Biofilm is a Major Virulence Determinant in Bacterial Colonization of Chronic Skin Ulcers Independently from the Multidrug Resistant Phenotype
Abstract
:1. Introduction
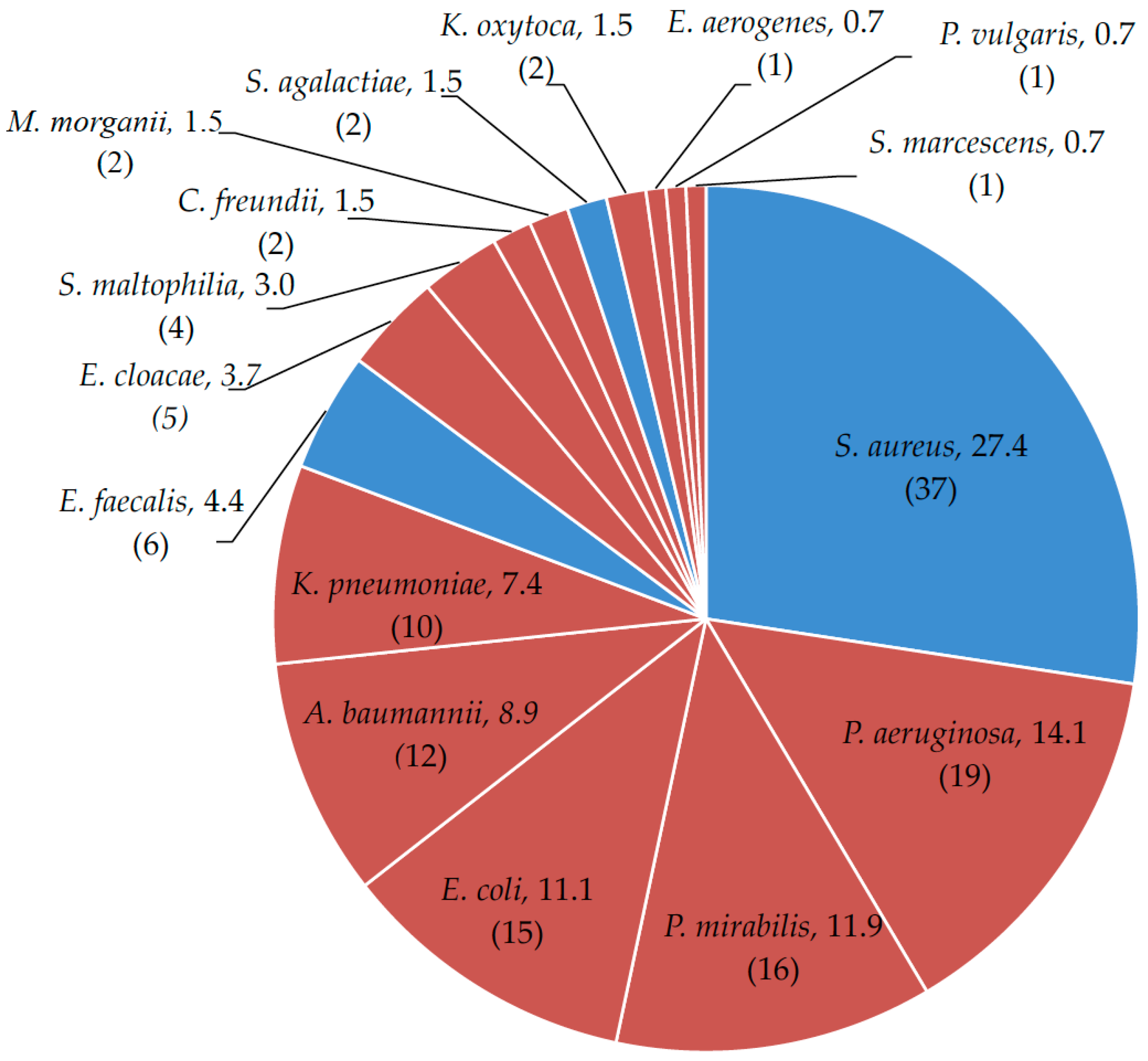
2. Results
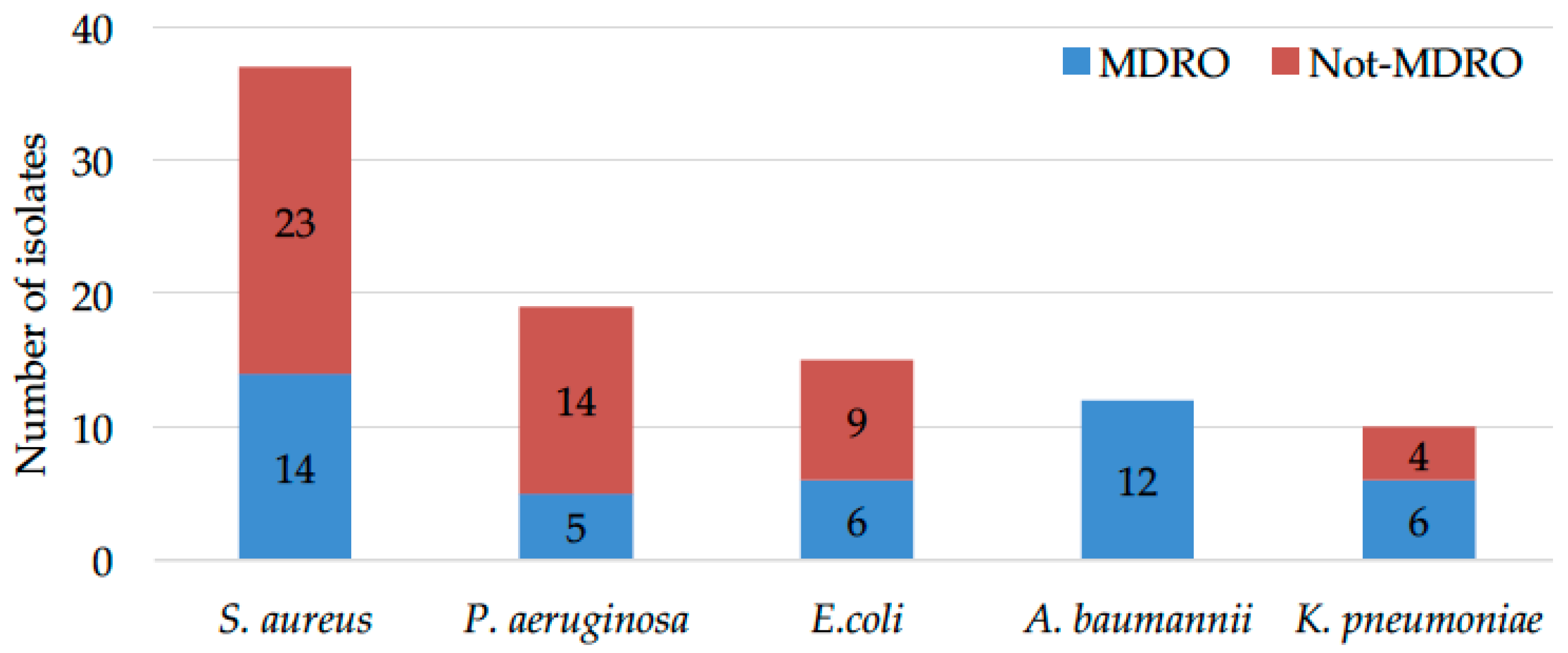
2.1. Microbial Drug-Resistance Profiles
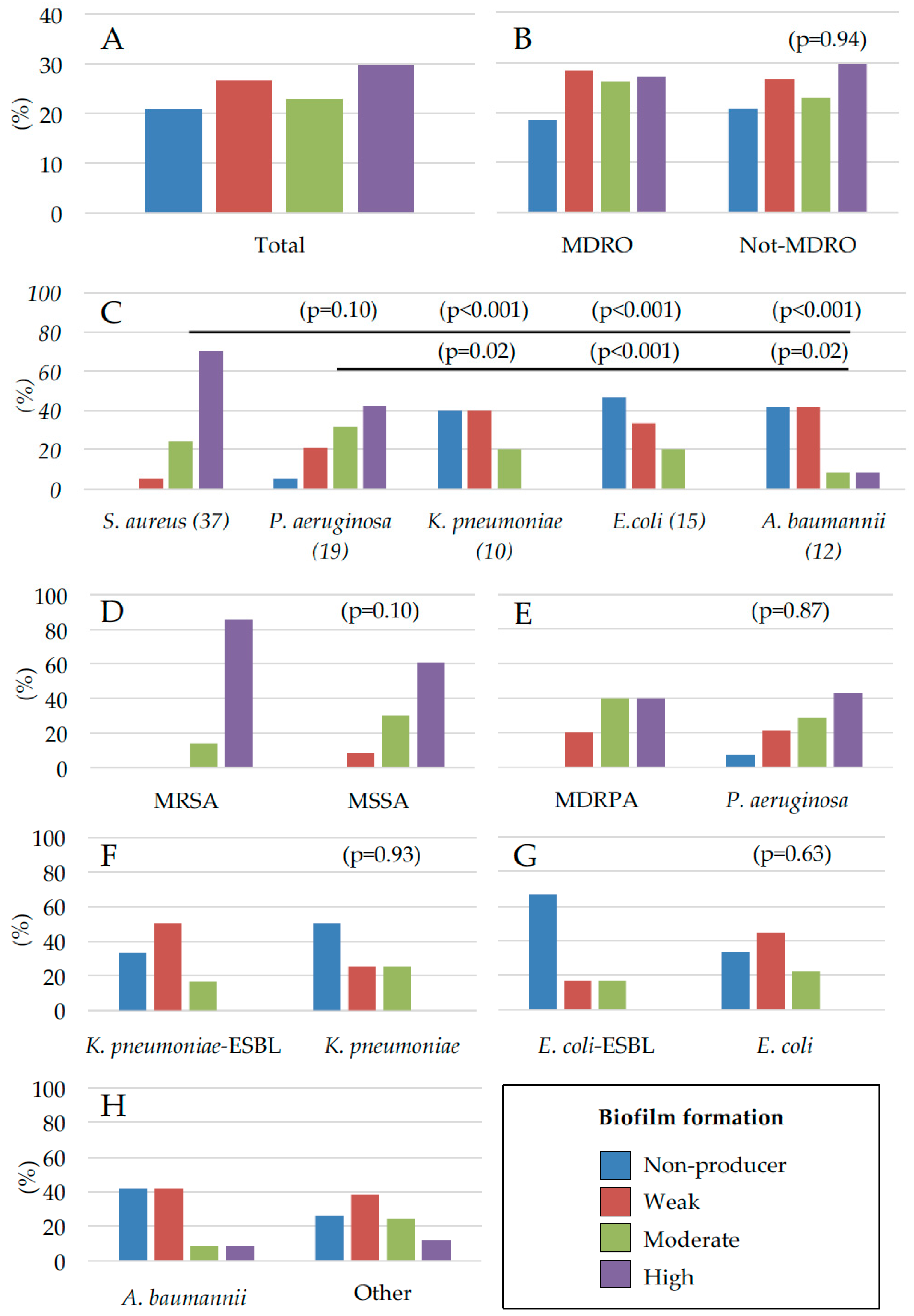
2.2. Assessment of Microbial Biofilm Production
3. Discussion
4. Materials and Methods
4.1. Inclusion Criteria
4.2. Sample Collection
4.3. Bacterial Identification
4.4. Evaluation of Biofilm Production
4.5. Statistic Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siegel, J.D.; Rhinehart, E.; Jackson, M.; Chiarello, L. Management of multidrug-resistant organisms in healthcare settings, 2006. Am. J. Infect. Control 2007, 35, 165–193. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, S.E. The relationship between antimicrobial resistance and patient outcomes: Mortality, length of hospital stay, and health care costs. Clin. Infect. Dis. 2006, 42, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Lautenbach, E.; Patel, J.B.; Bilker, W.B.; Edelstein, P.H.; Fishman, N.O. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: Risk factors for infection and impact of resistance on outcomes. Clin. Infect. Dis. 2001, 32, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.L.; Sotto, A.; Jourdan, N.; Combescure, C.; Vannereau, D.; Rodier, M.; Lavigne, J.P. Risk factors and healing impact of multidrug-resistant bacteria in diabetic foot ulcers. Diabete Metab. 2008, 34, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, U.; Parameswaran, S.; Armstrong, A.; Burgueno-Vega, D.; Griswold, J.; Dissanaike, S.; Rumbaugh, K.P. Prevalence of multiple antibiotic resistant infections in diabetic versus nondiabetic wounds. J. Pathog. 2014, 2014, 173053. [Google Scholar] [CrossRef] [PubMed]
- Gottrup, F. Optimizing wound treatment through health care structuring and professional education. Wound Repair Regen. 2004, 12, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Sen, C.K.; Gordillo, G.M.; Roy, S.; Kirsner, R.; Lambert, L.; Hunt, T.K.; Gottrup, F.; Gurtner, G.C.; Longaker, M.T. Human skin wounds: A major and snowballing threat to public health and the economy. Wound Repair Regen. 2009, 17, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Hartemann-Heurtier, A.; Robert, J.; Jacqueminet, S.; Van Ha, G.; Golmard, J.L.; Jarlier, V.; Grimaldi, A. Diabetic foot ulcer and multidrug-resistant organisms: Risk factors and impact. Diabet. Med. 2004, 21, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, T.C.; Sabel, A.L.; Sarcone, E.E.; Price, C.S.; Mehler, P.S.; Burman, W.J. Skin and soft-tissue infections requiring hospitalization at an academic medical center: Opportunities for antimicrobial stewardship. Clin. Infect. Dis. 2010, 51, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.R.; Bernstein, J.M. Chronic wound infection: Facts and controversies. Clin. Dermatol. 2010, 28, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Egbe, C.A.; Omoregie, R.; Igbarumah, I.O.; Onemu, S. Microbiology of wound infections among patients of a tertiary hospital in Benin City, Nigeria. J. Res. Health Sci. 2011, 11, 109–113. [Google Scholar] [PubMed]
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef] [PubMed]
