Nrf2 Inhibits Periodontal Ligament Stem Cell Apoptosis under Excessive Oxidative Stress
Abstract
:1. Introduction
2. Results
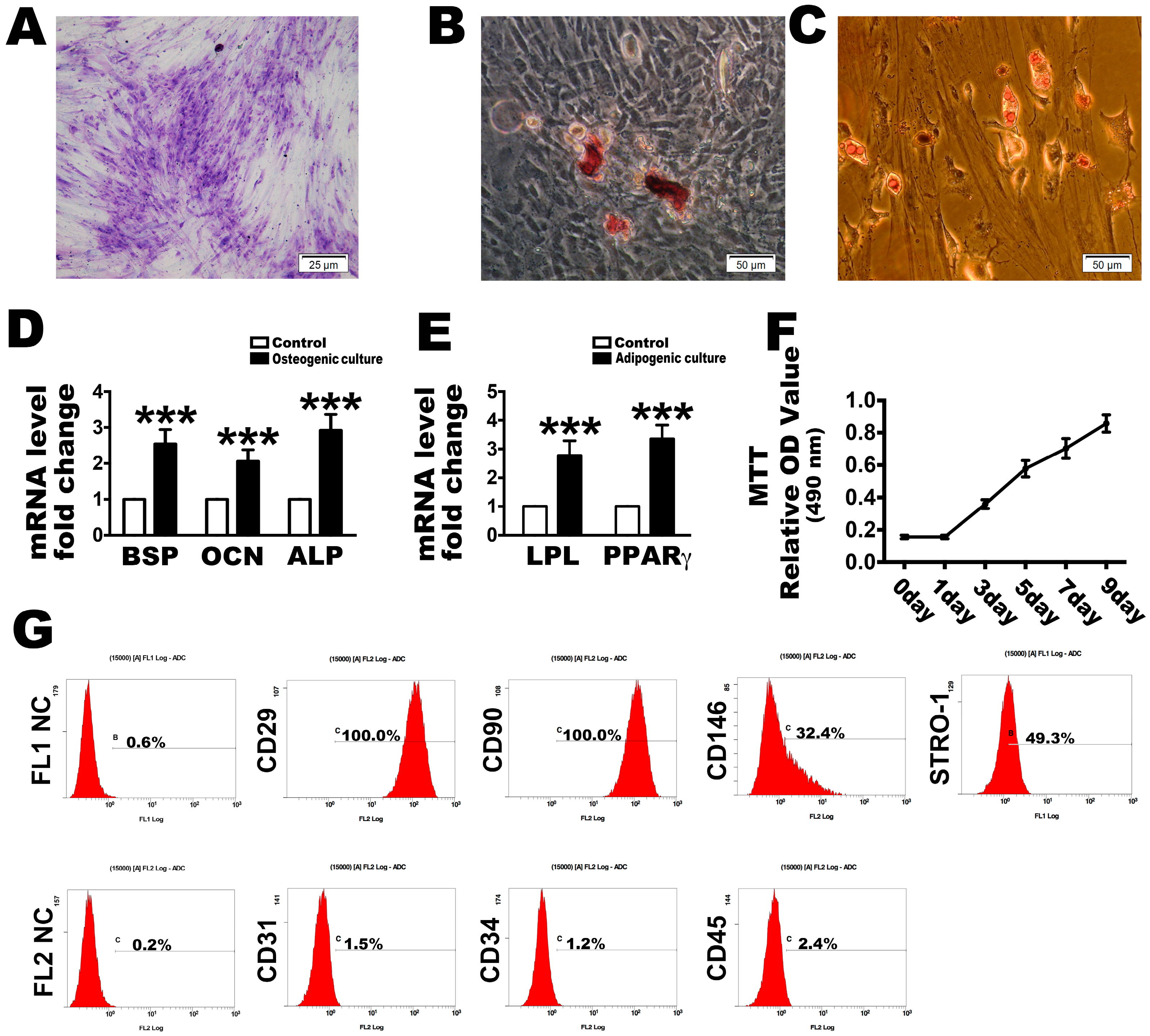
2.1. Culture and Identification of PDLSCs
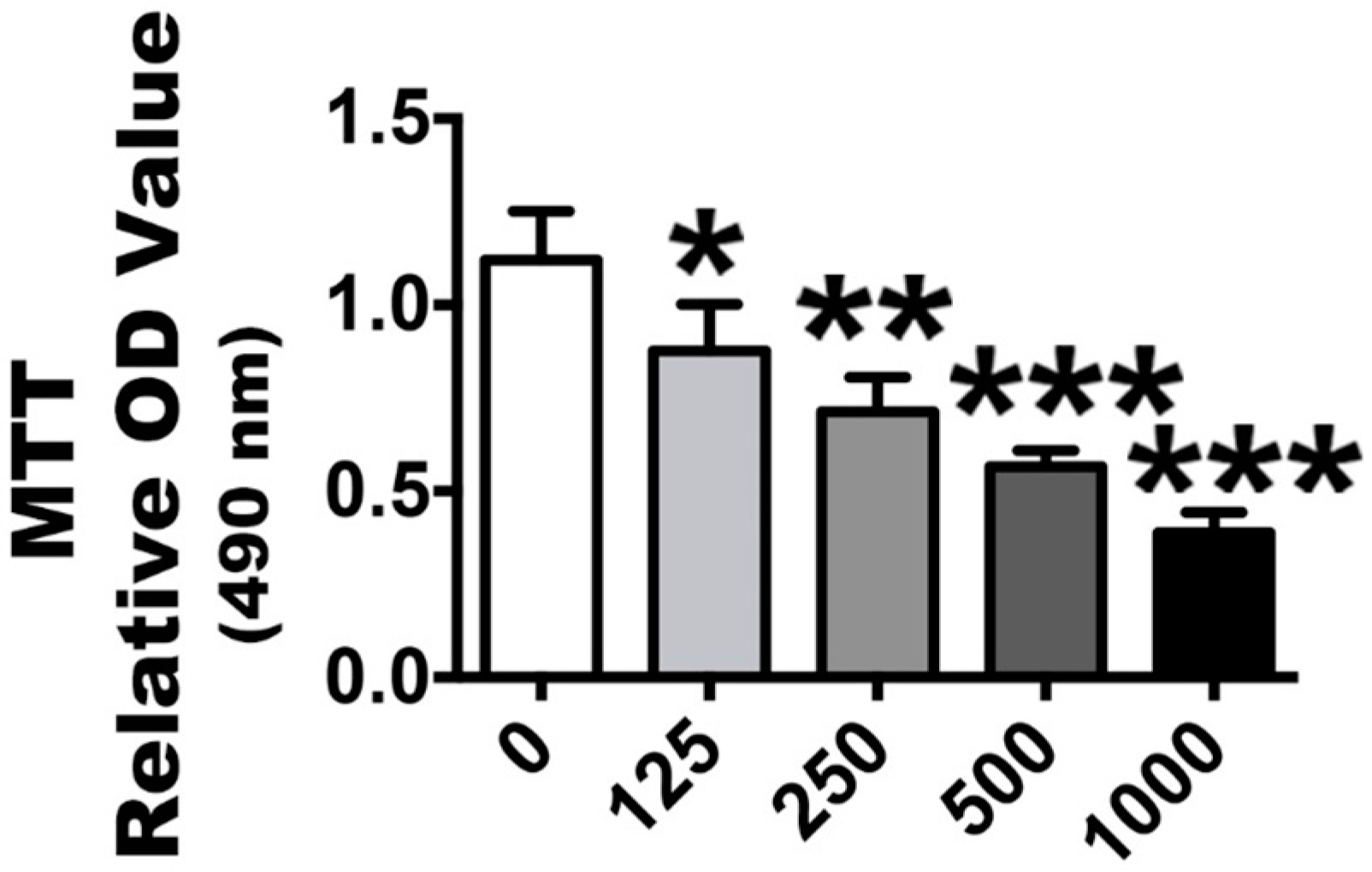
2.2. H2O2 Stimulation Leads to OS and Nrf2 Signaling Changes
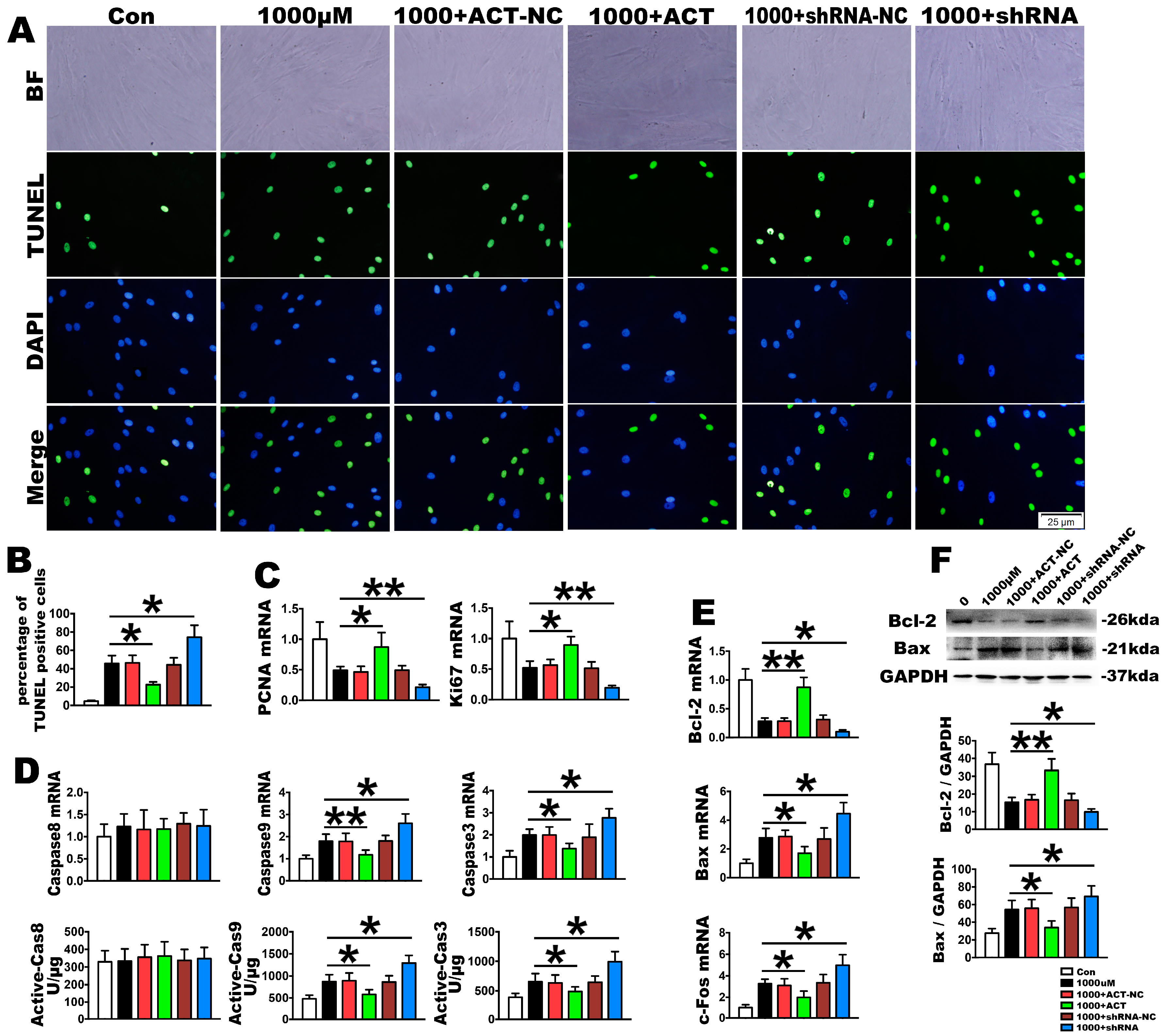
2.3. OS Induces PDLSC Apoptosis
2.4. Nrf2 Is a Key Molecule in PDLSC Apoptosis Protection during OS
3. Discussion
4. Methods
4.1. Isolation and Culture of Human PDLSCs
4.2. Colony-Forming and Cell Viability Assay
4.3. Flow Cytometry
4.4. Detection of the Multi-Lineage Differentiation Ability of PDLSCs
4.5. Establishment of an Oxidative Stress Model and Determinations
4.6. Plasmids and Lentiviruses
4.7. Fluorogenic Assay
4.8. TUNEL Staining
4.9. RNA Extraction, Reverse Transcriptase PCR and Real-Time PCR
4.10. Western Blotting
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of interest
References
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Pihlstrom, B.L.; Michalowicz, B.S.; Johnson, N.W. Periodontal diseases. Lancet. 2005, 366, 1809–1820. [Google Scholar] [CrossRef]
- Park, C.H.; Rios, H.F.; Jin, Q.; Sugai, J.V.; Padial-Molina, M.; Taut, A.D.; Flanagan, C.L.; Hollister, S.J.; Giannobile, W.V. Tissue engineering bone-ligament complexes using fiber-guiding scaffolds. Biomaterials 2012, 33, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Liang, M. Periodontal ligament stem cells: Current status, concerns, and future prospects. Stem Cells Int. 2015, 2015, 972313. [Google Scholar] [CrossRef] [PubMed]
- Panjamurthy, K.; Manoharan, S.; Ramachandran, C.R. Lipid peroxidation and antioxidant status in patients with periodontitis. Cell. Mol. Boil. Lett. 2005, 10, 255–264. [Google Scholar]
- Kimura, S.; Yonemura, T.; Kaya, H. Increased oxidative product formation by peripheral blood polymorphonuclear leukocytes in human periodontal diseases. J. Periodontal Res. 1993, 28, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Waddington, R.J.; Moseley, R.; Embery, G. Reactive oxygen species: A potential role in the pathogenesis of periodontal diseases. Oral Dis. 2000, 6, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; D’Aiuto, F.; Nibali, L.; Donald, A.; Storry, C.; Parkar, M.; Suvan, J.; Hingorani, A.D.; Vallance, P.; Deanfield, J. Treatment of periodontitis and endothelial function. N. Engl. J. Med. 2007, 356, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Dangaria, S.J.; Ito, Y.; Luan, X.; Diekwisch, T.G. Successful periodontal ligament regeneration by periodontal progenitor preseeding on natural tooth root surfaces. Stem Cells Dev. 2011, 20, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.I.; Yi, J.K.; Bae, W.J.; Lee, S.; Cha, H.J.; Kim, E.C. Thymosin β-4 suppresses osteoclastic differentiation and inflammatory responses in human periodontal ligament cells. PLoS ONE. 2016, 11, e0146708. [Google Scholar] [CrossRef] [PubMed]
- D'Aiuto, F.; Nibali, L.; Parkar, M.; Patel, K.; Suvan, J.; Donos, N. Oxidative stress, systemic inflammation, and severe periodontitis. J. Dent. Res. 2010, 89, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Helmersson, J.; Jarosinska, D.; Sallsten, G.; Mazzolai, B.; Barregard, L. Regulatory factors of basal F2-isoprostane formation: Population, age, gender and smoking habits in humans. Free Radic. Res. 2009, 43, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Blomgren, K. Mitochondrial cell death control in familial Parkinson disease. PLoS Biol. 2007, 5, e206. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jian, C.X.; Zhang, H.Y.; Zhou, Y.; Wu, X.; Zhang, G.; Tan, Y.H. Hypoxia enhances periodontal ligament stem cell proliferation via the MAPK signaling pathway. Genet. Mol. Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Granville, D.J.; Gottlieb, R.A. Mitochondria: Regulators of cell death and survival. Sci. World J. 2002, 2, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Sykiotis, G.P.; Habeos, I.G.; Samuelson, A.V.; Bohmann, D. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Pantano, C.; Reynaert, N.L.; van der Vliet, A.; Janssen-Heininger, Y.M. Redox-sensitive kinases of the nuclear factor-κB signaling pathway. Antioxid. Redox Signal. 2006, 8, 1791–1806. [Google Scholar] [CrossRef] [PubMed]
