 1.97 ± 0.02 |
| EF1α | 1.98 ± 0.01 | 1.93 ± 0.01 | 1.96 ± 0.03 | 1.93 ± 0.01 | 1.94 ± 0.04 | 1.94 ± 0.02 | 1.96 ± 0.02 | 1.95 ± 0.03 |
| FBOX | 1.93 ± 0.02 | 1.94 ± 0.03 | 1.97 ± 0.03 | 1.96 ± 0.02 | 1.95 ± 0.02 | 1.93 ± 0.02 | 1.94 ± 0.02 | 1.93 ± 0.02 |
| GADPH | 1.91 ± 0.02 | 1.95 ± 0.02 | 1.93 ± 0.02 | 1.96 ± 0.02 | 1.92 ± 0.02 | 1.94 ± 0.03 | 1.91 ± 0.02 | 1.97 ± 0.02 |
| UPL | 1.93 ± 0.02 | 1.95 ± 0.02 | 1.95 ± 0.03 | 1.97 ± 0.02 | 1.95 ± 0.03 | 1.92 ± 0.02 | 1.94 ± 0.03 | 1.93 ± 0.03 |
| SAND | 1.94 ± 0.02 | 1.95 ± 0.02 | 1.94 ± 0.01 | 1.96 ± 0.03 | 1.93 ± 0.01 | 1.95 ± 0.02 | 1.94 ± 0.02 | 1.96 ± 0.01 |
| TUB | 1.96 ± 0.01 | 1.95 ± 0.01 | 1.94 ± 0.02 | 1.97 ± 0.02 | 1.94 ± 0.03 | 1.96 ± 0.03 | 1.95 ± 0.03 | 1.97 ± 0.02 |
| TIP41 | 1.96 ± 0.02 | 1.91 ± 0.02 | 1.90 ± 0.02 | 1.91 ± 0.02 | 1.95 ± 0.02 | 1.90 ± 0.03 | 1.93 ± 0.03 | 1.91 ± 0.02 |
| CYP | 1.93 ± 0.02 | 1.94 ± 0.01 | 1.94 ± 0.01 | 1.96 ± 0.03 | 1.93 ± 0.01 | 1.95 ± 0.02 | 1.94 ± 0.02 | 1.96 ± 0.01 |
| PP2A | 1.95 ± 0.02 | 1.96 ± 0.02 | 1.95 ± 0.02 | 1.96 ± 0.02 | 1.93 ± 0.03 | 1.97 ± 0.03 | 1.89 ± 0.03 | 1.96 ± 0.02 |
| U2AF | 1.96 ± 0.01 | 1.93 ± 0.01 | 1.89 ± 0.02 | 1.92 ± 0.02 | 1.94 ± 0.02 | 1.91 ± 0.03 | 1.91 ± 0.02 | 1.92 ± 0.01 |
| Total | Stability | CdL | Stability | CdR | Stability | PL | Stability | PR | Stability | SL | Stability | SR | Stability | CL | Stability | CR | Stability |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FBOX | 0.519 | U2AF | 0.374 | U2AF | 0.223 | UPL | 0.391 | U2AF | 0.392 | SAND | 0.461 | U2AF | 0.383 | U2AF | 0.326 | U2AF | 0.294 |
| ACT | 0.604 | GAPDH | 0.444 | FBOX | 0.37 | PP2A | 0.402 | GAPDH | 0.397 | CACS | 0.54 | FBOX | 0.455 | PP2A | 0.392 | TIP41 | 0.458 |
| U2AF | 0.67 | ACT | 0.471 | TIP41 | 0.376 | EF1α | 0.466 | EF1α | 0.522 | TUB | 0.594 | CYP | 0.512 | FBOX | 0.443 | GAPDH | 0.465 |
| PP2A | 0.673 | UPL | 0.506 | EF1α | 0.441 | SAND | 0.474 | FBOX | 0.535 | PP2A | 0.618 | TUB | 0.519 | GAPDH | 0.48 | CYP | 0.525 |
| TIP41 | 0.703 | PP2A | 0.529 | GAPDH | 0.474 | TUB | 0.563 | TIP41 | 0.55 | EF1α | 0.669 | TIP41 | 0.553 | CYP | 0.529 | TUB | 0.528 |
| CYP | 0.773 | SAND | 0.53 | CYP | 0.478 | FBOX | 0.583 | ACT | 0.592 | FBOX | 0.674 | CACS | 0.562 | SAND | 0.535 | FBOX | 0.555 |