- Ramakant, P.; Verma, A.K.; Misra, R.; Prasad, K.N.; Chand, G.; Mishra, A.; Agarwal, G.; Agarwal, A.; Mishra, S.K. Changing microbiological profile of pathogenic bacteria in diabetic foot infections: Time for a rethink on which empirical therapy to choose? Diabetologia 2011, 54, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Lipsky, B.A.; Berendt, A.R.; Cornia, P.B.; Pile, J.C.; Peters, E.J.; Armstrong, D.G.; Deery, H.G.; Embil, J.M.; Joseph, W.S.; Karchmer, A.W.; et al. 2012 Infectious diseases society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin. Infect. Dis. 2012, 54, e132–e173. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.K.; Geringer, M.R.; Hong, S.J.; Leung, K.P.; Mustoe, T.A.; Galiano, R.D. In vivo modelling of biofilm-infected wounds: A review. J. Surg. Res. 2012, 178, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; McCarty, S.M.; Lipsky, B. Biofilms and wounds: An overview of the evidence. Adv. Wound Care (New Rochelle) 2015, 4, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, E.G. Microbial biofilms in dermatology: A matter of skin. Esperienze Dermatologiche 2015, 17, 163–166. [Google Scholar]
- Kingsley, A. A proactive approach to wound infection. Nurs. Stand. 2001, 15, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Harding, K.G. Bacteria and wound healing. Curr. Opin. Infect. Dis. 2004, 17, 91–96. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Cutting, K.F. Critical colonisation—The concept under scrutiny. Ostomy Wound Manag. 2006, 52, 50–56. [Google Scholar]
- Zhao, G.; Usui, M.L.; Lippman, S.I.; James, G.A.; Stewart, P.S.; Fleckman, P.; Olerud, J.E. Biofilms and inflammation in chronic wounds. Adv. Wound Care (New Rochelle) 2013, 2, 389–399. [Google Scholar] [CrossRef] [PubMed]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Hengzhuang, W.; Wu, H.; Ciofu, O.; Song, Z.; Høiby, N. In vivo pharmacokinetics/pharmacodynamics of colistin and imipenem in Pseudomonas aeruginosa biofilm infection. Antimicrob. Agents Chemother. 2012, 56, 2683–2690. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; Ghigo, J.M. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005, 13, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Reiter, K.C.; Da Silva Paim, T.G.; De Oliveira, C.F.; D’Azevedo, P.A. High biofilm production by invasive multiresistant staphylococci. APMIS 2011, 119, 776–781. [Google Scholar] [CrossRef] [PubMed]
- Kwon, A.S.; Park, G.C.; Ryu, S.Y.; Lim, D.H.; Lim, D.Y.; Choi, C.H.; Park, Y.; Lim, Y. Higher biofilm formation in multidrug-resistant clinical isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents 2008, 32, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.S.; Karthika, R.U.; Singh, S.P.; Shashikala, P.; Kanungo, R.; Jayachandran, S.; Prashanth, K. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J. Med. Microbiol. 2008, 26, 333–337. [Google Scholar] [PubMed]
- Sanchez, C.J., Jr.; Mende, K.; Beckius, M.L.; Akers, K.S.; Romano, D.R.; Wenke, J.C.; Murray, C.K. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect. Dis. 2013, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Kirketerp-Møller, K.; Jensen, P.Ø.; Madsen, K.G.; Phipps, R.; Krogfelt, K.; Høiby, N.; Givskov, M. Why chronic wounds will not heal: A novel hypothesis. Wound Repair Regen. 2008, 16, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Macià, M.D.; Rojo-Molinero, E.; Oliver, A. Antimicrobial susceptibility testing in biofilm—Growing bacteria. Clin. Microbiol. Infect. 2014, 20, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.A.; Bjarnsholt, T.; Alhede, M. Biofilms in wounds: A review of present knowledge. J. Wound Care 2014, 23, 570–582. [Google Scholar] [CrossRef] [PubMed]
