Pnserpin: A Novel Serine Protease Inhibitor from Extremophile Pyrobaculum neutrophilum
Abstract
:1. Introduction
2. Results
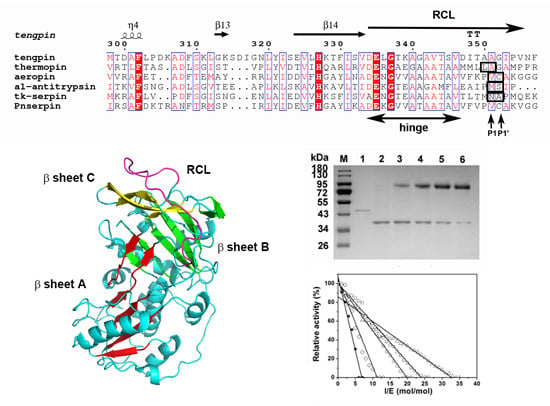
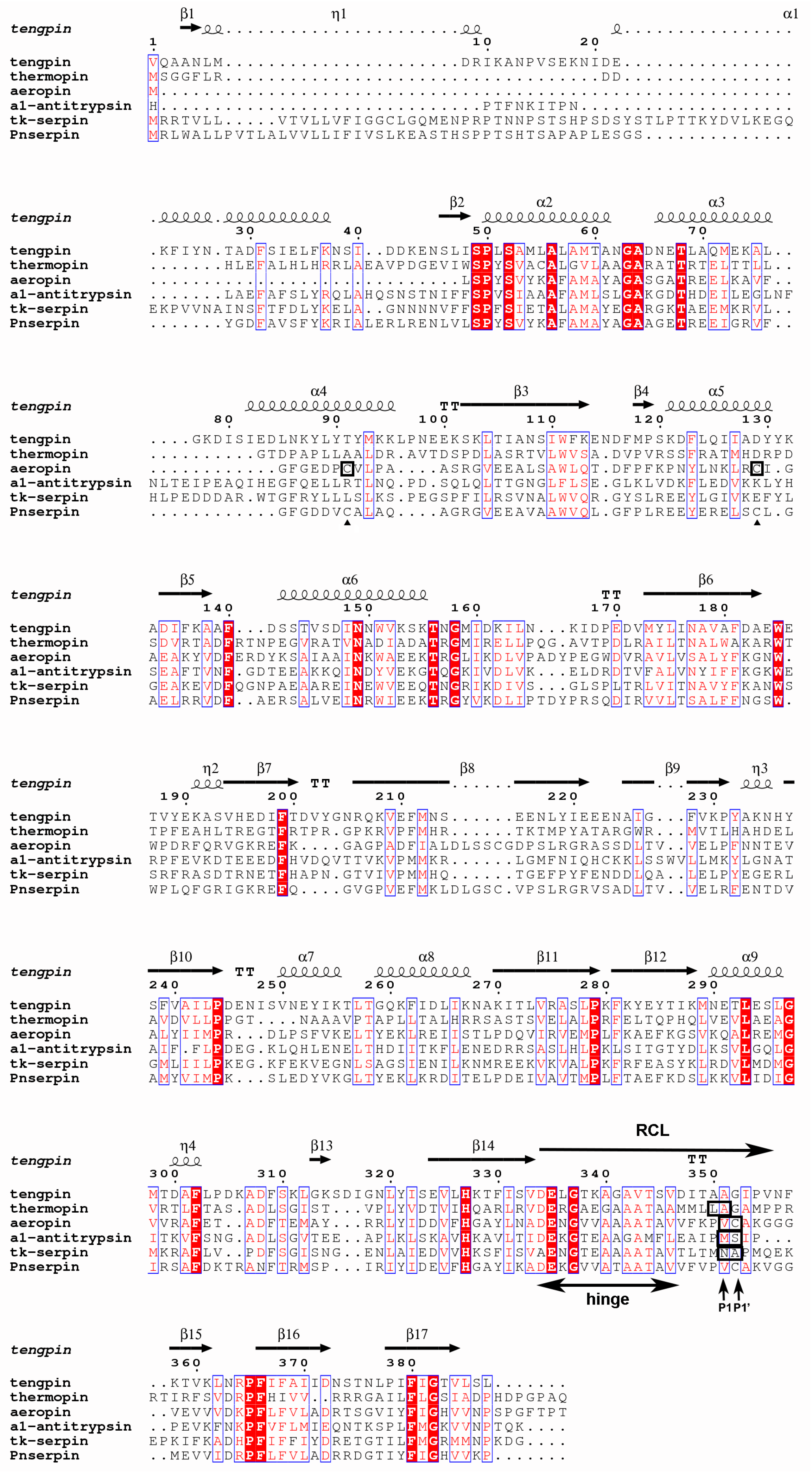
2.1. Sequence Analysis of Pnserpin
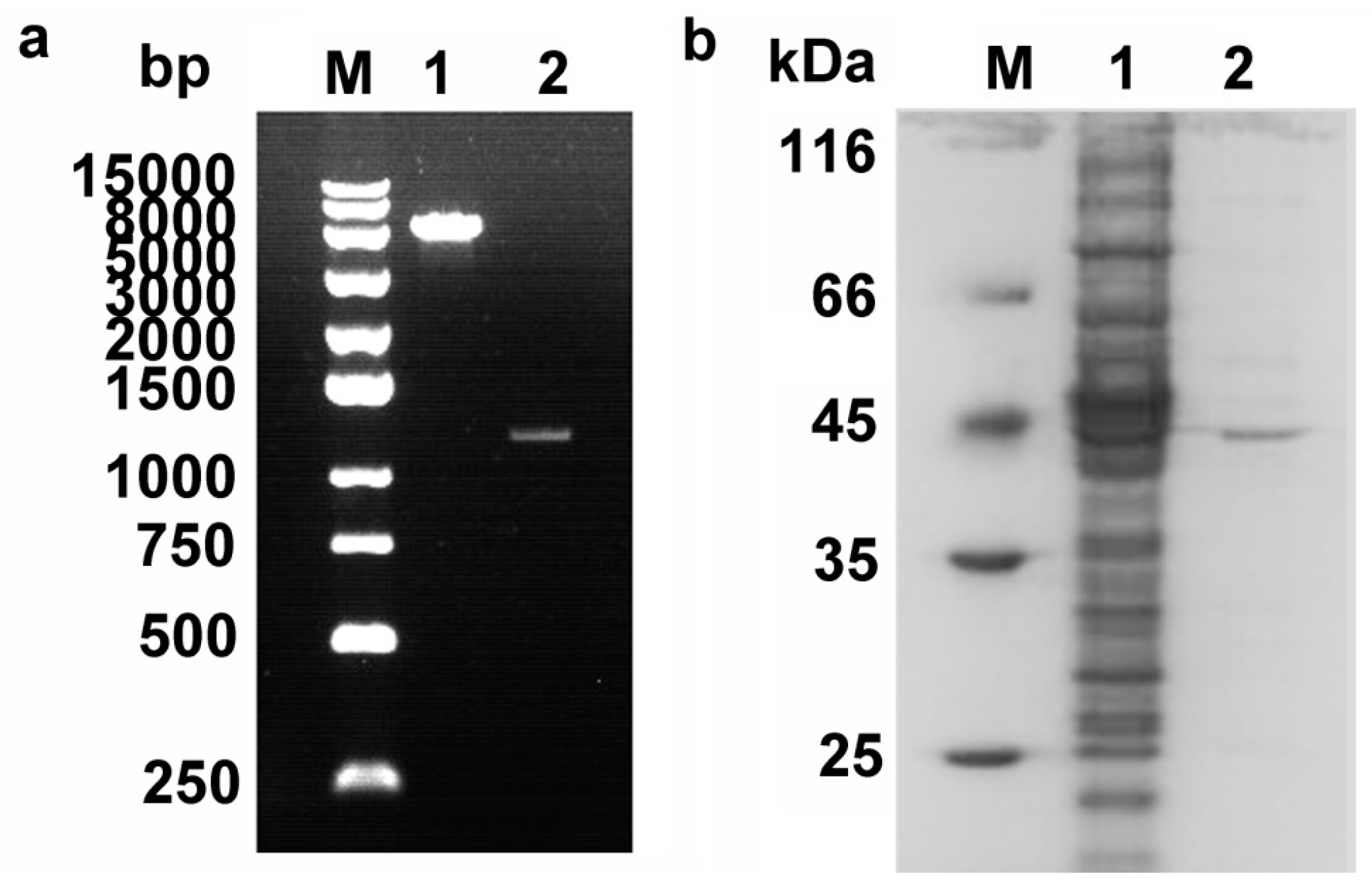
2.2. Cloning, Expression, and Purification of Pnserpin
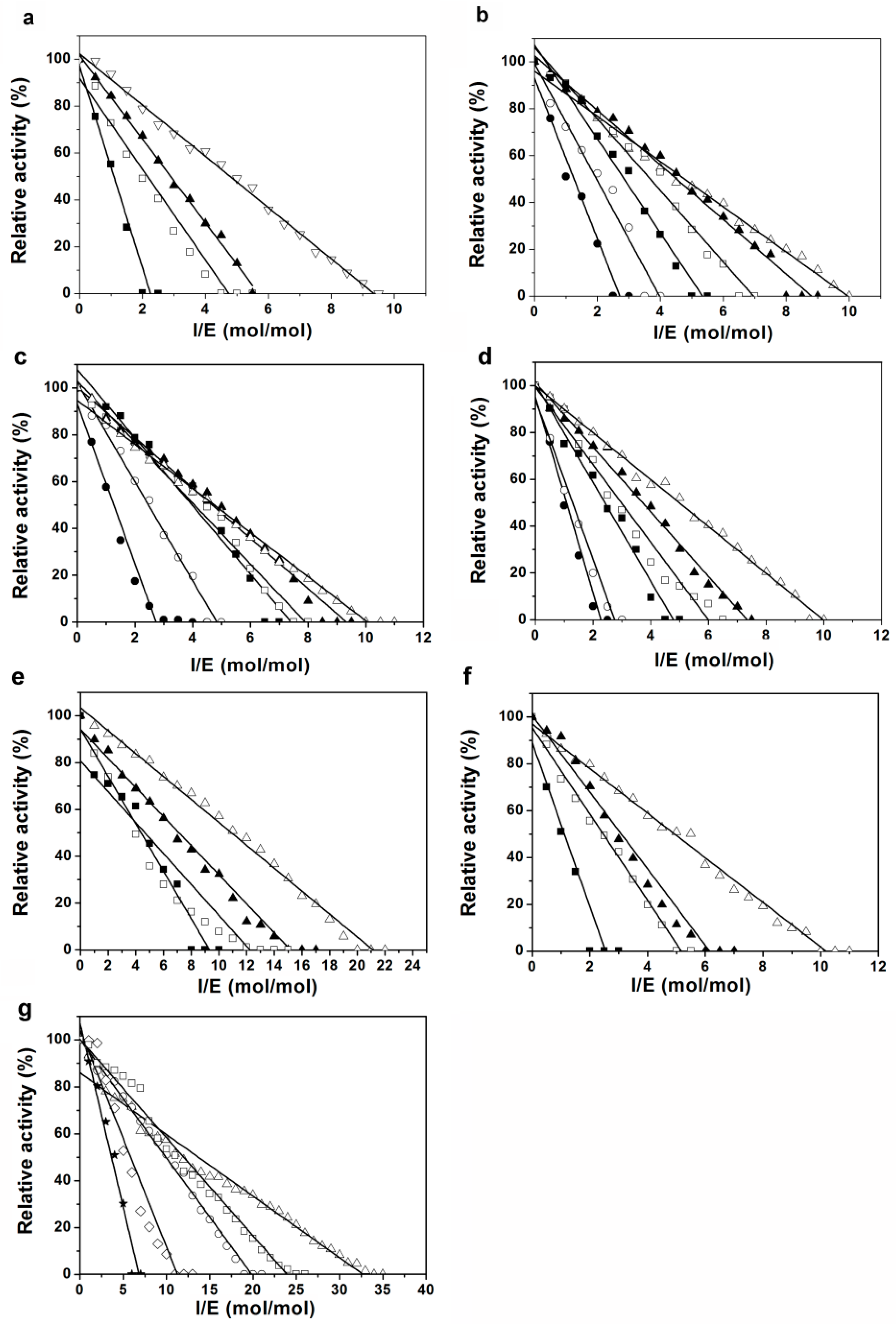
2.3. Inhibition of Proteases by Pnserpin
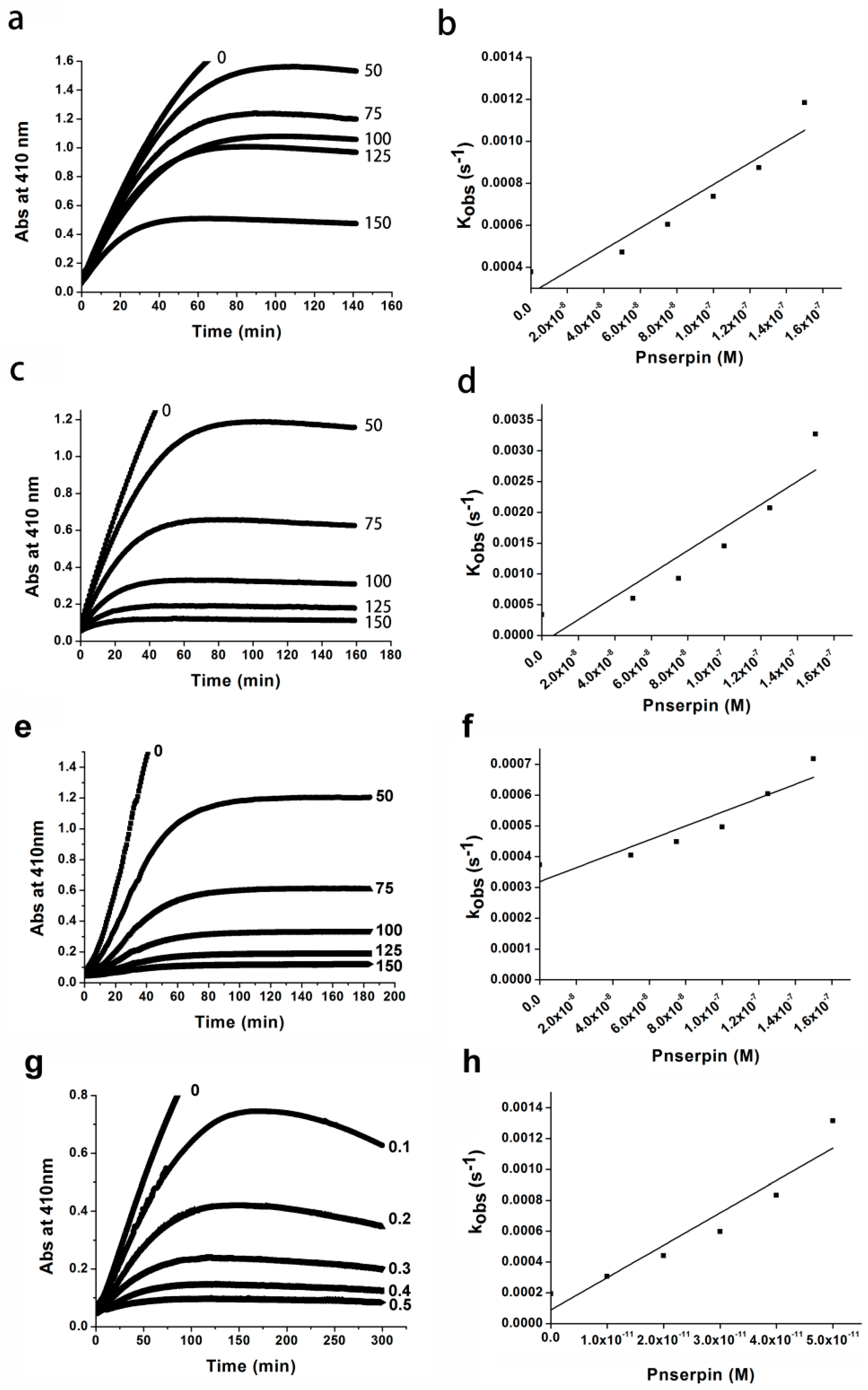
2.4. Association Rate Constant of Pnserpin
2.5. The Temperature and pH Stabilities of Pnserpin
2.6. Formation of Covalent Complex between Pnserpin and Target Proteases
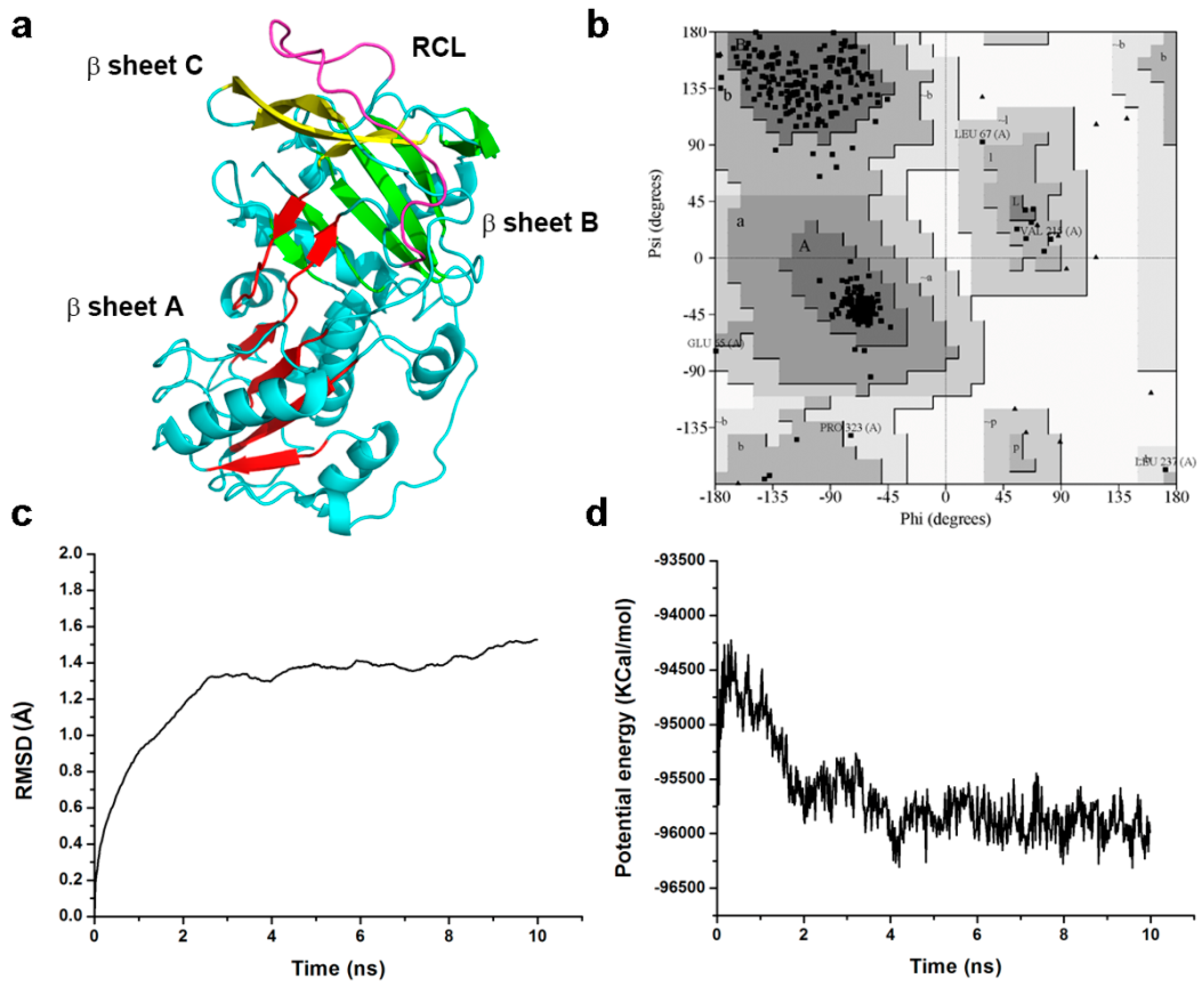
2.7. Homology Modeling and Molecular Dynamic Simulation
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression and Purification of Pnserpin and PnCHT Genes in E. coli
4.2. Stoichiometry of Inhibition
4.3. Kinetics of Inhibition
4.4. Temperature and pH Stabilities of Pnserpin
4.5. SDS-PAGE Analysis of the Interaction between Pnserpinand Target Proteases