| GAPDH | 0.778 | TIP41 | 0.594 | TUB | 0.484 | CACS | 0.618 | CYP | 0.687 | CYP | 0.738 | GAPDH | 0.656 | ACT | 0.577 | ACT | 0.555 |
| CACS | 0.811 | CYP | 0.606 | ACT | 0.519 | ACT | 0.638 | CACS | 0.687 | ACT | 0.755 | EF1α | 0.657 | TIP41 | 0.651 | CACS | 0.589 |
| SAND | 0.963 | FBOX | 0.619 | SAND | 0.527 | U2AF | 0.724 | TUB | 0.689 | UPL | 0.775 | ACT | 0.681 | EF1α | 0.755 | PP2A | 0.694 |
| TUB | 1.073 | CACS | 0.689 | CACS | 0.553 | GAPDH | 0.857 | PP2A | 0.73 | TIP41 | 0.9 | PP2A | 0.769 | CACS | 0.776 | EF1α | 0.869 |
| EF1α | 1.131 | TUB | 0.747 | PP2A | 0.815 | TIP41 | 0.953 | SAND | 0.999 | U2AF | 0.948 | SAND | 1.059 | UPL | 0.886 | SAND | 1.036 |
| UPL | 1.496 | EF1α | 0.833 | UPL | 1.163 | CYP | 1.09 | UPL | 1.612 | GAPDH | 1.042 | UPL | 1.355 | TUB | 1.497 | UPL | 1.154 |
| Rank | Total | CV ± SD | CdL | CV ± SD | CdR | CV ± SD | PL | CV ± SD | PR | CV ± SD | SL | CV ± SD | SR | CV ± SD | CL | CV ± SD | CR | CV ± SD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | FBOX | 2.95 ± 0.80 | FBOX | 1.61 ± 0.44 | FBOX | 1.36 ± 0.35 | UPL | 1.42 ± 0.41 | U2AF | 2.62 ± 0.67 | PP2A | 2.43 ± 0.54 | U2AF | 2.28 ± 0.55 | FBOX | 1.89 ± 0.52 | FBOX | 1.09 ± 0.28 |
| 2 | U2AF | 3.15 ± 0.79 | U2AF | 1.73 ± 0.44 | U2AF | 1.58 ± 0.38 | TUB | 1.51 ± 0.36 | FBOX | 3.67 ± 1.03 | ACT | 2.62 ± 0.53 | ACT | 2.44 ± 0.47 | CACS | 1.92 ± 0.48 | CACS | 1.62 ± 0.37 |
| 3 | ACT | 3.39 ± 0.68 | UPL | 1.85 ± 0.54 | TUB | 1.96 ± 0.42 | FBOX | 1.52 ± 0.41 | CACS | 3.75 ± 0.91 | CYP | 3.63 ± 0.81 | PP2A | 2.49 ± 0.53 | U2AF | 1.95 ± 0.51 | EF1α | 1.88 ± 0.35 |
| 4 | TIP41 | 3.67 ± 1.04 | CACS | 1.89 ± 0.47 | GAPDH | 2.02 ± 0.39 | EF1α | 1.86 ± 0.41 | ACT | 3.79 ± 0.79 | TUB | 2.00 ± 0.48 | FBOX | 2.54 ± 0.67 | PP2A | 2.15 ± 0.50 | TUB | 2.13 ± 0.46 |
| 5 | SAND | 3.77 ± 0.99 | SAND | 1.92 ± 0.50 | TIP41 | 2.04 ± 0.56 | SAND | 1.92 ± 0.49 | TUB | 3.81 ± 0.89 | CACS | 2.84 ± 0.69 | CACS | 2.56 ± 0.58 | TIP41 | 2.32 ± 0.67 | U2AF | 2.32 ± 0.58 |
| 6 | UPL | 4.10 ± 1.22 | ACT | 1.97 ± 0.40 | CACS | 2019 ± 0.49 | U2AF | 1.95 ± 0.49 | TIP41 | 4.02 ± 1.17 | EF1α | 3.46 ± 0.74 | TUB | 2.75 ± 0.61 | CYP | 2.33 ± 0.55 | ACT | 2.47 ± 0.48 |
| 7 | GAPDH | 4.18 ± 0.84 | GAPDH | 2.02 ± 0.40 | ACT | 2.44 ± 0.47 | CACS | 2.04 ± 0.49 | EF1α | 4.17 ± 0.85 | FBOX | 1.55 ± 0.42 | EF1α | 2.75 ± 0.53 | EF1α | 2.47 ± 0.54 | GAPDH | 2.78 ± 0.56 |