- Grigsby, B.; Rodriguez-Rilo, H.; Khan, K. Antioxidants and chronic pancreatitis: Theory of oxidative stress and trials of antioxidant therapy. Dig. Dis. Sci. 2012, 57, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liu, Z.; Xie, Y.; Hu, J.; Wang, H.; Fan, Z.; Zhang, C.; Wang, J.; Wu, C.T.; Wang, S. Adenovirus-mediated transfer of hepatocyte growth factor gene to human dental pulp stem cells under good manufacturing practice improves their potential for periodontal regeneration in swine. Stem. Cell Res. Ther. 2015, 6, 249. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, H.; Katsuoka, F.; Engel, J.D.; Yamamoto, M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 6379–6384. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Johnson, J.A. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J. Biochem. Mol. Biol. 2004, 37, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Haines, D.D.; Lekli, I.; Teissier, P.; Bak, I.; Tosaki, A. Role of haeme oxygenase-1 in resolution of oxidative stress-related pathologies: Focus on cardiovascular, lung, neurological and kidney disorders. Acta Physiol. 2012, 204, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.; Jaiswal, A.K. NAD(P)H: Quinone oxidoreductase1 (DT diaphorase) specifically prevents the formation of benzo[a]pyrene quinone-DNA adducts generated by cytochrome P4501A1 and P450 reductase. Proc. Natl. Acad. Sci. USA 1994, 91, 8413–8417. [Google Scholar] [CrossRef] [PubMed]
- Gebicki, J.M.; Nauser, T.; Domazou, A.; Steinmann, D.; Bounds, P.L.; Koppenol, W.H. Reduction of protein radicals by GSH and ascorbate: Potential biological significance. Amino Acids 2010, 39, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, R.T.; Wartman, M.A.; Bailey, H.H.; Gipp, J.J. Constitutive and β-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J. Biol. Chem. 1997, 272, 7445–7454. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.M.; Zhang, J.; Zhang, M.; An, Y.; Chen, F.; Wu, Z.F. A review on endogenous regenerative technology in periodontal regenerative medicine. Biomaterials 2010, 31, 7892–7927. [Google Scholar] [CrossRef] [PubMed]
