Identifying the Genes Regulated by AtWRKY6 Using Comparative Transcript and Proteomic Analysis under Phosphorus Deficiency
Abstract
:1. Introduction
2. Results
2.1. Measurements of Biochemical Data in Three Plant Samples
2.2. 2-D DIGE Determination and MS (Mass spectrometry) Identification of Phosphorus Responsive Proteins in 35S:WRKY6-9 and wrky6-1
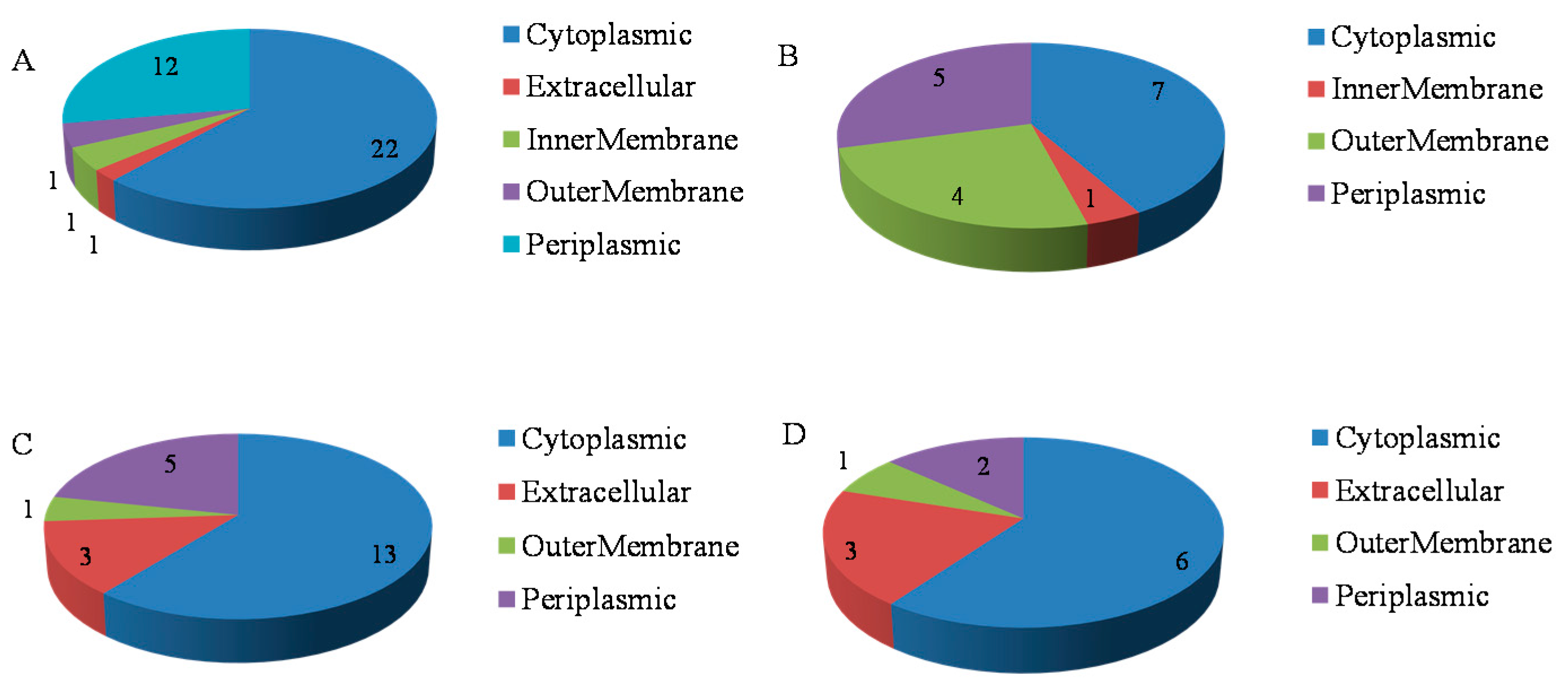
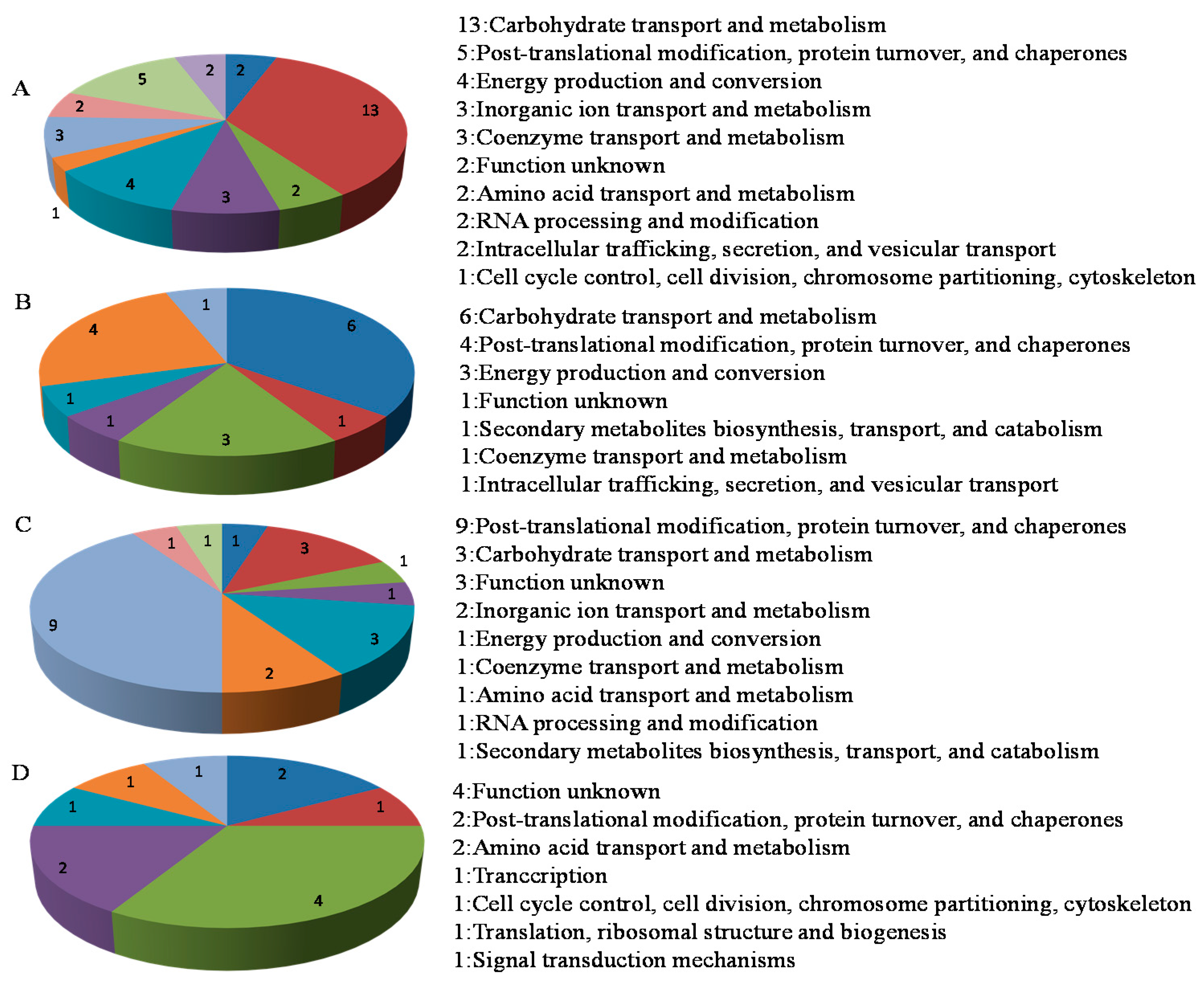
2.3. Functional Analysis of the Phosphorus Responsive Proteins
2.4. Analysis of the Differentially Abundant Proteins in wrky6-1
2.5. Analysis of the Differentially Abundant Proteins in 35S:WRKY6-9
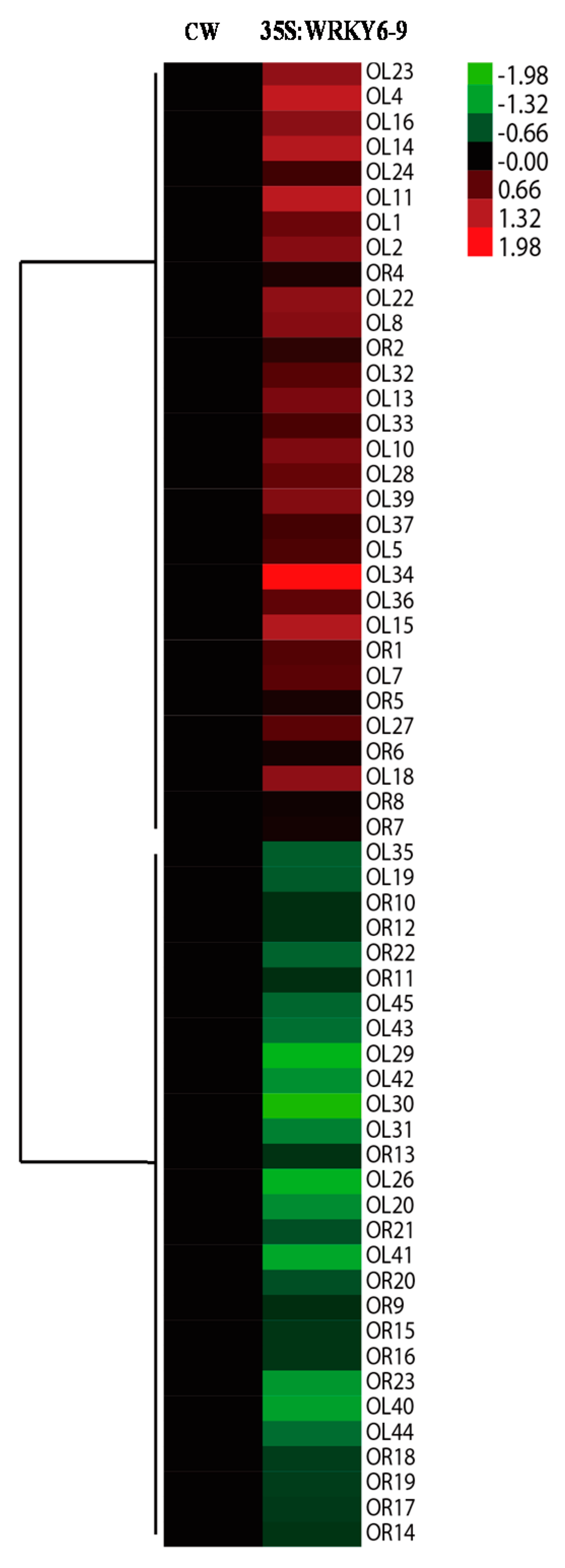
2.6. Potential Target Genes Regulated by WRKY6 in the Leaves and Roots
3. Discussion
3.1. Physiological Changes Were Induced after Phosphorus Deprivation
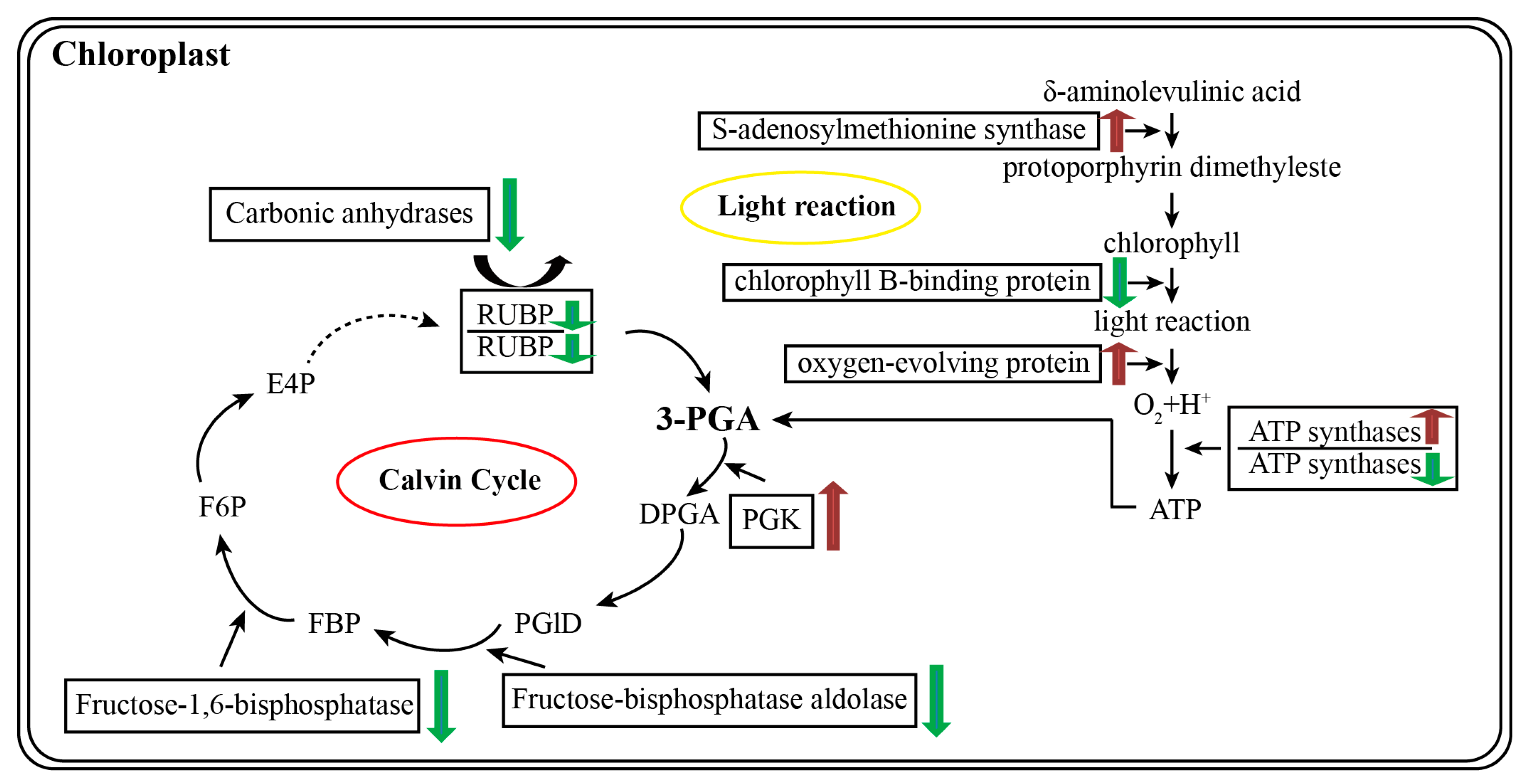
3.2. Proteomics Revealed That the Enzymes Primarily Involved in Photosynthesis Are Differentially Abundant in 35S:WRKY6-9 and wrky6-1
3.3. Proteins Related to Energy Metabolism and Stress Were Identified in 35S:WRKY6-9
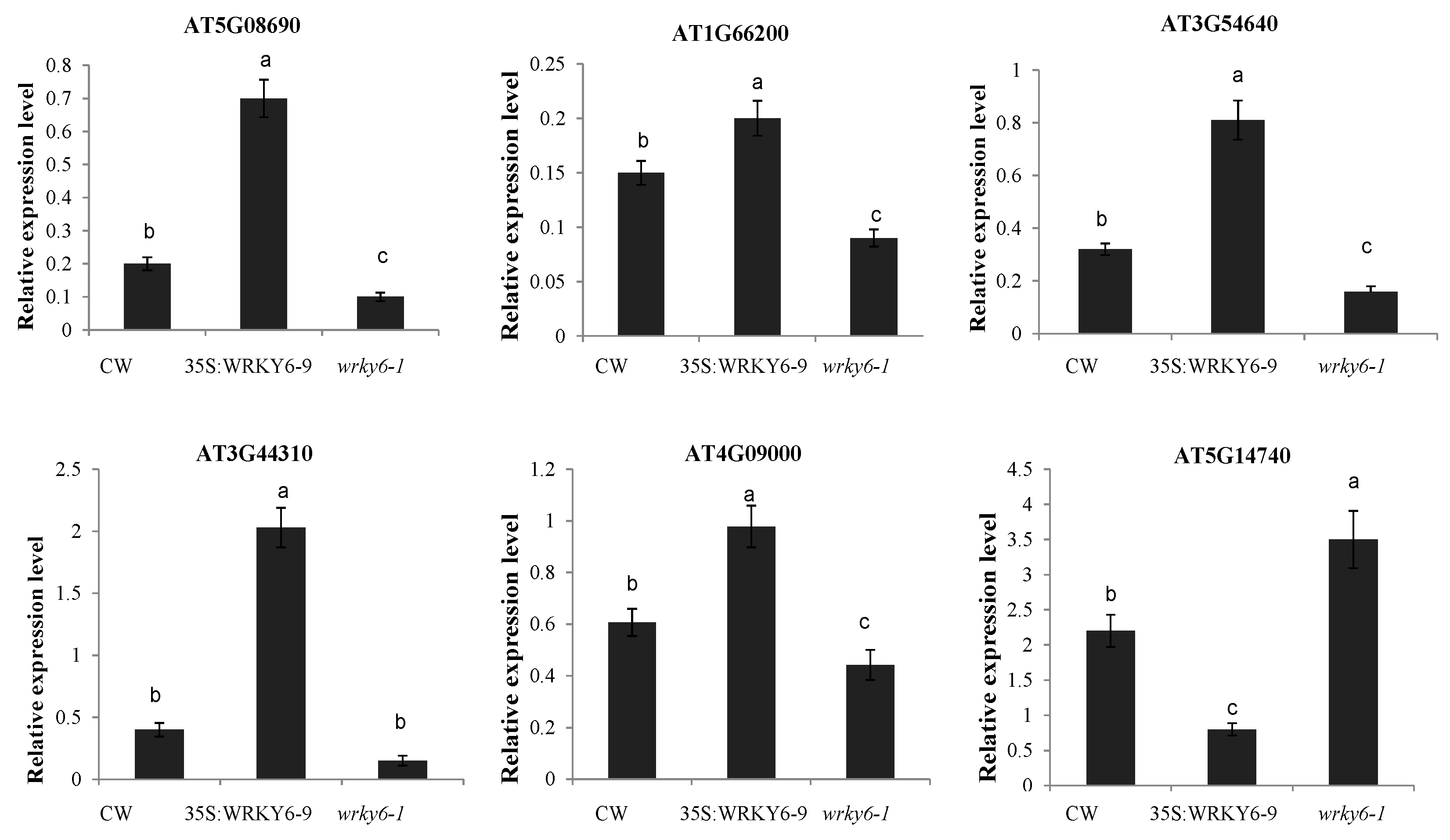
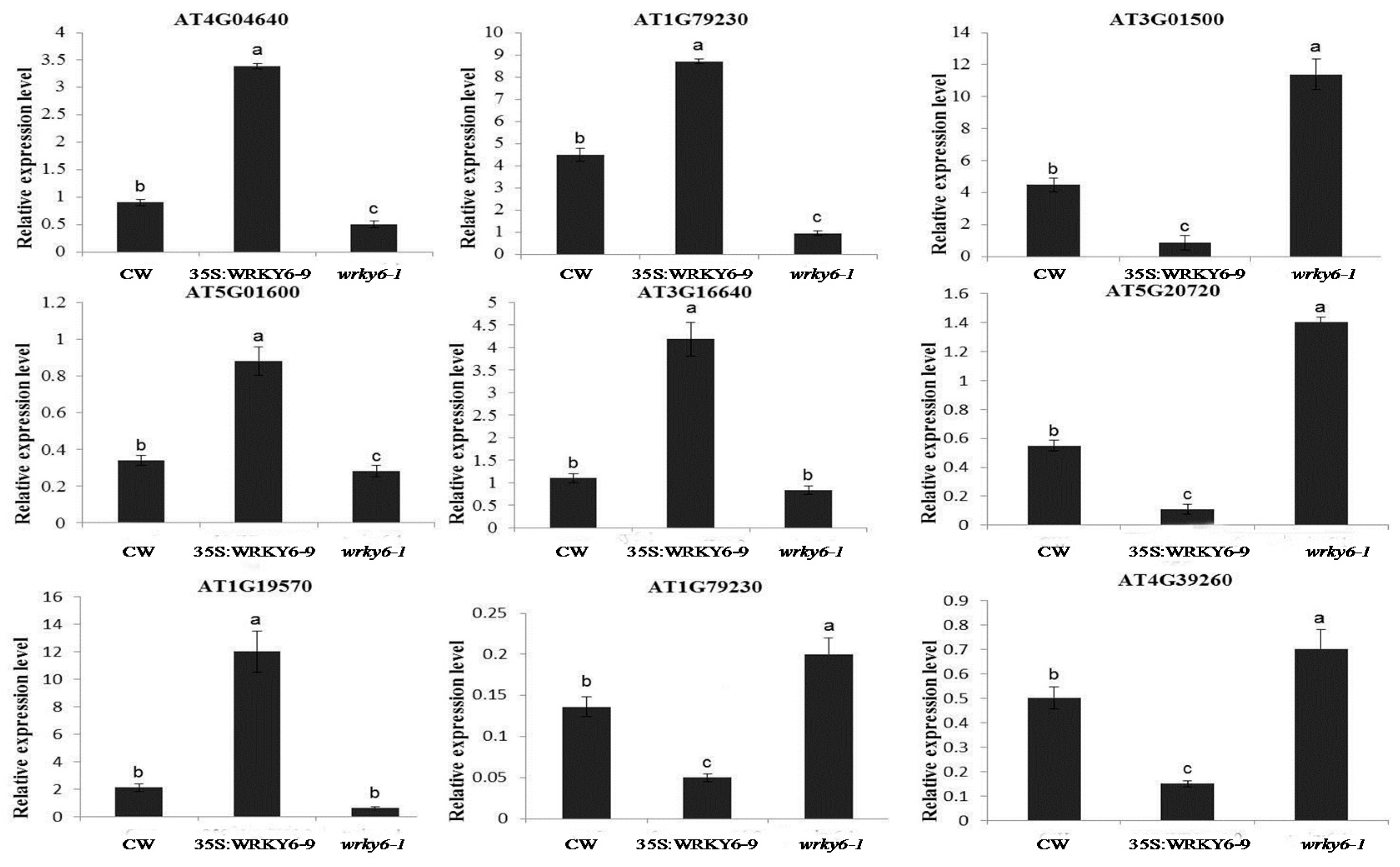
3.4. Potential Target Genes of WRKY6 Identified Using Proteomics and qRT-PCR Analysis
4. Materials and Methods
4.1. Plant Materials and Low Phosphorus Treatment
4.2. Protein Extraction and 2-D DIGE Analysis
4.3. Protein Identification via Mass Spectrometry and Bioinformatics Analysis
4.4. Gene Expression Analysis by qRT-PCR
4.5. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wissuwa, M. How do plants achieve tolerance to phosphorus deficienby? Small causes with big effects. Plant Physiol. 2003, 133, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, F. Phosphorus dynamics: From soil to plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P. Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Tran, H.T. Metabolic adaptations of phosphate starved plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, Q.; Kong, Y.H.; Chen, Y.; Duan, J.Y.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY45 transcription factor activates Phosphate transporter 1;1 expression in response to phosphate starvation. Plant Physiol. 2014, 164, 2020–2029. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Xu, Q.; Zhang, F.C.; Chen, Y.; Li, L.Q.; Wu, W.H.; Chen, Y.F. WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 2015, 167, 1579–1591. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Lee, J.; Gong, Q.; Ma, S.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Sato, A.; Bohnert, H.J.; Hasegawa, P.M. SIZ1 Regulation of phosphate starvation induced root architecture remodeling involves the control of auxin accumulation. Plant Physiol. 2011, 155, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.P.; Jain, A.; Deal, R.B.; Nagarajan, V.K.; Poling, M.D.; Raghothama, K.G.; Meagher, R.B. Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 2010, 152, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Bucciarelli, B.; Liu, J.; Zinn, K.; Miller, S.; Patton, V.J.; Allan, D.; Shen, J.; Vance, C.P. White lupin cluster root acclimation to phosphorus deficiency and root hair development involve unique glycerophosphodiester phosphodiesterases. Plant Physiol. 