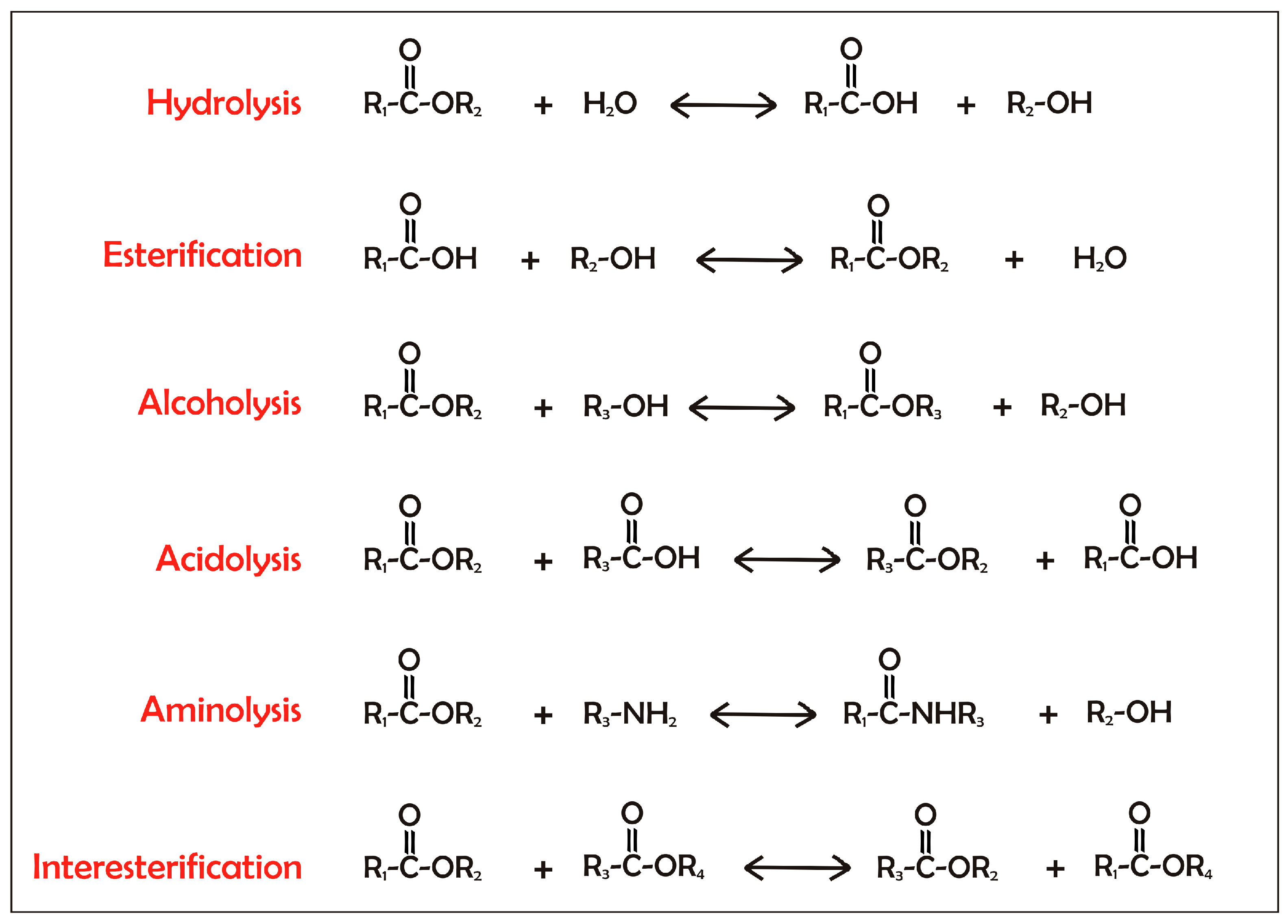
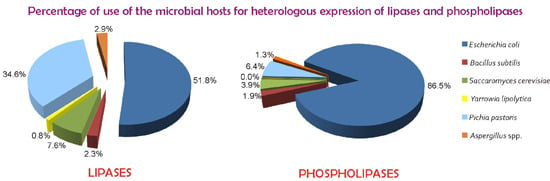
Over the last two decades, there has been a tremendous increase in the significance of biotechnological applications of lipases and phospholipases. The use of these enzymes in industrial processes has grown hand-in-hand with the ability to clone and express their genes in heterologous hosts so as to obtain commercially relevant amounts of these enzymes. In addition, the industrial use of both lipases and phospholipases is increasing due to the optimization of their properties through protein engineering.