| 8 | PP2A | 4.32 ± 0.97 | PP2A | 2.04 ± 0.46 | PP2A | 2.46 ± 0.52 | PP2A | 2.10 ± 0.47 | GAPDH | 4.20 ± 0.90 | SAND | 1.80 ± 0.47 | CYP | 2.86 ± 0.60 | ACT | 2.56 ± 0.52 | TIP41 | 2.85 ± 0.79 |
| 9 | CACS | 4.50 ± 1.07 | CYP | 2.17 ± 0.49 | EF1α | 2.54 ± 0.49 | ACT | 2.36 ± 0.48 | CYP | 4.83 ± 1.12 | TIP41 | 3.45 ± 0.96 | TIP41 | 3.44 ± 0.95 | SAND | 2.58 ± 0.68 | CYP | 3.13 ± 0.67 |
| 10 | TUB | 4.70 ± 1.08 | TIP41 | 2.54 ± 0.72 | CYP | 2.60 ± 0.54 | GAPDH | 3.41 ± 0.69 | PP2A | 4.89 ± 1.16 | GAPDH | 4.66 ± 0.92 | SAND | 3.83 ± 0.99 | GAPDH | 2.96 ± 0.59 | UPL | 3.35 ± 1.01 |
| 11 | CYP | 4.74 ± 1.06 | TUB | 2.7 ± 0.66 | SAND | 2.65 ± 0.67 | TIP41 | 3.58 ± 1.01 | UPL | 5.33 ± 1.67 | UPL | 2.39 ± 0.68 | GAPDH | 4.03 ± 0.79 | UPL | 3.09 ± 0.90 | PP2A | 3.63 ± 0.81 |
| 12 | EF1α | 5.74 ± 1.18 | EF1α | 3.22 ± 0.69 | UPL | 3.04 ± 0.89 | CYP | 3.89 ± 0.90 | SAND | 5.42 ± 1.49 | U2AF | 2.89 ± 0.75 | UPL | 4.08 ± 1.22 | TUB | 4.21 ± 0.97 | SAND | 4.47 ± 1.17 |
| Experimental Treatments | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | CdL | CdR | PL | PR | SL | SR | CL | CR | |||||||||
| Most | Least | Most | Least | Most | Least | Most | Least | Most | Least | Most | Least | Most | Least | Most | Least | Most | Least |
| FBOX | UPL | U2AF | EF1a | U2AF | UPL | UPL | CYP | U2AF | UPL | SAND | GAPDH | U2AF | UPL | U2AF | TUB | U2AF | UPL |
| ACT | GAPDH | FBOX | PP2A | ACT | CACS | CYP | PP2A | GAPDH | |||||||||
| U2AF | EF1a | FBOX | |||||||||||||||
| PP2A | |||||||||||||||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Liu, J.; Xu, L.; Lai, H.; Chen, Y.; Yang, Z.; Huang, B. Identification and Validation of Reference Genes for Seashore Paspalum Response to Abiotic Stresses. Int. J. Mol. Sci. 2017, 18, 1322. https://doi.org/10.3390/ijms18061322
Liu Y, Liu J, Xu L, Lai H, Chen Y, Yang Z, Huang B. Identification and Validation of Reference Genes for Seashore Paspalum Response to Abiotic Stresses. International Journal of Molecular Sciences. 2017; 18(6):1322. https://doi.org/10.3390/ijms18061322
Chicago/Turabian StyleLiu, Yu, Jun Liu, Lei Xu, Hui Lai, Yu Chen, Zhimin Yang, and Bingru Huang. 2017. "Identification and Validation of Reference Genes for Seashore Paspalum Response to Abiotic Stresses" International Journal of Molecular Sciences 18, no. 6: 1322. https://doi.org/10.3390/ijms18061322