- Bosshardt, D.D.; Sculean, A. Does periodontal tissue regeneration really work? Periodontology 2009, 51, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.S.; Feng, Z.H.; Wu, G.F.; Bai, S.Z.; Dong, Y.; Chen, F.M.; Zhao, Y.M. The use of platelet-rich fibrin combined with periodontal ligament and jaw bone mesenchymal stem cell sheets for periodontal tissue engineering. Sci. Rep. 2016, 21, 28126. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Liu, Y.; Wang, W.; Wei, F.; Liu, D.; Fan, Z.; An, Y.; Zhang, C.; Wang, S. Allogeneic periodontal ligament stem cell therapy for periodontitis in swine. Stem Cells 2010, 28, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Vaquette, C.; Fan, W.; Xiao, Y.; Hamlet, S.; Hutmacher, D.W.; Ivanovski, S. A biphasic scaffold design combined with cell sheet technology for simultaneous regeneration of alveolar bone/periodontal ligament complex. Biomaterials 2012, 33, 5560–5573. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Kroemer, G. The pathophysiology of mitochondrial cell death. Science 2004, 305, 626–629. [Google Scholar] [CrossRef] [PubMed]
- Krammer, P.H. CD95's deadly mission in the immune system. Nature 2000, 407, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazi, A.; Dixit, V.M. Death receptors: Signaling and modulation. Science 1998, 281, 1305–1308. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Du, Q.; Yin, X.; Li, J.; Wang, T.; Zhang, L. Agonist antibody activates death receptor 6 downstream signaling involving TRADD recruitment. FEBS Lett. 2014, 588, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.; Esposti, M.D.; Gilmore, A.P. Bcl-2 proteins and mitochondria—Specificity in membrane targeting for death. Biochim. Biophys. Acta 2011, 1813, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Henkler, F.; Behrle, E.; Dennehy, K.M.; Wicovsky, A.; Peters, N.; Warnke, C.; Pfizenmaier, K.; Wajant, H. The extracellular domains of FasL and Fas are sufficient for the formation of supramolecular FasL-Fas clusters of high stability. J. Cell Biol. 2005, 168, 1087–1098. [Google Scholar] [CrossRef] [PubMed]