2011, 156, 1131–1148. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.J.; Lin, S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Alexova, R.; Millar, A.H. Proteomics of phosphate use and deprivation in plants. Proteomics 2013, 13, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xu, C.; Li, Z.; Zhang, K.; Yang, A.; Zhang, J. Comparative proteome analyses of phosphorus responses in maize (Zea mays L.) roots of wild-type and a low-P-tolerant mutant reveal root characteristics associated with phosphorus efficiency. Plant J. 2008, 55, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Li, W.; Schmidt, W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteom. 2012, 11, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Wang, Y.; Lee, C.H.; Mun, B.G.; Kim, P.J.; Lee, S.Y.; Kim, Y.C.; Kang, K.Y.; Rakwal, R.; Agrawal, G.K.; et al. A comparative proteomics survey of proteins responsive to phosphorous starvation in roots of hydroponically-grown rice seedlings. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 667–677. [Google Scholar] [CrossRef]
- Chen, Z.; Cui, Q.; Liang, C.; Sun, L.; Tian, J.; Liao, H. Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomic analysis. Proteomics 2011, 11, 4648–4659. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F.; Rossignol, M. Proteomic analysis of Arabidopsis thaliana ecotypes with contrasted root architecture in response to phosphate deficiency. J. Plant Physiol. 2011, 168, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY Superfamily of Plant Transcription Factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evolut. Biol. 2005, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001, 28, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Li, L.Q.; Xu, Q.; Kong, Y.H.; Wang, H.; Wu, W.H. The WRKY6 transcription factor modulates PHOSPHATE1 Expression in Response to Low Pi Stress in Arabidopsis. Plant Cell 2009, 21, 3554–3566. [Google Scholar] [CrossRef] [PubMed]
- Kasajima, I.; Ide, Y.; Yokota, H.M.; Fujiwara, T. WRKY6 is involved in the response to boron deficiencyin Arabidopsis thaliana. Physiol. Plant. 2010, 139, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Castrillo, G.; Eduardo, S.B.; Lorenzo, L.D.; Crevillén, P.; Ana, F.E.; Mohan, T.C.; Mouriz, A.; Catarecha, P.; Juan, S.P.; Olsson, S.; et al. WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 2013, 25, 2944–2957. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Feng, C.Z.; Ye, Q.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 2016, 12, e1005833. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.P.; Broadley, M.R.; White, P.J. Genetic responses to phosphorus deficiency. Ann. Bot. 2004, 94, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.P.; White, P.J. Sugar signaling in root responses to low phosphorus availability. Plant Physiol. 2011, 156, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Hermans, C.; Hammond, J.P.; White, P.J.; Verbruggen, N. How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci. 2006, 11, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Poling, M.D.; Karthikeyan, A.S.; Blakeslee, J.J.; Peer, W.A.; Titapiwatanakun, B.; Murphy, A.S.; Raghothama, K.G. Differential effects of sucrose and auxin on localized Pi–deficiency induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol. 2007, 144, 232–247. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Liu, Y.; Zhang, B.; Zhao, Y.; Wang, X.; Zhou, Y.; Raghothama, K.G.; Liu, D. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 2011, 156, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Suriyagoda, L.D.; Lambers, H.; Ryan, M.H.; Renton, M. Effects of leaf development and phosphorus supply on the photosynthetic characteristics of perennial legume species with pasture potential: Modelling photosynthesis with leaf development. Funct. Plant Biol. 2010, 37, 713–725. [Google Scholar] [CrossRef]
- Hammond, J.P.; White, P.J. Sucrose transport in the phloem: Integrating root responses to phosphorus starvation. J. Exp. Bot. 2008, 59, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.W.; Liu, H.H.; Tao, P.L.; Chen, H. Comparative proteomic analyses provide new insights into low phosphorus stress responses in maize leaves. PLoS ONE 2014, 9, e98215. [Google Scholar] [CrossRef] [PubMed]
- Chivasa, S.; Tomé, D.F.; Hamilton, J.M.; Slabas, A.R. Proteomic analysis of extracellular ATP-regulated proteins identifies ATP synthase beta-subunit as a novel plant cell death regulator. Mol. Cell. Proteom. 2011, 10, M110.003905. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.C.; Zakhleniuk, O.V. Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J. Exp. Bot. 2004, 55, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Morcuende, R.; Bari, R.; Gibon, Y.; Zheng, W.M.; Pant, B.D.; Bla¨sing, O.; Usadel, B.; Czechowski, T.; Udvardi, M.K.; Stitt, M.; et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007, 30, 85–112. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.M.; Li, A.; Feissner, R.; Ahmad, T. RT-qPCR demonstrates light-dependent AtRBCS1A and AtRBCS3B mRNA expressions in Arabidopsis thaliana leaves. Biochem. Mol. Biol. Educ. 2016, 44, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Zhan, G.M.; Li, R.J.; Hu, Z.Y.; Liu, J. Cosuppression of RBCS3B in Arabidopsis leads to severe photoinhibition caused by ROS accumulation. Plant Cell Rep. 2014, 33, 1091–1108. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, M.A.; Maldonado, A.M.; Ogueta, S.; Jorrı´n, J.V. Proteomic analysis of responses to drought stress in sunflower (Helianthus annuus) leaves by 2DE gel electrophoresis and mass spectrometry. Open Proteom. J. 2008, 1, 59–71. [Google Scholar] [CrossRef]
- Singh, R.; Jwa, N.S. Understanding the responses of rice to environmental stress using proteomics. J. Proteome Res. 2013, 12, 4652–4669. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, A.; Rehman, S.; Hiraga, S.; Makino, T.; Komatsu, S. Organ-specific proteomics analysis forresponse mechanism in soybean seedlings under flooding stress. J. Proteom. 2012, 75, 5706–5723. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Hossain, Z. Organ-specific proteome analysis for identification of abiotic stress response mechanism in crop. Front. Plant Sci. 2013, 4, 71. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Karan, R.; Singla-pareek, S.L.; Pareek, A. Ectopic expression of Pokkali phosphoglycerate kinase-2 (OsPGK2-P) improves yield in tobacco plants under salinity stress. Plant Cell Rep. 2016, 35, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Hajano, J.D.; Ren, Y.D.; Lu, C.T.; Wang, X.F. iTRAQ-based quantitative proteomics analysis of rice leaves infected by rice stripe virus reveals several proteins involved in symptom formation. Virol. J. 2015, 12, 99–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cheng, Y.; Zhang, X.; Guan, Q.; Nishiuchi, S.; Hase, K.; Takano, T. Expression of an NADP- malic enzyme gene in rice (Oryza sativa L.) is induced by environmental stresses: Over-expression of the gene in Arabidopsis confers salt and osmotic stress tolerance. Plant Mol. Biol. 2007, 64, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Papenbrock, J.; Schmidt, A. Characterization of two sulfurtransferase isozymes from Arabidopsis thaliana. Eur. J. Biochem. 2000, 267, 5571–5579. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.; Meike, B.; Michael, B.; Jutta, P. Arabidopsis sulfurtransferases: Investigation of their function during senescence and in cyanide detoxification. Planta 2003, 217, 1–10. [Google Scholar] [PubMed]
- Sharma, S.; Mustafiz, A.; Singla-Pareek, S.L.; Srivastava, P.S.; Sopory, S.K. Characterization of stress and methylglyoxal inducible triose phosphate isomerase (OscTPI) from rice. Plant Signal. Behav. 2012, 7, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Sepideh, T.; Matthias, W.; Manzar, H.; Mohammad, R.N.; Salekdeh, G.H.; Komatsu, S. Crop proteomics: Aim at sustainable agriculture of tomorrow. Proteomics 2007, 7, 2976–2996. [Google Scholar]
- Cummins, I.; Cole, D.J.; Edwards, R. A role for glutathione transferases functioning as glutathione peroxidases in resistance to multiple herbicides in black-grass. Plant J. 1999, 18, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Torabi, S.; Wissuwa, M.; Heidari, M.; Naghavi, M.R.; Gilany, K.; Hajirezaei, M.R.; Omidi, M.; Yazdi, S.B.; Ismail, A.M.; Salekdeh, G.H. A comparative proteome approach to decipher the mechanism of rice adaptation to phosphorous deficiency. Proteomics 2009, 9, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Bournier, M.; Tissot, N.; Mari, S.; Boucherez, J.; Lacombe, E.; Briat, J.F.; Gaymard, F. Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 2013, 288, 22670–22680. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.J.; Cao, Y.R.; Chen, J.W.; Wei, W.; Li, Q.T.; Ma, B.; Zhang, W.K.; Chen, S.Y.; Zhang, J.S. Tobacco translationally controlled tumor protein interacts with ethylene receptor tobaccohistidine kinase1 and enhances plant growth through promotion of cell proliferation. Plant Physiol. 2015, 169, 96–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, L.; Chen, X.; Ye, T.; Liu, R.; Wu, Y.; Chan, Z. MLP-like protein 43 (MLP43) functions as a positive regulator during ABA responses and confers drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Zorb, C.; Schmitt, S.; Muhling, K.H. Proteomic changes in maize roots after short-term adjustment to saline growth conditions. Proteomics 2010, 10, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Mikkat, S.; Stegmann, H.; Horn, R. Physiology and proteomics of drought stress acclimation in sunflower (Helianthus annuus L.). Plant Biol. 2011, 13, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Baskin, T.I.; Brwon, K.M.; Lynch, J.P. Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol. 2013, 131, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Roldan, M.; Dinh, P.; Leung, S.; Mcmanus, M.T. Ethylene and the response of plants to phosphate deficiency. AOB Plants 2013, 5, 1–10. [Google Scholar] [CrossRef]
- Nagarajan, V.K.; Smith, A.P. Ethylene’s role in phosphate starvation signaling: More than just a root growth regulator. Plant Cell Physiol. 2012, 53, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Koumoto, Y.; Tsugeki, R.; Shimada, T.; Mori, H.; Kondo, M.; Hara, N.I.; Nishimura, M. Isolation and characterization of a cDNA encoding mitochondrial chaperonin 10 from Arabidopsis thaliana by functional complementation of an Escherichia coli groES mutant. Plant J. 1996, 10, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gallie, D.R. Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol. 2006, 142, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Manna, M.; Reddy, M.K. Glutathione peroxidase of Pennisetum glaucum (PgGPx) is a functional Cd2+ dependent peroxiredoxin that enhances tolerance against salinity and drought stress. PLoS ONE 2015, 23, e0143344. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Cai, X.F.; Xu, C.X.; Wang, Q.H.; Dai, S.J. Drought-responsive mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef] [PubMed]
- Prasad, T.K.; Hack, E.; Hallberg, R.L. Function of the maize mitochondrial chaperonin hsp60: Specific Association between hsp60 and newly synthesized F1-ATPase alpha Subunits. Mol. Cell. Biol. 1990, 10, 3979–3986. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, C.D.; Kreps, J.A.; Simon, A.E. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadianrhythm. Plant Physiol. 1994, 104, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.; Marnef, A.; Cheung, M.K.; Wilson, J.; Hancock, J.; Staiger, D.; Ladomery, M. A Proteomic analysis of oligo(dT)-bound mRNP containing oxidative stress-induced Arabidopsis thaliana RNA-binding proteins ATGRP7 and ATGRP8. Mol. Biol. Rep. 2010, 37, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, S.; Takano, T. Overexpression of a mitochondrial ATP synthase small subunit gene (AtMtATP6) confers tolerance to several abiotic stresses in Saccharomyces cerevisiae and Arabidopsis thaliana. Biotechnol. Lett. 2008, 30, 1289–1294. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.M.; Zhou, Y.; Xiao, J.H.; Li, X.H.; Zhang, Q.F.; Lian, X.M. Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 2009, 28, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Choidu, S.; Lim, C.W.; Hwang, B.K. Proteomics and functional analyses of Arabidopsis nitrilases involved in the defense response to microbial pathogens. Planta 2016, 244, 449–465. [Google Scholar]