4.6. Homology Modeling and Validation of the Model Structure
4.7. Molecular Dynamics Simulation
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Irving, J.A.; Pike, R.N.; Lesk, A.M.; Whisstock, J.C. Phylogeny of the serpin superfamily: Implications of patterns of amino acid conservation for structure and function. Genome Res. 2000, 10, 1845–1864. [Google Scholar] [CrossRef] [PubMed]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, G.M. Role of antithrombin concentrate in treatment of hereditary antithrombin deficiency. An update. Thromb. Haemost. 2009, 101, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.M.; Tuder, R. α 1 anti-trypsin: One protein, many functions. Curr. Mol. Med. 2012, 12, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Janciauskiene, S.M.; Bals, R.; Koczulla, R.; Vogelmeier, C.; Köhnlein, T.; Welte, T. The discovery of α1-antitrypsin and its role in health and disease. Respir. Med. 2011, 105, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Tong, Y.; Yu, D.; Mao, M. Tissue plasminogen activator-independent roles of neuroserpin in the central nervous system. Neural Regen. Res. 2012, 7, 146–151. [Google Scholar] [PubMed]
- Yepes, M.; Lawrence, D.A. Neuroserpin: A selective inhibitor of tissue-type plasminogen activator in the central nervous system. Thromb. Haemost. 2004, 91, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Chen, H.; Davids, J.; Bryant, M.; Lucas, A. Serpins for diagnosis and therapy in cancer. Cardiovasc. Hematol. Disord. Drug Targets 2013, 13, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Jaadane, I.; Nagbou, A.; Behar-Cohen, F.; Torriglia, A. Interaction of Leukocyte Elastase Inhibitor/L-DNase II with BCL-2 and BAX. Biochim. Biophys. Acta 2014, 1843, 2807–2815. [Google Scholar] [CrossRef] [PubMed]
- Morito, D.; Nagata, K. ER Stress Proteins in Autoimmune and Inflammatory Diseases. Front. Immunol. 2012, 3, 48. [Google Scholar] [CrossRef] [PubMed]