- Eum, S.; Bergsbaken, R.L.; Harvey, C.L.; Warren, J.B.; Rotschafer, J.C. Discrepancy in vancomycin AUC/MIC ratio targeted attainment based upon the susceptibility testing in Staphylococcus aureus. Antibiotics 2016, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, E.G.; Toma, L.; Provot, C.; Ascenzioni, F.; Sperduti, I.; Prignano, G.; Gallo, M.T.; Pimpinelli, F.; Bordignon, V.; Bernardi, T.; et al. Development of an in vitro assay, based on the Biofilm Ring Test®, for rapid profiling of biofilm—Growing bacteria. Front. Microbiol. 2016, 7, 1429. [Google Scholar] [CrossRef] [PubMed]
- Abdulrazak, A.; Bitar, Z.I.; Al-Shamali, A.A.; Mobasher, L.A. Bacteriological study of diabetic foot infections. J. Diabetes Complicat. 2005, 19, 138–141. [Google Scholar] [CrossRef] [PubMed]
- Körber, A.; Schmid, E.N.; Buer, J.; Klode, J.; Schadendorf, D.; Dissemond, J. Bacterial colonization of chronic leg ulcers: Current results compared with data 5 years ago in a specialized dermatology department. J. Eur. Acad. Dermatol. Venereol. 2010, 24, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Oien, R.F.; Akesson, N. Bacterial cultures, rapid strep test, and antibiotic treatment in infected hard-to-heal ulcers in primary care. Scand. J. Prim. Health Care 2012, 30, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, P.; Jeya, M.; Susan, S.L. The bacteriology of diabetic foot ulcers, with a special reference to multidrug resistant strains. J. Clin. Diagn. Res. 2013, 7, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Hoeger, P.H.; Lenz, W.; Boutonnier, A.; Fournier, J.M. Staphylococcal skin colonization in children with atopic dermatitis: Prevalence, persistence, and transmission of toxigenic and nontoxigenic strains. J. Infect. Dis. 1992, 165, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Kac, G.; Buu-Hoï, A.; Hérisson, E.; Biancardini, P.; Debure, C. Methicillin-resistent Staphylococcus aureus. Nosocomial acquisition and carrier state in a wound care center. Arch. Dermatol. 2000, 136, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Gjødsbøl, K.; Skindersoe, M.E.; Skov, R.L.; Krogfelt, K.A. Cross-Contamination: Comparison of nasal and chronic leg ulcer Staphylococcus aureus Strains isolated from the same patient. Open Microbiol. J. 2013, 7, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Stanaway, S.; Johnson, D.; Moulik, P.; Gill, G. Methicillin-Resistant Staphylococcus aureus (MRSA) isolation from diabetic foot ulcers correlates with nasal MRSA carriage. Diabetes Res. Clin. Pract. 2007, 75, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Demling, R.H.; Waterhouse, B. The increasing problem of wound bacterial burden and infection in acute and chronic soft-tissue wounds caused by methicillin-resistant Staphylococcus aureus. J. Burns Wounds 2007, 7, e8. [Google Scholar] [PubMed]