Lipases and phospholipases produced by the heterologous host can be used as free enzymes after purification from the producing organism, or they can be used while still contained within the producing organism. Both free enzymes and whole-cell systems can be immobilized on suitable matrices or supports. However, for some applications of immobilized lipases, the relatively high cost associated with the usage of an expensive support material becomes prohibitive for industrial uses. Therefore, further development of a less expensive immobilization process technology is necessary for the use within low margin applications, such as oils and fats industry, as compared to high margin applications, such as enantio-selective synthesis of pharmaceutical intermediates [
200]. On the other side, immobilization has the advantage to increase the chance of enzyme/whole cell re-usability, and eases the separation of the biocatalyst from the products. In some cases, this also leads to an improvement in the activity, specificity and stability of the enzyme, thereby allowing its application to industrial processes that are carried out under relatively harsh conditions [
201]. All these advantages may allow reducing the cost in the use of an immobilized biocatalyst; this, together with a low cost of recombinant enzyme production, can render immobilisation attractive for a wider range of process applications. In recent years, yeast cells that display the recombinant enzyme on their cell surface have been emerging as whole-cell systems that are more suitable for applications in biocatalysis. Indeed, enzyme-displaying yeast cells have the typical advantage of whole-cell systems, that is the avoidance of the need for enzyme purification from the producing microorganism, and also present the advantage to be directly used as biocatalysts without the additional step of immobilization, as displaying the enzyme on the cell surface of the host cell represents a type of self-immobilization.
An overview is given below relating to the efforts that have been made by various research groups to develop recombinant lipases and phospholipases that meet the specific needs of these enzymes in industrial processes. The picture that emerges from the findings is that despite the high potential of lipases and phospholipases, their large-scale application is to date mainly limited to the dairy, baking and detergent industries, with more than 50 enzymes fully developed and commercialized in the past 25 years. As a consequence, current research efforts in these application areas are limited. On the contrary, the large-scale application of recombinant lipases and phospholipases to other industrial processes, such as biodiesel production, oil degumming, and synthesis of flavor compounds and nutraceuticals, is still under development, and extensive research is in progress to develop full heterologous expression systems and design tailor-made enzymes that allow the drawbacks that limit their use in such relevant industrial processes to be overcome.
5.1. The Biodiesel Industry
Biodiesels can be produced from non-edible or low-value feedstocks, such as animal fats, vegetable and microalgal oils, waste products of vegetable oil refineries and animal rendering, and used cooking oils. They represent one of the most promising sustainable alternatives to decrease dependence on traditional fossil fuel [
202]. Biodiesels are a mixture of FAAEs that are usually produced by esterification of FFAs or transesterification of TAGs. Currently, the world supply of biodiesels comes almost exclusively from base-catalyzed transesterification of TAGs with short-chain alcohols, such as methanol and ethanol. However, this chemical process has some drawbacks, as it requires downstream purification to neutralize the catalyst and to separate the biodiesel and glycerol. This latter process is complicated by the large amount of soaps generated during the reaction. A downstream process is also needed for the treatment of the large quantities of wastewater produced from the process. Moreover, alkali-catalyzed transesterification is very sensitive with respect to the feedstock purity, as it needs raw materials that are essentially free of phosphatides, FFAs and water [
203]. For all of these reasons, this alkali-catalyzed transesterification represents an expensive and nonecologically friendly process.
Lipase-catalyzed transesterification is becoming more and more attractive, as this has several advantages compared to chemical catalysis. Indeed, lipases can catalyze not only TAG transesterification reactions, but also FFA esterification with an alcohol, thus making it possible to produce biodiesel from used oils or fats with high FFA levels. Moreover, lipases produce FAAEs without forming soaps, thus allowing easier glycerol recovery and fuel purification; they are also highly selective, require mild reaction conditions, and produce fewer side products and low amounts of wastewater. Among the commercially available lipases, the most widely used in biodiesel production are free lipases from
P. fluorescens (Lipase AK, Amano),
B. cepacia (Lipase PS, Amano), and
T. lanuginosus (Lipase LA201 and Lipopan 50BG, Novozymes), and immobilized lipases from
T. lanuginosus (Lipozyme TL IM, Novozymes) and
R. miehei (Lipozyme RM IM, Novozymes) [
88]. However, the high cost of lipase production is the major obstacle to the use of this enzyme for industrial-scale biodiesel production. As a consequence, biodiesel derived from chemical transformations still dominates the current global market, whereas the application of lipases on an industrial scale has been achieved only in China, where the first plant for biodiesel production using lipases came into operation in 2006. Recently, the successful application of lipases to biodiesel production was also demonstrated by a pilot-scale plant that came into operation in Brazil, which is considered a leader in the biodiesel economy [
204].
Among recombinant lipases, CALB expressed in
A. niger and immobilized onto an acrylic macroporous resin, that is commercially known as Novozym 435 (Novozymes), is the lipase that is most widely used for the production of biodiesel [
88]. This recombinant lipase has been successfully used in conventional biodiesel production, as well as in biodiesel production using isopropanolysis of soybean oil, simultaneous synthesis of biodiesel and glycerol carbonate from corn oil using dimethyl carbonate as the acyl acceptor, and production of so-called “Ecodiesel”, which contains a mixture of MAGs and FAAEs that are obtained through partial ethanolysis of TAGs in sunflower oil [
205].