- Ola, M.S.; Nawaz, M.; Ahsan, H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol. Cell. Biochem. 2011, 351, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Martinou, J.C.; Youle, R.J. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell 2011, 21, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Leonard, M.O.; Kieran, N.E.; Howell, K.; Burne, M.J.; Varadarajan, R.; Dhakshinamoorthy, S. Reoxygenation-specific activation of the antioxidant transcription factor Nrf2 mediates cytoprotective gene expression in ischemia-reperfusion injury. FASEB J. 2006, 20, 2624–2626. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.K.; Wakabayashi, N.; Itoh, K.; Motohashi, H.; Yamamoto, M.; Kensler, TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Boil. Chem. 2003, 278, 8135–8145. [Google Scholar] [CrossRef] [PubMed]
- Thimmulappa, R.K.; Mai, K.H.; Srisuma, S.; Kensler, T.W.; Yamamoto, M.; Biswal, S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002, 62, 5196–5203. [Google Scholar] [PubMed]
- Wang, S.; Zhang, C.; Niyazi, S.; Zheng, L.; Li, J.; Zhang, W.; Xu, M.; Rong, R.; Yang, C.; Zhu, T. A novel cytoprotective peptide protects mesenchymal stem cells against mitochondrial dysfunction and apoptosis induced by starvation via Nrf2/Sirt3/FoxO3a pathway. J. Transl. Med. 2017, 15, 33. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Shang, W.L.; Li, D.H.; Wu, W.W.; Hou, S.X. Simvastatin protects osteoblast against H2O2-induced oxidative damage via inhibiting the upregulation of Nox4. Mol. Cell Biochem. 2012, 360, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Zhaleh, F.; Amiri, F.; Mohammadzadeh-Vardin, M.; Bahadori, M.; Harati, M.D.; Roudkenar, M.H.; Saki, S. Nuclear factor erythroid-2 related factor 2 overexpressed mesenchymal stem cells transplantation, improves renal function, decreases injuries markers and increases repair markers in glycerol-induced Acute kidney injury rats. Iran. J. Basic Med. Sci. 2016, 19, 323–329. [Google Scholar] [PubMed]
- Kubben, N.; Zhang, W.; Wang, L.; Voss, T.C.; Yang, J.; Qu, J.; Liu, G.H.; Misteli, T. Repression of the Antioxidant Nrf2 Pathway in Premature Aging. Cell 2016, 165, 1361–1374. [Google Scholar] [CrossRef] [PubMed]



| Genes | Forward Primer (5′–3′) | Reverse Primer (3′–5′) |