- Jockenhöfer, F.; Gollnick, H.; Herberger, K.; Isbary, G.; Renner, R.; Stücker, M.; Valesky, E.; Wollina, U.; Weichenthal, M.; Karrer, S.; et al. Bacteriological pathogen spectrum of chronic leg ulcers: Results of a multicenter trial in dermatologic wound care centers differentiated by regions. J. Dtsch. Dermatol. Ges. 2013, 11, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; Hall, V.; Paull, A.; Morris, T.; Brown, S.; McCulloch, D.; Richardson, M.C.; Harding, K.G. Surface bacteriology of venous leg ulcers and healing outcome. J. Clin. Pathol. 2010, 63, 830–834. [Google Scholar] [CrossRef] [PubMed]
- Fazli, M.; Bjarnsholt, T.; Kirketerp-Møller, K.; Jørgensen, B.; Andersen, A.S.; Krogfelt, K.A.; Givskov, M.; Tolker-Nielsen, T. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 2009, 47, 4084–4089. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef] [PubMed]
- Kirketerp-Møller, K.; Jensen, P.Ø.; Fazli, M.; Madsen, K.G.; Pedersen, J.; Moser, C.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M.; Bjarnsholt, T. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 2008, 46, 2717–2722. [Google Scholar]
- Wang, J.T.; Chen, P.C.; Chang, S.C.; Shiau, Y.R.; Wang, H.Y.; Lai, J.F.; Huang, I.W.; Tan, M.C.; Lauderdale, T.L. Antimicrobial susceptibilities of Proteus mirabilis: A longitudinal nationwide study from the Taiwan surveillance of antimicrobial resistance (TSAR) program. BMC Infect. Dis. 2014, 14, 486. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Lee, S.; Kim, Y.R.; Oh, E.J.; Woo, G.J.; Lee, K. Occurence of extended-spectrum β-lactamases and plasmid-mediated AmpC β-lactamases among Korean isolates of Proteus mirabilis. J. Antimicrob. Chemother. 2006, 57, 156–158. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, M.M.; Nucleo, E.; Luzzaro, F.; Giani, T.; Migliavacca, R.; Vailati, F.; Kroumova, V.; Pagani, L.; Rossolini, G.M. CMY-16, a novel acquired AmpC-type β-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from Northern Italy. Antimicrob. Agents Chemother. 2006, 50, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Aragón, L.M.; Mirelis, B.; Miró, E.; Mata, C.; Gómez, L.; Rivera, A.; Coll, P.; Navarro, F. Increase in β-lactam-resistant Proteus mirabilis strains due to CTX-M- and CMY-type as well as new VEB- and inhibitor-resistant TEM-type β-lactamases. J. Antimicrob. Chemother. 2008, 61, 1029–1032. [Google Scholar] [CrossRef] [PubMed]
- Power, P.; Galleni, M.; Ayala, J.A.; Gutkind, G. Biochemical and molecular characterization of three new variants of AmpC β-lactamases from Morganella morganii. Antimicrob. Agents Chemother. 2006, 50, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Attinger, C.; Wolcott, R. Clinically addressing biofilm in chronic wounds. Adv. Wound Care (New Rochelle) 2012, 1, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Fazli, M.; Bjarnsholt, T.; Kirketerp-Møller, K.; Jørgensen, A.; Andersen, C.B.; Givskov, M.; Tolker-Nielsen, T. Quantitative analysis of the cellular inflammatory response against biofilm bacteria in chronic wounds. Wound Repair Regen. 2011, 19, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Ghigo, J.M. Natural conjugative plasmids induce bacterial biofilm development. Nature 2001, 412, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Ghigo, J.M.; Beloin, C. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, L.J.; Bouwer, E.J. RP4 plasmid transfer among species of Pseudomonas in a biofilm reactor. Water Sci. Technol. 1999, 39, 163–171. [Google Scholar] [CrossRef]