In addition to Novozym 435, several other recombinant lipases have been produced using different host organisms, and these have been successfully applied to biodiesel production (
Table 4). Lipases expressed in the
E. coli host system have been used almost exclusively in their purified form, as free or immobilized enzymes. Among these, the new variant of
Proteus mirabilis lipase is worth mention, which was developed by means of a directed evolution approach by Korman and coworkers [
206], and is called Dieselzyme 4. This enzyme was found to be able to convert canola oil into biodiesel with high yields (76% and 100% in the first and second cycle, respectively). Moreover, compared to the WT enzyme, the Dieselzyme 4 variant presented an improved methanol tolerance that was even more evident when the enzyme was immobilized and reused. Indeed, while the native
P. mirabilis lipase lost nearly all of its activity in the second cycle of transesterification, the new variant completely retained its activity even after a fourth cycle of transesterification.
Only in a few cases have
E. coli-expressing lipases been used in biodiesel production as a whole-cell biocatalyst (
Table 4).
E. coli expressing lipK107, which is a lipase produced from a soil-bacteria strain of
Proteus sp., represents the first example of an
E. coli-expressing lipase applied to biodiesel production as a whole-cell biocatalyst [
207]. This system produced yields of about 100% at a temperature of 15 °C, which is the lowest temperature among all of the known catalysts used in biodiesel production. An
E. coli whole-cell system that co-expresses CALB and TLL has also been used to produce biodiesel from waste grease, and in this case, FFA esterification and TAG transesterification were simultaneously achieved with a yield of 87% and 95% using wet and dry cells, respectively [
208].
Recombinant lipases produced by the yeasts
S. cerevisiae and
P. pastoris have been used as crude supernatants, purified enzymes, and whole-cell systems (
Table 4). An interesting application of crude lipases to directly catalyze biodiesel production is that reported by Guan and coworkers [
209]. In this study, the 1,3-specific RML and the MAG- and DAG-specific lipase from
P. cyclopium (MDL) were separately expressed in
P. pastoris, and the supernatant of the two fermentation broths was used to catalyze the transesterification of soybean oil with methanol. The results obtained demonstrated that the transesterification yield of RML assisted by MDL was higher than that of RML alone (95%
versus 68.5%, respectively), thus suggesting that MDL addition is helpful to assist RML in the hydrolysis of both MAGs and DAGs.
Recombinant lipases purified from their expressing yeast systems have been used as free or immobilized enzymes, and these include CALB, ROL, RML and lipZ03, where this last is a lipase that is encoded by a gene isolated from an oil-contaminated soil-derived metagenomic library. For CALB, different synthetic variants have been generated using automated PCR, and these have been tested for biodiesel production from corn and soybean oil. In particular, Hughes and coworkers [
210] fused the CALB gene with the sequence encoding the Lyt-1 peptide, to facilitate the movement of the enzyme out of the
S. cerevisiae cell. As expected, the supernatant of the yeast cells expressing this CALB Lyt-1 fusion protein had a specific activity almost 4-fold higher compared to the supernatant of the yeast cells expressing CALB alone. Both the purified CALB Lyt-1 immobilized on a resin support and the intact yeast cells expressing CALB Lyt-1 had specific activities greater than Novozym 435 (80 and 160 µg FAEEs/mg lipase
versus 40 µg FAEEs/mg lipase, respectively). With the same approach, Hughes and coworkers [
211] obtained a truncated variant of CALB that lacked the initial 34 amino-acid residues. This truncated CALB, once expressed and purified from
P. pastoris, was immobilized on a resin support and tested for biodiesel production from corn and soybean oils, and it was shown to have a specific activity 36% and 50% higher, respectively, than Novozym 435. High yields (>90%) have also been obtained using recombinant RML [
212,
213] and ROL [
214] expressed in
P. pastoris and used in their free and immobilized forms, respectively. This was achieved by inducing high expression levels; in the first case, with the introduction of two copies of the
RML gene and, in the second case, with the replacement of the prosequence of ROL with that of RCL.
Among the lipase-producing yeasts that have been used as whole-cell systems, of note, there is the dual biocatalytic system that couples lipase production and
in-situ biodiesel synthesis [
215]. This system was based on a recombinant
P. pastoris strain transformed with a heterologous lipase-encoding gene from
T. lanuginosus, which can simultaneously grow, overexpress the lipase, and efficiently catalyze biodiesel production from waste cooking oils with an 87% yield. However, among the yeast whole-cell systems, the most promising biocatalyst for the production of biodiesel on an industrial scale has been with those systems that have the lipase on the cell surface. Cell-surface-displayed lipases were used for the first time in biodiesel production by Matsumoto and coworkers [
216], who displayed ROL on the cell surface of
S. cerevisiae cells. This system successfully catalyzed the methanolysis reaction with yields higher than 78% after 72 h of reaction. A RML-displaying
P. pastoris whole-cell biocatalyst has also been developed and used for biodiesel production from soybean oil [
217]. This system reached a yield of 83% after 72 h, which was higher than that previously obtained using ROL-displaying
S. cereviasiae. Moreover, the RML displayed on
P. pastoris cells had good storage stability and maintained more than 80% of its original activity even after 10 cycles of transesterification. Recently, to improve biodiesel production, RML displayed on
P. pastoris cells has been used in combination with CALB, which was also on the
P. pastoris cells [
218]. When used for transesterification of different feedstocks, these two displayed lipases reached yields that were higher than those for the RML-displaying
P. pastoris cells alone, and the time to obtain maximum yield was shortened from 72 to 12 h. Compared to the lipase-displaying systems previously used, this new combined system showed an improved stability, as its activity remained almost unchanged after storage for 1 year at room temperature, and the yield remained >85% after 20 repeated batch cycles.