- Cao, A.; Jain, A.; Baldwin, J.C.; Raghothama, K.G. Phosphate differentially regulates 14-3-3 family membersand GRF9 plays a role in Pi-starvation induced responses. Planta 2007, 226, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.P.; Lin, H.X.; Chen, S.; Becker, K.; Yang, Y.Q.; Zhao, J.F.; Kudla, J.; Schumaker, K.S.; Guo, Y. Inhibition of the Arabidopsis salt overly sensitive Pathway by 14-3-3 Proteins. Plant Cell 2014, 26, 1166–1182. [Google Scholar] [CrossRef] [PubMed]
- Moroney, J.V.; Bartlett, S.G.; Samuelsson, G. Carbonic anhydrases in plant and algae. Plant Cell Environ. 2011, 24, 141–153. [Google Scholar] [CrossRef]
- Ames, B.N. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966, 8, 115–118. [Google Scholar]
- Solfanelli, C.; Poggi, A.; Loreti, E.; Alpi, A.; Perata, P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006, 140, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Caspar, T.; Lin, T.P.; Kakefuda, G.; Benbow, L.; Preiss, J.; Somerville, C. Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol. 1991, 95, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, G.A.; Knight, J.D.; Vance, C.P.; Allan, D.L. Acid phosphatase activity in phosphorus-deficient white lupin roots. Plant Cell Environ. 1999, 22, 801–810. [Google Scholar] [CrossRef]
- Wang, X.C.; Li, X.F.; Deng, X.; Han, H.P.; Shi, W.L.; Li, Y.X. A protein extraction method compatible with proteomic analysis for the euhalophyte Salicornia europaea. Electrophoresis 2007, 28, 3976–3987. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Xu, S.L.; Oses, J.A.; Putil, S.; Xu, P.; Wang, R.J.; Li, K.H.; Maltby, D.A.; An, L.H.; Burlingame, A.L.; et al. Proteomics analysis reveals post-translational mechanisms for cold-induced metabolic changes in Arabidopsis. Mol. Plant. 2011, 4, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
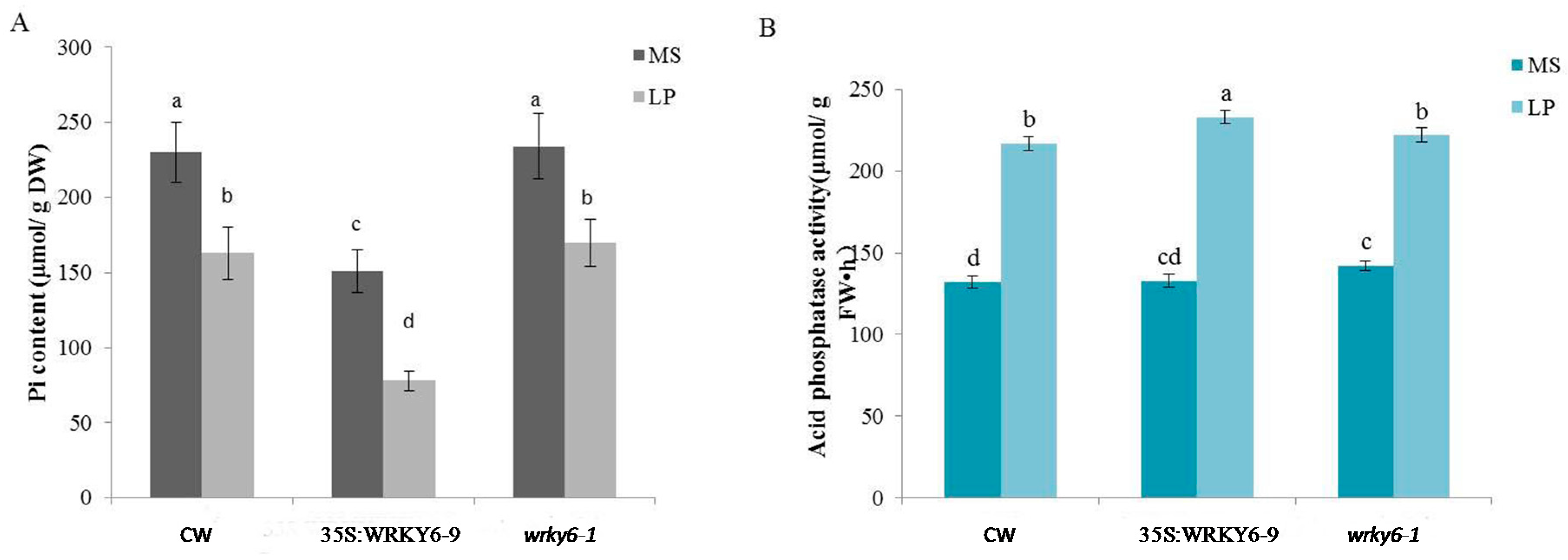
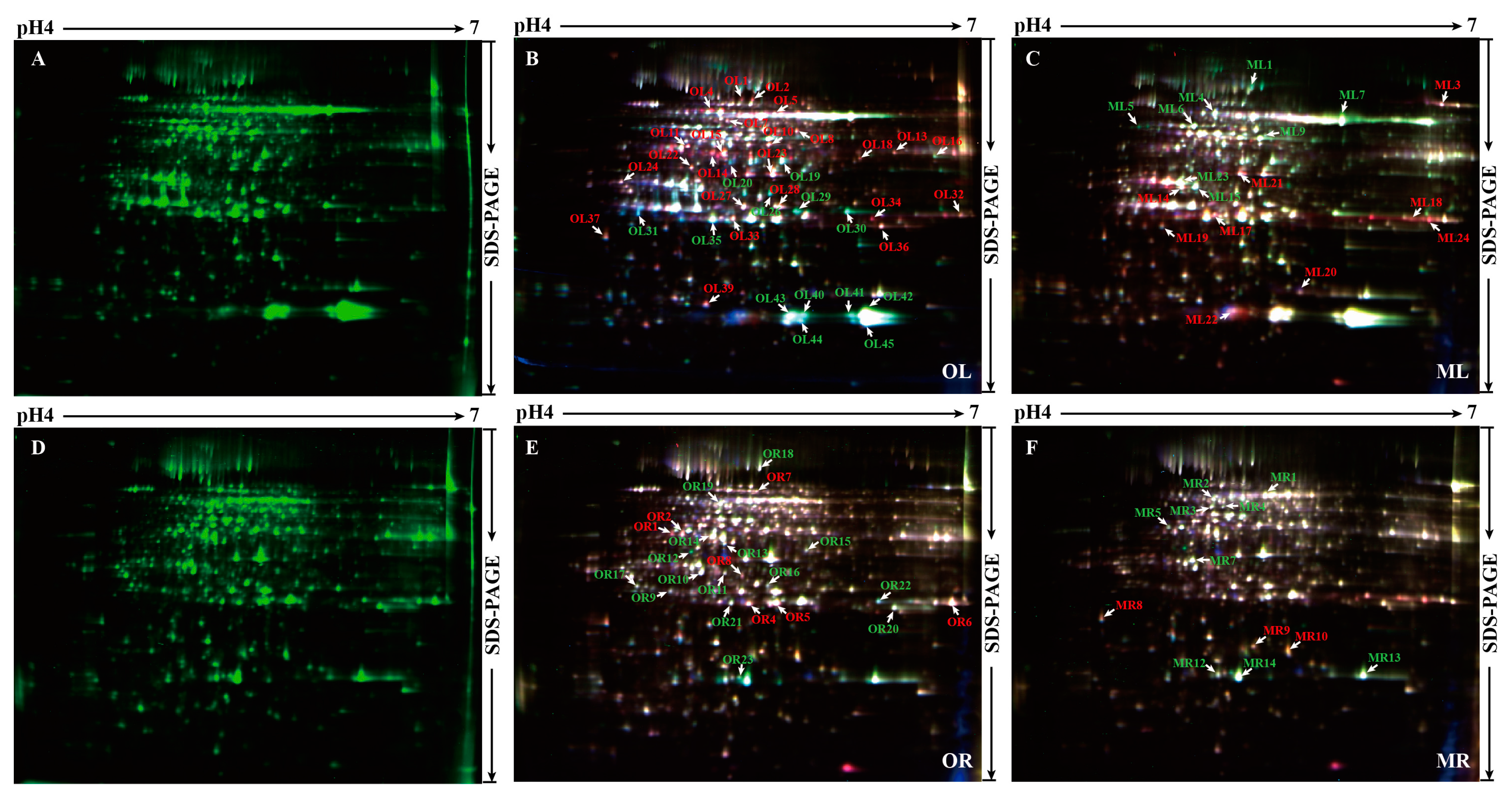

| Samples | Chlorophyll Content (μg/g FW) | Reducing Sugar Content (μg/g FW) | Sucrose Content (mg/g FW) |
|---|---|---|---|
| CW (MS) | 107 ± 8.7 c | 0.56 ± 0.05 b | 10.1 ± 0.98 c |
| 35S:WRKY6-9 (MS) | 132 ± 10.1 b | 0.78 ± 0.06 a | 13.2 ± 0.88 c |
| wrky6-1 (MS) | 113 ± 9.8 c | 0.61 ± 0.05 b | 11.5 ± 1.12 c |
| CW (LP) | 136 ± 10.6 b | 0.83 ± 0.07 a | 28.3 ± 2.22 b |
| 35S:WRKY6-9 (LP) | 155 ± 11.3 a | 0.88 ± 0.09 a | 40.6 ± 3.65 a |
| wrky6-1 (LP) | 142 ± 10.4 a,b | 0.86 ± 0.08 a | 27.8 ± 2.37 b |
| Spot | Name | Score | Version | Pi/Mr (kDa) | Ttest (P) | Cov. (%) | Matched | Change Fold | |
|---|---|---|---|---|---|---|---|---|---|
| Experimental | Theoretical | ||||||||
| OL1 | iPGAM | 105 | AAL09820.1 | 5.48/59 | 5.27/61 | 9.3 × 10−3 | 3 | 2 | 1.82 |
| OL2 | iPGAM | 150 | AAL09820.1 | 5.54/58 | 5.27/61 | 3.1 × 10−5 | 3 | 2 | 2.07 |
| OL4 | ATP synthase CF1 alpha subunit | 200 | NP_051044.1 | 5.29/56 | 5.19/55 | 1.5 × 10−3 | 7 | 3 | 2.82 |
| OL5 | Mitochondrial F1 ATP synthase beta subunit | 339 | CAC81058.1 | 5.65/54 | 6.52/64 | 2.4 × 10−4 | 10 | 5 | 1.58 |
| OL7 | AT4g23100/F7H19_290 | 228 | AAL08228.1 | 5.38/54 | 6.16/59 | 9.7 × 10−5 | 6 | 3 | 1.68 |
| OL8 | S-adenosylmethionine synthetase like protein | 291 | BAF01944.1 | 5.7/54 | 5.60/43 | 5.9 × 10−5 | 10 | 3 | 2.06 |