- Stalder, T.; Top, E. Plasmid transfer in biofilms: A perspective on limitations and opportunities. NPJ Biofilms Microbiomes 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Driffield, K.; Miller, K.; Bostock, J.M.; O’Neill, A.J.; Chopra, I. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 2008, 61, 1053–1056. [Google Scholar] [CrossRef] [PubMed]
- Allegrucci, M.; Sauer, K. Formation of Streptococcus pneumoniae non-phase variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J. Bacteriol. 2008, 190, 6330–6339. [Google Scholar] [CrossRef] [PubMed]
- Ryder, V.J.; Chopra, I.; O’Neill, A.J. Increased mutability of Staphylococci in biofilms as a consequence of oxidative stress. PLoS ONE 2012, 7, e47695. [Google Scholar] [CrossRef] [PubMed]
- Madsen, J.S.; Burmølle, M.; Sørensen, S.J. A spatiotemporal view of plasmid loss in biofilms and planktonic cultures. Biotechnol. Bioeng. 2014, 110, 3071–3074. [Google Scholar] [CrossRef] [PubMed]
- Patel, R. Biofilms and antimicrobial resistance. Clin. Orthop. Relat. Res. 2005, 437, 41–47. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, F.J.; Liu, Y.; Xiong, L.R.; Xie, L.L.; Xia, P.Y. Enhancement of biofilm formation by subinhibitory concentrations of macrolides in icaADBC-positive and -negative clinical isolates of Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2010, 54, 2707–2711. [Google Scholar] [CrossRef] [PubMed]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Vrany, J.D.; Stewart, P.S.; Suci, P.A. Comparison of recalcitrance to ciprofloxacin and levofloxacin exhibited by Pseudomonas aeruginosa bofilms displaying rapid-transport characteristics. Antimicrob. Agents Chemother. 1997, 41, 1352–1358. [Google Scholar] [PubMed]
- Stone, G.; Wood, P.; Dixon, L.; Keyhan, M.; Matin, A. Tetracycline rapidly reaches all the constituent cells of uropathogenic Escherichia coli biofilms. Antimicrob. Agents Chemother. 2002, 46, 2458–2461. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Stewart, P.S. Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2002, 4, 900–903. [Google Scholar] [CrossRef]
- Darouiche, R.O.; Dhir, A.; Miller, A.J.; Landon, G.C.; Raad, I.I.; Musher, D.M. Vancomycin penetration into biofilm covering infected prostheses and effect on bacteria. J. Infect. Dis. 1994, 170, 720–723. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.L.; Patel, R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 2007, 82, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Antimicrobial Tolerance in Biofilms. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Smith, K.; Perez, A.; Ramage, G.; Lappin, D.; Gemmell, C.G.; Lang, S. Biofilm formation by Scottish clinical isolates of Staphylococcus aureus. J. Med. Microbiol. 2008, 57, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Karauzum, H.; Ferry, T.; de Bentzmann, S.; Lina, G.; Bes, M.; Vandenesch, F.; Schmaler, M.; Berger-Bächi, B.; Etienne, J.; Landmann, R. Comparison of Adhesion and Virulence of Two Predominant Hospital-Acquired Methi-cillin-Resistant Staphylococcus aureus Clones and Clonal Methicillin-Susceptible Staphylococcus aureus Isolates. Infect. Immun. 2008, 76, 5133–5138. [Google Scholar] [CrossRef] [PubMed]