The lipase-expressing
A. oryzae has been used in biodiesel production exclusively as a whole-cell biocatalyst immobilized on biomass support particles. For the first time Hama and coworkers [
219] applied immobilized
A. oryzae cells transformed with the gene that encodes the FHL in the enzymatic production of biodiesel. Since then, there have been several reports on the use of immobilized
A. oryzae cells that express FHL in biodiesel production, using oils from different sources [
220,
221,
222]. To improve the efficiency of the expression system, different strategies have been used, such as multiple copy insertion of the expression cassette [
220] and the use of the improved enolase promoter (P-enoA124) together with the 5′-UTR of a heat-shock protein [
221]. Thus, high yields (90%–100%) have been achieved in biodiesel production, and these systems have also shown to maintain high lipase activity upon reuse. Adachi and coworkers [
223,
224] also immobilized
A. oryzae cells that expressed CALB and a thermostable and solvent-tolerant lipase from
Geobacillus thermocatenulatus; this latter system reached nearly 100% yield at high temperatures (40–50 °C) and high methanol concentrations.
Table 4.
Biodiesel production by recombinant lipases.
Table 4.
Biodiesel production by recombinant lipases.
| Host Strain | Donor Species | Lipase Name | Feedstock | Alcohol | Alcohol:Oil Ratio | Lipase Form | Temp. (°C) | Yield (%) | Refs. |
|---|
| Escherichia coli | Pseudomonas aeruginosa | Lipd25A | Jatropha seed oil | Methanol | 3:1 (n:n) | Purified and immobilized | 33 | 75 | [225] |
| Bacillus amyloliquefaciens | LipBA | Olive oil | Methanol | 3:1 (n:n) | Purified and immobilized | 35 | ~100 | [226] |
| Bacillus subtilis | Lipase I4-2 | Soybean oil | Methanol | 3:1 (n:n) | Purified | 37 | 98 | [227] |
| Proteus mirabilis | Dieselzyme 4 | Canola oil | Methanol | 5.1 (n:n) | Purified and immobilized | 25 | 76, 100 | [206] |
| Staphylococcus haemolyticus | Lipase L62 | Olive oil | Methanol | 6:1 (n:n) | Purified and immobilized | 30 | 90 | [228] |
| Candida antarctica/Thermomyces lanuginosus | CALB/TLL | Waste grease | Methanol | 1:1 (n:n) | Whole-cell (wet and dry) | 30 | 87, 95 | [208] |
| Bacillus thermocatenulatus | BTL2 | Soybean oil | Methanol | 2 mL in 100 g | Purified | 45 | 94.8 | [229] |
| Proteus vulgaris | Lipase K80 | Plant and waste oil | Methanol | 3:1 (n:n) | Purified and immobilized | 30 | 78-100 | [230] |
| Proteus sp. (soil sample) | LipK107 | Olive oil | Methanol | 5:1 (n:n) | Whole-cell | 15 | ~100 | [207] |
| Pseudomonas fluorescens | LipB68 | Soybean oil | Methanol | 3:1 (n:n) | Purified and immobilized | 20 | 92 | [231] |
| Saccharomyces cerevisiae | Candida sp. | Lip2 | S. cerevisiae lipids | Ethanol | 4% (v/v) | Whole-cell | 30 | 11.4 mg/g cell dry weight | [232] |
| Candida antarctica | truncated CALB | Corn and soybean oil | Ethanol Butanol | 5:1 (n:n) | Purified and immobilized | 37 | 894–1394 µg/mg of lipase | [211] |
| Candida antarctica | CALB Lyt-1 | Corn and soybean oil | Ethanol | 15:1 (v:v) | Whole-cell; Purified and immobilized | 50 | 160 µg/mg of lipase, 80 µg/mg of lipase | [210] |
| Rhizopus oryzae | ROL | Soybean oil | Methanol | 1:1 (n:n) | Whole-cell surface-displayed | 37 | >78 | [216] |
| Rhizopus oryzae | ROL | Soybean oil | Methanol | 1:1 (n:n) | Whole-cell | 37 | 71 | [233] |
| Pichia pastoris | Rhizomucor miehei | GH2 | Microalgae oil | Methanol | 30:0.1 (µL:g) | Purified | 30 | >90% | [212,213] |
| Thermomyces lanuginosus | Tll | Waste cooking oil | Methanol | 3:1–9:1 (n:n) | Whole-cell | 30 | 50%–87% | [215] |
| Thermomyces lanuginosus | Tll | Waste cooking oil | Methanol | 4:1 (n:n) | Whole-cell | 40 | 82% | [234] |
| Oil contaminated soil-derived metagenomic library | lipZ01 | Olive oil | Methanol | 3:1 (n:n) | Crude supernatant | 37 | ~92% | [235] |
| Rhizopus oryzae/Rhizopus chinensis | Chimera lipase | Tung oil | Methanol | 3.88:1 (n:n) | Purified and immobilized | 40 | 92% | [214] |