- Huntington, J.A. Serpin structure, function and dysfunction. J. Thromb. Haemost. 2011, 9, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Devlin, G.L.; Bottomley, S.P. A protein family under “stress”—Serpin stability, folding and misfolding. Front. Biosci. 2005, 10, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Whisstock, J.C.; Bottomley, S.P. Molecular gymnastics: Serpin structure, folding and misfolding. Curr. Opin. Struct. Biol. 2006, 16, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.L.; Chae, Y.K.; Jung, C.H.; Kim, M.J.; Na, Y.R.; Kim, Y.H.; Kang, S.J.; Im, H. The native metastability and misfolding of serine protease inhibitors. Protein Pept. Lett. 2005, 12, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Irving, J.A.; Cabrita, L.D.; Rossjohn, J.; Pike, R.N.; Bottomley, S.P.; Whisstock, J.C. The 1.5 Å crystal structure of a prokaryote serpin: Controlling conformational change in a heated environment. Structure 2003, 11, 387–397. [Google Scholar] [CrossRef]
- Fulton, K.F.; Buckle, A.M.; Cabrita, L.D.; Irving, J.A.; Butcher, R.E.; Smith, I.; Reeve, S.; Lesk, A.M.; Bottomley, S.P.; Rossjohn, J.; et al. The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition. J. Biol. Chem. 2005, 280, 8435–8442. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Buckle, A.M.; Law, R.H.; Pearce, M.C.; Cabrita, L.D.; Lloyd, G.J.; Irving, J.A.; Smith, A.I.; Ruzyla, K.; Rossjohn, J.; et al. The N terminus of the serpin, tengpin, functions to trap the metastable native state. EMBO Rep. 2007, 8, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Law, R.H.; Bottomley, S.P.; Whisstock, J.C.; Buckle, A.M. A structural basis for loop C-sheet polymerization in serpins. J. Mol. Biol. 2008, 376, 1348–1359. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Koga, Y.; Takano, K.; Kanay, S. Inhibition of chymotrypsin- and subtilisin-like serine proteases with Tk-serpin from hyperthermophilicarchaeon Thermococcus kodakaraensis. Biochim. Biophys. Acta 2011, 1814, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, L.D.; Irving, J.A.; Pearce, M.C.; Whisstock, J.C.; Bottomley, S.P. Aeropin from the extremophile Pyrobaculum aerophilum bypasses the serpin misfolding trap. J. Biol. Chem. 2007, 282, 26802–26809. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Cozen, A.E.; Lowe, T.M. Reclassification of Thermoproteus neutrophilus Stetter and Zillig 1989 as Pyrobaculum neutrophilum comb. nov. based on phylogenetic analysis. Int. J. Syst. Evol. Microbiol. 2013, 63, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, P.C.; Carrel, R.W.; Stone, S.R. Effects of mutations in the hinge region of serpins. Biochemistry 1993, 32, 7650–7657. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new END script server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Chmelar, J.; Oliveira, C.J.; Rezacova, P.; Francischetti, I.M.; Kovarova, Z.; Pejler, G.; Kopacek, P.; Ribeiro, J.M.; Mares, M.; Kopecky, J.; et al. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood 2011, 117, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thorntonl, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Luthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Profile-3D User Guide; Accelrys Inc.: San Diego, CA, USA, 1999.