- Doulgeraki, A.I.; Di Ciccio, P.; Ianieri, A.; Nychas, G.E. Methicillin-resistant food-related Staphylococcus aureus: A review of current knowledge and biofilm formation for future studies and applications. Res. Microbiol. 2017, 168, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mirzaee, M.; Najar-Peerayeh, S.; Behmanesh, M.; Forouzandeh, M. Relationship between adhesin genes and biofilm formation in vancomycin-intermediate Staphylococcus aureus clinical isolates. Curr. Microbial. 2015, 70, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, C.; Waters, E.M.; Rudkin, J.K.; Schaeffer, C.R.; Lohan, A.J.; Tong, P.; Loftus, B.J.; Pier, G.B.; Fey, P.D.; Massey, R.C.; et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012, 8, e1002626. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, H.; Rudkin, J.K.; Black, N.S.; Gallagher, L.; O’Neill, E.; O’Gara, J.P. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2015, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Rudkin, J.K.; Laabei, M.; Edwards, A.M.; Joo, H.; Otto, M.; Lennon, K.L.; O’Gara, J.P.; Waterfield, N.R.; Massey, R.C. Oxacillin alters the toxin expression profile of community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarzadeh Corehtash, Z.; Khorshidi, A.; Firoozeh, F.; Akbari, H.; Mahmoudi Aznaveh, A. Biofilm formation and virulence factors among Pseudomonas aeruginosa isolated from burn patients. Jundishapur J. Microbiol. 2015, 8, e22345. [Google Scholar] [CrossRef] [PubMed]
- Abidi, S.H.; Sherwani, S.K.; Siddiqui, T.R.; Bashir, A.; Kazmi, S.U. Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 2013, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Pastar, I.; Nusbaum, A.G.; Gil, J.; Patel, S.B.; Chen, J.; Valdes, J.; Stojadinovic, O.; Plano, L.R.; Tomic-Canic, M.; Davis, S.C. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 2013, 8, e56846. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, K.J.; Burd, T.A.; Anglen, J.O.; Simpson, A.W.; Christensen, G.D.; Gainor, BJ. Synergy between Staphylococcus aureus and Pseudomonas aeruginosa in a rat model of complex orthopaedic wounds. J. Bone Jt. Surg. Am. 2001, 83, 855–861. [Google Scholar] [CrossRef]
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 2014, 82, 4718–4728. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluth, D.B.; Wilson, K.; Ferkol, T.; Schuster, D.P. Lung function decline in cystic fibrosis patients and timing for lung transplantation referral. Chest 2004, 126, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.E.; Richet, H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 2006, 42, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Bano, J.; Marti, S.; Soto, S.; Fernandez-Cuenca, F.; Cisneros, J.M.; Pachon, J.; Pascual, A.; Martínez-Martínez, L.; McQueary, C.; Actis, L.A.; et al. Biofilm formation in Acinetobacter baumannii: Associated features and clinical implications. Clin. Microbiol. Infect. 2008, 14, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Marti, S.; Rodriguez-Bano, J.; Catel-Ferreira, M.; Jouenne, T.; Vila, J.; Seifert, H.; Dé, E. Biofilm formation at the solid-liquid and air-liquid interfaces by Acinetobacter species. BMC Res. Notes 2011, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; He, L.; Tao, X.; Meng, F.; Zhang, J. Biofilm may not be necessary for the epidemic spread of Acinetobacter baumannii. Sci. Rep. 2016, 6, 32066. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.R. Acinetobacter baumannii displays inverse relationship between meropenem resistance and biofilm production. J. Chemother. 