| Candida antarctica/Rhizomucor miehei | CALB/RML | Different feedstocks | Methanol | 3:1–7:1 (n:n) | Whole-cell surface-displayed | 50 | >90% | [218] |
| Oil contaminated soil-derived metagenomic library | LipZ03 | Olive oil | Methanol | 3:1 (n:n) | Purified | 37 | 74% | [236] |
| Rhizomucor miehei | RML | Soybean oil | Methanol | 3:1–9:1 (n:n) | Whole-cell surface-displayed | 55 | 83% | [217] |
| Rhizopus oryzae | ROL | Soybean oil, Jatropha curcas and Pistacia chinensis seed oil | Methanol | 0.35:5 (g:g) | Crude supernatant | 35 | 92%–95% | [237] |
| Rhizomucor miehei/Penicillium cyclopium | RML/MDL | Soybean oil | Methanol | 4:1 (n:n) | Crude supenatant | 25 | >95% | [209] |
| Aspergillus oryzae | Candida antarctica | CALB | Soybean and palm oil | Methanol | 1.74:200 (g:g) | Whole-cell immobilized | 30, 50 | 90% | [223] |
| Geobacillus thermocatenulatus | BTL2 | Palm oil | Methanol | 0.37:9.63 (g:g) | Whole-cell immobilized | 40-50 | ~100% | [224] |
| Fusarium heterosporum | FHL | Shirashime oil | Methanol | 4.25:1 (n:n) | Whole-cell immobilized | r.t. | >96% | [220] |
| Fusarium heterosporum | FHL | Soybean oil | Methanol | 0.35:9.65 (g:g) | Whole-cell immobilized | 30 | 95% | [221] |
| Fusarium heterosporum | FHL | Rapeseed oil | Ethanol | 0.5:9.65 (g:g) | Whole-cell immobilized | 30 | 94% | [222] |
| Fusarium heterosporum | FHL | Soybean oil | Methanol | 0.35:9.65 (g:g) | Whole-cell immobilized | 30 | 94% | [219] |
5.2. The Food and Nutraceutical Industries
In the food industry, lipases are used for the synthesis of esters, including short-chain esters, as flavor compounds, and sugar esters as emulsifiers. Other classes of emulsifiers include partial glycerides produced by lipases, such as MAGs and DAGs, and lysolecithin produced by phospholipases. Lipases and phospholipases are used in the dairy industry to accelerate cheese ripening and to improve cheese yield, respectively, and in the baking industry to improve volume, softness and shelf-life of bread and other baked products. Both of these enzymes are important biocatalysts for the synthesis of nutraceuticals, which are bioactive compounds that can provide beneficial effects for human health.
The production of flavors by extraction from the fruit is preferred to chemical synthesis, as the product can then be labelled as “natural”; however, the extraction process entails high cost. At the same time, the use of microbial lipases for the synthesis of flavor compounds represents a good solution, as this is not so expensive and the product obtained can still be labelled as “natural” [
89]. As most of these compounds are chiral, flavor synthesis using enantio-selective lipases is preferable to non-selective chemical synthesis. For this reason, the enantio-selectivity of CALB towards secondary alcohols has made this the most widely used lipase in the synthesis of short-chain flavor esters [
238]. This is followed by CRL and
Rhizopus lipases, both in their free and immobilized forms [
89]. As the lipase enantio-selectivity depends on efficient immobilization [
239], this is strictly required when the flavor compounds being synthesized are chiral.
Recently, lipase-displaying yeast-cell biocatalysts are emerging as an alternative to immobilized lipases. Thus, as shown in
Table 5, recombinant CALB displayed on the surface of
S. cerevisiae cells [
240,
241] or of
P. pastoris cells [
242,
243,
244,
245] is the form of recombinant lipase that is most widely used in the synthesis of short-chain flavor esters. In this form, the enzyme can produce short-chain esters at high rates, which in some cases were even higher than the rates measured for the commercial Novozym 435 [
245]. Interestingly, Jin and coworkers [
243] used CALB-displaying
P. pastoris cells to scale-up the synthesis of 12 kinds of flavor esters in a 5 L batch-stirred reactor. The synthesis yield of 10 of these compounds exceeded 95% after reactions of 4 h, which thus demonstrated the potential commercial applications of this biocatalyst for large-scale production.