| OL10 | Phosphoglycerate kinase, putative | 519 | AAM61185.1 | 5.62/41 | 5.49/42 | 5.6 × 10−11 | 24 | 6 | 1.99 |
| OL11 | Glutamine synthetase like protein | 180 | BAE98486.1 | 5.11/44 | 5.14/39 | 5.0 × 10−9 | 3 | 2 | 2.73 |
| OL13 | AT1G04410 | 50 | BAH20341.1 | 5.79/53 | 6.59/26 | 3.4 × 10−8 | 8 | 1 | 1.96 |
| OL14 | Annexin-like protein | 270 | AAC49472.1 | 5.28/41 | 5.34/36 | 6.0 × 10−5 | 17 | 5 | 2.63 |
| OL15 | Annexin, partial | 350 | CAA67608.1 | 5.35/40 | 5.19/36 | 1.6 × 10−4 | 18 | 4 | 2.6 |
| OL16 | AT4G04640 | 299 | BAH20433.1 | 6.68/35 | 5.06/17 | 1.4 × 10−3 | 31 | 3 | 2.12 |
| OL18 | AT3g44310/T10D17_100 | 149 | AAL16248.1 | 6.2/35 | 5.78/39 | 3.6 × 10−3 | 7 | 3 | 2.16 |
| OL19 | AT4g38970/F19H22_70 | 133 | AAL16224.1 | 5.68/36 | 6.79/43 | 7.6 × 10−5 | 7 | 2 | −1.53 |
| OL20 | AT2G21330 | 111 | BAH56881.1 | 5.39/37 | 6.34/41 | 7.3 × 10−8 | 6 | 2 | −2.02 |
| OL22 | Mercaptopyruvate sulfurtransferase 1 | 197 | NP_565203.1 | 5.17/37 | 5.95/42 | 2.1 × 10−1 | 9 | 3 | 2.15 |
| OL23 | AT3G16420 | 576 | BAH57171.1 | 5.64/31 | 5.44/30 | 3.9 × 10−7 | 28 | 6 | 2.2 |
| OL24 | GF14chi isoform | 354 | AAA96254.1 | 4.68/30 | 4.68/30 | 5.8 × 10−8 | 22 | 6 | 1.5 |
| OL26 | AT5G14740 | 367 | BAH20249.1 | 5.61/27 | 5.94/31 | 3.0 × 10−6 | 22 | 4 | −2.69 |
| OL27 | Cytosolic triose phosphate isomerase | 363 | AAA03449.1 | 5.46/23 | 5.24/27 | 3.6 × 10−6 | 20 | 5 | 1.68 |
| OL28 | Cytosolic triose phosphate isomerase | 513 | AAA03449.1 | 5.66/22 | 5.24/27 | 8.0 × 10−7 | 20 | 5 | 1.77 |
| OL29 | AT3G01500 | 388 | BAH56801.1 | 5.75/21 | 6.14/32 | 1.2 × 10−4 | 22 | 5 | −2.98 |
| OL30 | AT3G01500 | 344 | BAH56801.1 | 6.05/20 | 6.14/32 | 5.0 × 10−6 | 22 | 5 | −4.04 |
| OL31 | AT5G54270 | 63 | BAH56768.1 | 4.8/22 | 4.76/18 | 4.5 × 10−5 | 6 | 2 | −1.89 |
| OL32 | Glutathione transferase | 68 | CAA72973.1 | 6.72/19 | 7.03/24 | 3.5 × 10−7 | 5 | 1 | 1.65 |
| OL33 | Ferritin 1 | 78 | BAD94306.1 | 5.39/21 | 4.74/6 | 5.6 × 10−6 | 18 | 1 | 1.55 |
| OL34 | Glutathione S-transferase | 297 | AAG30126.1 | 6.35/20 | 6.31/24 | 4.6 × 10−3 | 18 | 4 | 3.89 |
| OL35 | Ferritin 1 | 107 | AAM61077.1 | 5.27/21 | 5.73/28 | 4.1 × 10−5 | 7 | 2 | −1.55 |
| OL36 | Fe-superoxide dismutase | 159 | AAA32791.1 | 6.48/19 | 6.30/25 | 4.2 × 10−6 | 15 | 3 | 1.71 |
| OL37 | Translationally-controlled tumor protein | 115 | AAM66134.1 | 4.62/19 | 4.52/19 | 2.0 × 10−5 | 11 | 2 | 1.52 |
| OL39 | MLP-like protein 423 | 281 | NP_173813.1 | 5.2/16 | 5.1/17 | 1.3 × 10−8 | 29 | 3 | 2.04 |
| OL40 | Ribulose bisphosphate carboxylase | 261 | CAA32701.1 | 5.72/15 | 7.59/21 | 1.4 × 10−9 | 19 | 3 | −2.26 |
| OL41 | Ribulose bisphosphate carboxylase | 133 | CAA32701.1 | 6.17/15 | 7.59/21 | 5.4 × 10−7 | 14 | 2 | −2.37 |
| OL42 | Ribulose bisphosphate carboxylase | 197 | CAA32701.1 | 5.72/15 | 7.59/21 | 7.7 × 10−6 | 14 | 2 | −2.07 |
| OL43 | Ribulose bisphosphate carboxylase | 68 | CAA32701.1 | 5.76/15 | 7.59/21 | 7.0 × 10−9 | 6 | 1 | −1.71 |
| OL44 | Ribulose bisphosphate carboxylase | 303 | CAA31948.1 | 5.77/15 | 7.59/21 | 1.8 × 10−8 | 19 | 4 | −1.7 |
| OL45 | Ribulose bisphosphate carboxylase | 324 | CAA31948.1 | 6.2/15 | 7.59/21 | 4.6 × 10−8 | 19 | 4 | −1.63 |