2015, 27, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Li, H.; Zhang, C.; Liang, B.; Li, J.; Wang, L.; Du, X.; Liu, X.; Qiu, S.; Song, H. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 2016, 7, 483. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, Z. Biofilm-forming Klebsiella pneumoniae strains have greater likelihood of producing extended-spectrum β-lactamases. J. Hosp. Infect. 2008, 68, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Vuotto, C.; Longo, F.; Balice, M.P.; Donelli, G.; Varaldo, P.E. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens 2014, 3, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Lipsky, B.A.; Pecoraro, R.E.; Wheat, L.J. The diabetic foot: Soft tissue and bone infection. Infect. Dis. N. Am. 1990, 4, 409–432. [Google Scholar]
- Dowd, S.E.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Smith, E.; Rhoads, D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS ONE 2008, 3, e3326. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.C.; Vearncombe, M.; Sibbald, R.G. High bacterial load in asymptomatic diabetic patients with neurotrophic ulcers retards wound healing after application of Dermagraft. Ostomy Wound Manag. 2001, 47, 44–49. [Google Scholar]
- Ovington, L. Bacterial toxins and wound healing. Ostomy Wound Manag. 2003, 49, 8–12. [Google Scholar]
- Cutting, K.F.; White, R.J. Criteria for identifying wound infection-revisited. Ostomy Wound Manag. 2005, 51, 28–34. [Google Scholar]
- White, R.; Cutting, K.F.; Kingsley, A. Critical colonisation: Clinical reality or myth? Wounds UK 2005, 1, 94–95. [Google Scholar]
- Dissemond, J. When is a wound chronic? Hautarzt 2006, 57, 55. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, E.G.; Toma, L.; Prignano, G.; Pelagalli, L.; Police, A.; Cavallotti, C.; Torelli, R.; Sanguinetti, M.; Ensoli, F. Misidentification of Streptococcus uberis as a human pathogen: A casa report and literature review. Int. J. Infect. Dis. 2015, 33, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Healthcare-Associated Infections. Available online: http://www.cdc.gov/ncidod/dhqp/pdf/ar/mdroGuideline2006.pdf (accessed on 15 November 2016).
- De Gheldre, Y.; Avesani, V.; Berhin, C.; Delmée, M.; Glupczynski, Y. Evaluation of Oxoid combination discs for detection of extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2003, 52, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Moremi, N.; Mushi, M.F.; Fidelis, M.; Chalya, P.; Mirambo, M.; Mshana, S.E. Predominance of multi-resistant gram-negative bacteria colonizing chronic lower limb ulcers (CLLUs) at Bugando Medical Center. BMC Res. Notes 2014, 7, 211. [Google Scholar] [CrossRef] [PubMed]
| Drug | E. coli n = 9 | K. pneumoniae n = 4 | P. mirabilis n = 16 | E. cloacae n = 5 | C. freundii n = 2 | M. morganii n = 2 | P. aeruginosa n = 14 |
|---|---|---|---|---|---|---|---|
| Amikacin | 0 | 0 | 43.8 | 0 | 0 | 0 | 0 |
| Gentamicin | 0 | 0 | 75 | 0 | 0 | 50 | 14.3 |
| Tobramycin | 50 | 11.1 | |||||
| Ertapenem | 0 | 0 | 0 | 0 | |||
| Imipenem | 0 | 0 | 0 | 0 | 0 | ||
| Meropenem | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cefepime | 0 | 0 | 25 | 0 | 0 | 0 | 7.1 |
| Cefotaxime | 0 | 0 | 62.5 | 0 | 0 | 100 | |
| Ceftazidime | 0 | 0 | 62.5 | 0 | 0 | 100 | 0 |
| Nitrofurantoin | 0 | 100 | |||||