Emulsifiers can be endogenously produced or externally added to the processed food. An example of endogenously produced emulsifiers is lysolecithin produced from lecithin hydrolysis after treatment of egg yolk with phospholipases, which is intended for the mayonnaise and sauce industry. Commercial porcine pancreas PLA
2 (Lecitase
® 10L, Novozymes) and
A. niger PLA
2 (Maxapal
® A2, DSM) have been successfully used for this purpose [
75]. For externally added emulsifiers, the most widely used in the food industry are MAGs and DAGs produced by lipase-catalyzed esterification of glycerol with FFAs. The synthesis of MAGs and DAGs is carried out with lipases that show preference for these partial glycerides. In particular, a lipase from
P. camembertii (Lipase G, Amano) that shows selectivity for MAGs has been commercialized and used for this purpose [
246]. Recently, MDL with selectivity for DAGs and MAGs was recombinantly expressed in
P. pastoris. This enzyme was first used in biodiesel production in association with RML [
209], and then it was applied by the same authors to the production of MAGs and DAGs [
247] (
Table 5). This enzyme was used in the form of a fermentation broth supernatant, and was shown to provide high yields of MAG and DAG production, and to be more efficient than the commercial Lipase G50 when oleic acid was used as the acyl donor. Sugar esters are another class of compounds that are used in food processing for their emulsifying and antifungal properties [
73]. Sugar esters can be produced by lipase-catalyzed esterification of a sugar with a FFA, and also in this case, the most commonly used lipase is CALB [
248]. In particular, there is evidence that recombinant
P. pastoris with CALB displayed on its cell surface can act as an efficient whole-cell biocatalyst for sugar monoester production. Indeed, this system has been successfully used for the synthesis of both glucose laurate [
249] and fructose laurate [
250] (
Table 5).
Lipases are extensively used in the dairy industry for selective hydrolysis of milkfat TAGs during cheese ripening. The TAGs of ruminant milkfat are rich in short-chain FAs, and when these are released, they contribute significantly to the flavor of many cheese varieties [
77]. The traditional commercial lipases used in the dairy industry are from bovine and porcine pancreas or from pre-gastric tissues of young ruminants (P300, P400 and P600, GetCulture), although many microbial lipases have also been used efficiently, which have included lipases from
A. niger (Lipase A, Amano),
C. rugosa (Lipase AY, Amano),
P. roqueforti (Lipase R, Amano; Lipomod™, Biocatalysts),
P. camemberti (Lipase G, Amano),
R. javanicus (Lipase M, Amano),
R. miehei (Piccnate, Gist-Brocades), and
R. oryzae (Lipase DF, Amano). A lipase from
R. miehei that was recombinantly expressed in
A. oryzae (Palatase
® M, Novozymes) has also been commercialized for application in the dairy industry, to improve cheese flavor. Very recently, Peng and coworkers [
251] used a novel lipase (Est_p6) from a metagenomic library that they recombinantly expressed in
E. coli for the hydrolysis of milkfat and to impart a distinctive and desirable flavor to milk products (
Table 5). In recent years, there has been increasing interest in phospholipase modifications in the cheese industry, with the discovery that partial hydrolysis of phospholipids results in increased cheese yield. Indeed, a commercial PLA
1 from
A. oryzae (YieldMAX
® PL, Novozymes), a PLC from
B. cereus, and a PLD from
Streptomyces chromofuscus have been used to increase cheese yields [
75]. For recombinant phospholipases, Lilbaek and coworkers [
252] applied a commercial PLA
1 from
Fusarium venenatum expressed in
A. oryzae (Novozym 46016, Novozymes) in the dairy industry to improve process efficiencies and cheese yields (
Table 5).
Lipases and phospholipases have been used in the baking industry since 1990. Their addition to dough allows the hydrolysis of both nonpolar and polar lipids, which results in enhancement of flavor development and
in-situ generation of emulsifiers. These emulsifiers provide increased stability of the dough to mechanical stress, and thus improve the dough machinability; they also provide increased loaf volume, which results in an improved, more uniform and softer, crumb structure [
253]. Currently, various commercial lipases and phospholipases are available for bread baking. These include lipases from
A. niger (Lipase A, Amano) and
R. oryzae (Lipase DF, Amano), and PLA
2s from microbial sources (Lysomax R
®, Danisco; Maxapal
®, CakeZyme
® and BakeZyme
®, DSM Food Specialties). Examples of recombinant enzymes that have been commercialized for application in the baking industry include the 1,3-specific lipase from
T. lanuginosus (Lipopan 50BG
®, Novozymes) and the lipase/phospholipase from
F. oxysporum (Lipopan F
®, Novozymes), both of which are expressed in
A. oryzae. A new variant called Lipopan
® Xtra was recently developed by protein engineering, to complement Lipopan F
® [
75]. Other applications of recombinant lipases in the baking process are given in
Table 5. These include a lipase from
Geotrichum sp. (LIP 2) [
254], whereby dough fermented with
S. cerevisiae expressing LIP2 provides bread with greater loaf volume and more uniform crumb structure. Similar effects on the rheological properties of bread were observed by Li and coworkers [
255], with the addition of dough with a recombinant lipase from RCL in combination with transglutaminase. This combination also provided a product with improved sensorial properties. Recombinant RCL has also been shown to positively affect the thermomechanical properties of dough [
256].