| OR1 | PfkB-like carbohydrate kinase family protein | 343 | NP_199996.1 | 5.04/43 | 4.99/37 | 1.31 × 10−2 | 16 | 4 | 1.62 |
| OR2 | ACC oxidase | 254 | AAC27484.1 | 5.13/45 | 4.97/36 | 1.4 × 10−2 | 15 | 3 | 1.37 |
| OR4 | Chaperonin 10 | 111 | AAC14026.1 | 5.52/26 | 8.86/27 | 1.42 × 10−2 | 7 | 2 | 1.27 |
| OR5 | Dehydroascorbate reductase | 356 | NP_173387.1 | 5.68/25 | 5.56/24 | 2.04 × 10−2 | 36 | 5 | 1.25 |
| OR6 | Glutathione S-transferase PHI 9 | 432 | NP_180643.1 | 6.78/25 | 6.17/24 | 2.66 × 10−2 | 23 | 6 | 1.23 |
| OR7 | Mitochondrial chaperonin (HSP60) | 260 | AAC04902.1 | 5.55/61 | 5.30/56 | 2.66 × 10−2 | 11 | 4 | 1.23 |
| OR8 | TSA1 | 219 | OAP06349.1 | 5.47/32 | 7.68/33 | 2.96 × 10−2 | 13 | 2 | 1.21 |
| OR9 | Putative cysteine proteinase AALP | 161 | AAN31820.1 | 5.03/28 | 6.26/39 | 3.22 × 10−2 | 9 | 3 | −1.21 |
| OR10 | Putative lactoylglutathione lyase | 292 | AAL07227.1 | 5.21/32 | 5.11/32 | 3.26 × 10−2 | 16 | 5 | −1.22 |
| OR11 | Glyoxalase II | 455 | AAB17995.1 | 5.37/33 | 5.58/28 | 3.27 × 10−2 | 24 | 5 | −1.22 |
| OR12 | Mercaptopyruvate sulfurtransferase 1 | 343 | NP_565203.1 | 5.19/39 | 5.95/42 | 3.37 × 10−2 | 12 | 4 | −1.22 |
| OR13 | Transketolase family protein | 409 | NP_199898.1 | 5.36/40 | 5.67/39 | 3.61 × 10−2 | 18 | 5 | −1.24 |
| OR14 | MLP-like protein 34 | 329 | NP_850976.1 | 5.3/43 | 5.14/36 | 3.61 × 10−2 | 20 | 6 | −1.25 |
| OR15 | NmrA-like negative transcriptional regulator | 336 | NP_565107.1 | 5.85/38 | 5.66/34 | 3.61 × 10−2 | 18 | 4 | −1.26 |
| OR16 | GSTU26 | 106 | OAP16650.1 | 5.63/30 | 5.53/26 | 3.62 × 10−2 | 8 | 2 | −1.26 |
| OR17 | RCI1B | 58 | CAA52238.1 | 4.8/29 | 4.97/28 | 4.46 × 10−2 | 6 | 1 | −1.29 |
| OR18 | Mannose-binding lectin superfamily protein | 560 | NP_188267.1 | 5.58/74 | 5.31/72 | 4.59 × 10−2 | 11 | 6 | −1.31 |
| OR19 | F23N19.3 | 334 | AAF19535.1 | 5.33/55 | 7.26/82 | 4.64 × 10−2 | 7 | 5 | −1.31 |
| OR20 | GSTF8 | 341 | OAP11759.1 | 6.39/24 | 8.50/29 | 4.72 × 10−2 | 22 | 6 | −1.44 |
| OR21 | Ferritin 1 | 384 | AAM61077.1 | 5.4/26 | 5.73/28 | 4.8 × 10−2 | 14 | 5 | −1.44 |
| OR22 | Glutathione S transferase | 526 | AAM63854.1 | 6.28/26 | 6.08/24 | 4.81 × 10−2 | 36 | 6 | -1.6 |
| OR23 | Glycine-rich RNA-binding protein 8 | 528 | NP_195637.1 | 5.46/15 | 5.58/17 | 4.82 × 10−2 | 37 | 4 | −2.16 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.-Q.; Huang, L.-P.; Pan, G.; Liu, L.; Wang, X.-Y.; Lu, L.-M. Identifying the Genes Regulated by AtWRKY6 Using Comparative Transcript and Proteomic Analysis under Phosphorus Deficiency. Int. J. Mol. Sci. 2017, 18, 1046. https://doi.org/10.3390/ijms18051046
Li L-Q, Huang L-P, Pan G, Liu L, Wang X-Y, Lu L-M. Identifying the Genes Regulated by AtWRKY6 Using Comparative Transcript and Proteomic Analysis under Phosphorus Deficiency. International Journal of Molecular Sciences. 2017; 18(5):1046. https://doi.org/10.3390/ijms18051046
Chicago/Turabian StyleLi, Li-Qin, Lu-Ping Huang, Gang Pan, Lun Liu, Xi-Yao Wang, and Li-Ming Lu. 2017. "Identifying the Genes Regulated by AtWRKY6 Using Comparative Transcript and Proteomic Analysis under Phosphorus Deficiency" International Journal of Molecular Sciences 18, no. 5: 1046. https://doi.org/10.3390/ijms18051046