| Amoxicillin/clavulanic acid | 22.2 | 0 | 100 | 100 | 100 | ||
| Piperacillin/Tazobactam | 0 | 0 | 0 | ||||
| Colistin | 0 | 0 | 100 | 0 | 100 | 7.1 | |
| Ciprofloxacin | 22.2 | 0 | 68.8 | 0 | 0 | 66.6 | 14.3 |
| Levofloxacin | 50 | ||||||
| Trimethoprim/sulfamethoxazole | 55.5 | 0 | 75 | 20 | 0 | 50 | 100 |
| Tigecycline | 100 | 100 |
| Drug | MSSA n = 23 | E. faecalis n = 6 | S. agalactiae n = 2 |
|---|---|---|---|
| Gentamicin | 18.2 | ||
| Gentamicin High Level Resistance | 40 | ||
| Streptomycin High Level Resistance | 80 | ||
| Imipenem | 0 | ||
| Teicoplanin | 4.3 | 0 | 0 |
| Vancomycin | 0 | 0 | 0 |
| Clindamycin | 26.1 | 100 | 100 |
| Daptomycin | 0 | ||
| Erythromycin | 31.8 | 66.7 | |
| Nitrofurantoin | 0 | 0 | |
| Linezolid | 0 | 0 | 0 |
| Ampicillin/sulbactam | 0 | ||
| Benzylpenicillin | 69.6 | 0 | |
| Oxacillin | 0 | ||
| Levofloxacin | 8.7 | 50 | 0 |
| Moxifloxacin | 0 | ||
| Trimethoprim/sulfamethoxazole | 0 | 100 | 0 |
| Tetracyclin | 8.7 | 100 | |
| Fusidic Acid | 0 | ||
| Tigecycline | 0 | 0 | 0 |
| Drug | MRSA n = 14 | A. baumannii n = 12 | E. coli ESBL n = 6 | K. pneumoniae ESBL n = 6 | MDRPA n = 5 |
|---|---|---|---|---|---|
| Amikacin | 16.7 | 33.3 | 80 | ||
| Gentamicin | 50 | 75 | 100 | 66.7 | 100 |
| Tobramycin | 100 | ||||
| Ertapenem | 0 | ||||
| Imipenem | 75 | 0 | 33.3 | 20 | |
| Meropenem | 0 | 33.3 | 40 | ||
| Cefepime | 100 | 100 | 100 | 80 | |
| Cefotaxime | 100 | 100 | 100 | ||
| Ceftazidime | 100 | 100 | 100 | 60 | |
| Teicoplanin | 7.1 | ||||
| Vancomycin | 0 | ||||
| Clindamycin | 42.9 | ||||
| Daptomycin | 7.7 | ||||
| Erythromycin | 50 | ||||
| Nitrofurantoin | 0 | ||||
| Linezolid | 0 | ||||
| Amoxicillin/clavulanic acid | 100 | 33.3 | 83.3 | 100 | |
| Benzylpenicillin | 100 | ||||
| Oxacillin | 100 | ||||
| Piperacillin/Tazobactam | 20 | 83.3 | |||
| Colistin | 0 | 0 | 50 | 0 | |
| Ciprofloxacin | 91 | 100 | 83.3 | 80 | |
| Levofloxacin | 64.3 | ||||
| Moxifloxacin | 15.4 | ||||
| Trimethoprim/sulfamethoxazole | 14.3 | 75 | 66.7 | 83.3 | 100 |
| Tetracyclin | 28.8 | ||||
| Fosfomycin | 100 | 0 | 66.7 | ||
| Fusidic Acid | 7.1 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Domenico, E.G.; Farulla, I.; Prignano, G.; Gallo, M.T.; Vespaziani, M.; Cavallo, I.; Sperduti, I.; Pontone, M.; Bordignon, V.; Cilli, L.; et al. Biofilm is a Major Virulence Determinant in Bacterial Colonization of Chronic Skin Ulcers Independently from the Multidrug Resistant Phenotype. Int. J. Mol. Sci. 2017, 18, 1077. https://doi.org/10.3390/ijms18051077
Di Domenico EG, Farulla I, Prignano G, Gallo MT, Vespaziani M, Cavallo I, Sperduti I, Pontone M, Bordignon V, Cilli L, et al. Biofilm is a Major Virulence Determinant in Bacterial Colonization of Chronic Skin Ulcers Independently from the Multidrug Resistant Phenotype. International Journal of Molecular Sciences. 2017; 18(5):1077. https://doi.org/10.3390/ijms18051077
Chicago/Turabian StyleDi Domenico, Enea Gino, Ilaria Farulla, Grazia Prignano, Maria Teresa Gallo, Matteo Vespaziani, Ilaria Cavallo, Isabella Sperduti, Martina Pontone, Valentina Bordignon, Laura Cilli, and et al. 2017. "Biofilm is a Major Virulence Determinant in Bacterial Colonization of Chronic Skin Ulcers Independently from the Multidrug Resistant Phenotype" International Journal of Molecular Sciences 18, no. 5: 1077. https://doi.org/10.3390/ijms18051077