Lipases are used to synthesize nutritionally important structured lipids from low-cost oils. These include low-calorie TAGs with short-chain or medium-chain FAs at the sn-1 and sn-3 positions and long chain FAs at the sn-2 position (SLS-TAGs, MLM-TAGs), and TAGs with only medium-chain FAs (MMM-TAGs); TAGs that are rich in ω-3 PUFAs; anti-obesity DAG oil; and human milkfat substitutes.
Table 5.
Applications of recombinant lipases in food and nutraceutical industry.
Table 5.
Applications of recombinant lipases in food and nutraceutical industry.
| Application | Donor Species | Enzyme Name | Host Strain | Lipase Form | Application | Refs. |
|---|
| Flavors | Candida antarctica | CALB | Saccharomyces cerevisiae | Whole-cell surface-displayed | Ethyl lactate synthesis | [241] |
| Candida antarctica | CALB | Saccharomyces cerevisiae | Whole-cell surface-displayed | Ethyl hexanoate synthesis | [240] |
| Candida antarctica | CALB | Pichia pastoris | Whole-cell surface-displayed | Short chain ester synthesis | [242,243,244] |
| Candida antarctica | CALB | Pichia pastoris | Whole-cell surface-displayed | Short chain ester synthesis | [245] |
| Rhizopus oryzae | ROL | Pichia pastoris | Purified and immobilized | Ethyl butyrate synthesis | [257] |
| Metagenomic library | LipIAF5-2 | Escherichia coli | Whole-cell | Short chain ester synthesis | [258] |
| Emulsifiers | Penicillium cyclopium | MDL | Pichia pastoris | Crude supernatant | Synthesis of MAGs and DAGs | [247] |
| Candida antarctica | CALB | Pichia pastoris | Whole-cell surface-displayed | Synthesis of fructose laurate | [250] |
| Candida antarctica | CALB | Pichia pastoris | Whole-cell surface-displayed | Synthesis of glucose laurate | [249] |
| Dairy | Metagenomic library | Lipase Est_p6 | Escherichia coli | Purified | Milk flavor production | [251] |
| Fusarium venenatum | PLA1 | Aspergillus oryzae | Purified | Mozzarella cheese yield improvement | [252] |
| Bakery | Geotrichum sp. | LIP1 and LIP2 | Saccharomyces cerevisiae | Whole-cell | Increase in loaf volume and in crumb structure uniformity | [254] |
| Rhizopus chinensis | RCL | Pichia pastoris | Purified | Increase in baking properties of dough | [256] |
| Rhizopus chinensis | RCL | Pichia pastoris | Purified | Increase in loaf volume and in crumb structure uniformity | [255] |
| Nutraceuticals | Geobacillus zalihae | Lipase T1 | Escherichia coli | Purified | DAG production from palm stearin | [259] |
| Rhizopus oryzae | ROL | Pichia pastoris | Purified | DAG production from soybean oil | [260] |
| Rhizopus oryzae | ROL | Pichia pastoris | Purified and immobilized | HMFS production | [261] |
| Rhizopus oryzae | ROL | Pichia pastoris | Purified and immobilized | HMFS production | [262] |
| Rhizopus oryzae | ROL | Pichia pastoris | Purified and immobilized | Production of MLM-TAGs | [263,264] |
| Geotrichum sp. | GSL | Pichia pastoris | Whole-cell surface-displayed | EPA and DHA enrichment in fish oil | [265] |
| Candida antarctica | | Pichia pastoris | Purified and immobilized | Vitamin A ester synthesis | [266] |
| Candida antarctica | CALA | Escherichia coli | Crude supernatant | Removal of trans FAs from PHVO | [175] |
| Streptomyces chromofuscus | PLD | Pichia pastoris | Whole-cell surface-displayed | PS production from PC | [267] |
| Streptomyces chromofuscus | PLD | Escherichia coli | Purified | PS production from PC | [268] |
| Streptomyces racemochromogenes | PLD | Streptomyces lividans | Purified | PS and PG production from lecithin | [269] |
Lipases have also been used in selective removal of unhealthy
trans-FAs from partially hydrogenated vegetable oils, and in the synthesis of vitamins and phenolic esters that present higher antioxidant activities and stabilities compared to the corresponding free molecules [
89]. Similar to lipases, PLAs and PLBs can be used to incorporate medium-chain and long-chain FAs and PUFAs into phospholipids. The transphosphatidylation reaction catalyzed by PLD is used to produce nonabundant phospholipids with bioactive properties, such as PS, which has an important role in supporting mental functions, and phospholipid-vitamin derivatives that are obtained by introducing water-soluble vitamins as polar head-groups in phospholipids. In this form, vitamins show higher affinities for their biological membranes, where they can exert their antioxidant activities [
75].