|---|---|---|
| BSP | GGCCACGATATTATCTTTACAAGCA | TCAGCCTCAGAGTCTTCATCTTCA |
| OPG | CTGCAGTACGTCAAGCAGGAGTG | TTTGCAAACTGTATTTCGCTCTGG |
| ALP | GGACCATTCCCACGTCTTCAC | CCTTGTAGCCAGGCCCATTG |
| LPL | CCAAACTGGTGGGACAGGATG | GCTCCAAGGCTGTATCCCAAGA |
| PPAR-γ | TGGAATTAGATGACAGCGACTTGG | TTGAATGTCTTCAATGGGCTTCAC |
| PCNA | GGCCGAAGATAACGCGGATAC | GGCATATACGTGCAAATTCACCA |
| Ki67 | TGGGTCTGTTATTGATGAGCC | TGACTTCCTTCCATTCTGAAGAC |
| Bax | GCGAGTGTCTCAAGCGCATC | CCAGTTGAAGTTGCCGTCAGAA |
| Bcl-2 | TCGCCCTGTGGATGACTGAG | CAGAGTCTTCAGAGACAGCCAGGA |
| c-Fos | TCTTACTACCACTCACCCGCAGAC | GGAATGAAGTTGGCACTGGAGAC |
| Caspase3 | GACTCTGGAATATCCCTGGACAACA | AGGTTTGCTGCATCGACATCTG |
| Caspase8 | CCAGAGACTCCAGGAAAAGAGA | GATAGAGCATGACCCTGTAGGC |
| Caspase9 | TTCCCAGGTTTTGTCTCCTG | GGGACTGCAGGTCTTCAGAG |
| Nrf2 | GATTCTGACTCCGGCATTTC | TCCCCAGAAGAATGTACTGG |
| NQO1 | GGATTGGACCGAGCTGGAA | GAAACACCCAGCCGTCAGCTA |
| HO-1 | TTGCCAGTGCCACCAAGTTC | TCAGCAGCTCCTGCAACTCC |
| γ-GCS | TTGCAGGAAGGCATTGATCA | GCATCATCCAGGTGTATTTTCTCTT |
| GAPDH | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Yang, H.; Wen, Y.; Li, B.; Zhao, Y.; Xing, J.; Zhang, M.; Chen, Y. Nrf2 Inhibits Periodontal Ligament Stem Cell Apoptosis under Excessive Oxidative Stress. Int. J. Mol. Sci. 2017, 18, 1076. https://doi.org/10.3390/ijms18051076
Liu Y, Yang H, Wen Y, Li B, Zhao Y, Xing J, Zhang M, Chen Y. Nrf2 Inhibits Periodontal Ligament Stem Cell Apoptosis under Excessive Oxidative Stress. International Journal of Molecular Sciences. 2017; 18(5):1076. https://doi.org/10.3390/ijms18051076
Chicago/Turabian StyleLiu, Yanli, Hongxu Yang, Yi Wen, Bingyi Li, Yinhua Zhao, Jing Xing, Min Zhang, and Yongjin Chen. 2017. "Nrf2 Inhibits Periodontal Ligament Stem Cell Apoptosis under Excessive Oxidative Stress" International Journal of Molecular Sciences 18, no. 5: 1076. https://doi.org/10.3390/ijms18051076
APA StyleLiu, Y., Yang, H., Wen, Y., Li, B., Zhao, Y., Xing, J., Zhang, M., & Chen, Y. (2017). Nrf2 Inhibits Periodontal Ligament Stem Cell Apoptosis under Excessive Oxidative Stress. International Journal of Molecular Sciences, 18(5), 1076. https://doi.org/10.3390/ijms18051076