- Jaenicke, R. Do ultrastable proteins from hyperthermophiles have high or low conformational rigidity? Proc. Natl. Acad. Sci. USA 2000, 97, 2962–2964. [Google Scholar] [CrossRef] [PubMed]
- Fields, P.A. Protein function at thermal extremes: Balancing stability and flexibility. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2001, 129, 417–431. [Google Scholar] [CrossRef]
- Radestock, S.; Gohlke, H. Exploiting the link between protein rigidity and thermostability for data-driven protein engineering. Eng. Life Sci. 2008, 8, 507–522. [Google Scholar] [CrossRef]
- Ksiazek, M.; Mizgalska, D.; Enghild, J.J.; Scavenius, C.; Thogersen, I.B.; Potempa, J. Miropin, a novel bacterial serpin from the Periodontopathogen Tannerella forsythia, inhibits a broad range of proteases by using different peptide bonds within the reactive center loop. J. Biol. Chem. 2015, 290, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Schechter, N.M.; Plotnick, M.I. Measurement of the kinetic parameters mediating protease-serpin inhibition. Methods 2004, 32, 159–168. [Google Scholar] [CrossRef]
- Morrison, J.F.; Walsh, C.T. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988, 61, 201–301. [Google Scholar] [PubMed]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]



| Temperature | ||||||||
|---|---|---|---|---|---|---|---|---|
| Enzyme | 20 °C | 30 °C | 40 °C | 50 °C | 60 °C | 70 °C | 80 °C | 100 °C |
| CHT | 9.49 | 5.80 | 4.76 | 2.21 | - a | - a | - a | - a |
| SUC | 10.04 | 8.76 | 6.95 | 5.34 | 3.97 | 2.67 | - a | - a |
| elastase | 10.06 | 9.33 | 7.9 | 7.41 | 4.84 | 2.74 | - a | - a |
| PRK | 9.99 | 7.39 | 5.99 | 4.69 | 2.75 | 2.28 | - a | - a |
| thrombin | 21.13 | 15.16 | 12.22 | 9.31 | - a | - a | - a | - a |
| trypsin | 10.19 | 6.14 | 5.16 | 2.53 | - a | - a | - a | - a |
| PnCHT | 32.72 | - a | 23.92 | - a | 19.83 | - a | 11.26 | 6.82 |
| Program | Results | |
|---|---|---|
| PROCHECK | Residues in most favored regions | 92.3% |
| Residues in additional allowed regions | 6.4% | |
| Residues in generously allowed regions | 1.2% | |
| Residues in disallowed regions | 0% | |
| Profile_3D | Verify score | 169.8 |
| Verify expected high score | 170.9 | |
| Verify expected low score | 76.9 | |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Fei, R.; Xue, B.; Yu, S.; Zhang, Z.; Zhong, S.; Gao, Y.; Zhou, X. Pnserpin: A Novel Serine Protease Inhibitor from Extremophile Pyrobaculum neutrophilum. Int. J. Mol. Sci. 2017, 18, 113. https://doi.org/10.3390/ijms18010113
Zhang H, Fei R, Xue B, Yu S, Zhang Z, Zhong S, Gao Y, Zhou X. Pnserpin: A Novel Serine Protease Inhibitor from Extremophile Pyrobaculum neutrophilum. International Journal of Molecular Sciences. 2017; 18(1):113. https://doi.org/10.3390/ijms18010113
Chicago/Turabian StyleZhang, Huan, Rui Fei, Baigong Xue, Shanshan Yu, Zuoming Zhang, Sheng Zhong, Yuanqi Gao, and Xiaoli Zhou. 2017. "Pnserpin: A Novel Serine Protease Inhibitor from Extremophile Pyrobaculum neutrophilum" International Journal of Molecular Sciences 18, no. 1: 113. https://doi.org/10.3390/ijms18010113