As shown in
Table 5, ROL is the recombinant lipase that has been most widely used in the production of nutraceuticals. For instance, ROL recombinantly expressed in
P. pastoris and immobilized on macroporous beads has been used to produce low calorie MLM-TAGs, by acidolysis of virgin olive oil with caprylic and capric acids [
263,
264]. Also, ROL has been successfully applied as a biocatalyst in the production of human milkfat substitutes, by acidolysis of tripalmitine with oleic acid [
262], and of lard with FFAs from a fish oil concentrate rich in docosahexaenoic acid [
261]. When compared to different commercial enzymes, recombinant ROL has been shown to have levels of incorporation of FFAs similar to those achieved by Novozym 435 and Lipozyme RM IM, and even higher than those obtained with Lipozyme TL [
261]. Very recently, Li and coworkers [
260] also used recombinant ROL to hydrolyze soybean oil for the production of DAG, a functional oil with benefits for human health. DAG production has also been achieved from hydrolysis of palm stearine using a thermostable lipase (T1 lipase) recombinantly expressed in
E. coli [
259]. Other examples of recombinant lipases that have been used successfully in the synthesis of nutraceuticals include
Geotrichum sp. lipase (GSL) displayed on the cell surface of
P. pastoris, which was successfully used to enrich eicosapentaenoic acid and docosahexaenoic acid in fish oil on the basis of selective hydrolysis [
265], and a lipase from C
. antarctica ZJB09193 that was used in high-yield production of vitamin A esters [
266]. Also of note, there is the use of engineered CALA variants with acquired
trans-FA selectivity (see
Section 4.2.2) in large-scale removal of
trans-FAs from partially hydrogenated vegetable oils [
175].
Among the recombinant phospholipases, those evaluated for the production of nutraceutical compounds are almost exclusively PLDs from
Streptomyces spp. For instance, recombinant PLD from
S. chromofuscus has been used for the conversion of PC and
l-serine to PS. This enzyme has been used both in its purified form [
268] and displayed on the surface of
P. pastoris cells [
267]; in this latter case, the enzyme remained almost stable after several rounds of use, thus suggesting the potential use of this displaying system for viable industrial production of PS. Similarly, PLD from
Streptomyces racemochromogenes expressed in the host
Streptomyces lividans was shown to promote a high rate of conversion of the PC and phosphatidylethanolamine (PE) in soybean lecithin into phosphatidylglycerol and PS, in the presence of glycerol and
l-serine, respectively [
269]. The conditions to produce this enzyme on a large scale were assessed to define its use for industrial production of phosphatidyl derivatives from lecithin.
5.3. Oil Degumming
During storage, vegetable oils form a deposit of so-called gums, which include a variety of compounds, with the main form being phospholipids. These compounds pose many problems for the processing and storage of oils for food and nonfood applications. Indeed, the presence of phospholipids in edible oils can lead to dark-colored oils and the generation of off-flavors [
270]. In biodiesel production, a high content of phospholipids in the oil leads to loss of yield, as the FAs enclosed in the phospholipid molecules are not accessible to the lipase for transesterification [
271]. Therefore, the removal of nearly all of the phospholipids is essential for the production of high-quality finished oil. For successful oil refining, the phosphorus content should be reduced to less than 10 mg/kg [
272]. Hydratable phospholipids can be removed by water degumming [
273], whereas the non-hydratable forms are removed by either chemical (
i.e., acid or alkali addition) or enzymatic refining processes. Recently, enzymatic oil degumming has been attracting a lot of attention and, to date, enzymatic degumming has already been applied to soybean, sunflower, canola, palm, and several other vegetable oils, with highly satisfactory results. The enzymes most commonly used in degumming of vegetable oils are PLA
1, PLA
2 and PLC. All of these phospholipases can reduce the amount of nonhydratable phospholipids, as they cleave them into water-soluble and oil-soluble fragments. The first enzymatic degumming process was developed by the German Company Roehm and Lurgi in the 1990s. The process used, known as the “EnzyMax
® process” was based on the use of porcine PLA
2 (Lecitase
® 10L, Novozymes) [
274]. More recently, microbial phospholipases suitable for oil degumming have been commercialized both as native enzymes purified from their producing microorganisms, and as recombinant enzymes overexpressed in heterologous hosts. These include two PLA
1s from
F. oxysporum (Lecitase
® Novo, Novozymes) and
T. lanuginosus/
F. oxysporum (Lecitase
® Ultra, Novozymes), two PLA
2s from
T. reesei (Rohalase
® MPL, AB Enzymes) and
A. niger (Gumzyme™, DSM Food Specialties), a lipid acyl transferase with PLA
2 activity from
S. violaceoruber (Lysomax R
®, Danisco) and a PLC from a
B. anthracis-like strain (Purifine™, Verenium) [
9].
Among the commercial PLAs, Lecitase
® Ultra is of particular interest, as a recombinant enzyme that is obtained by fusion of homologous genes that encode
F. oxysporum phospholipase and
T. lanuginosus lipase. The chimeric gene was expressed in
A. niger, and it showed the thermal and pH stability of the
T. lanuginosus lipase, and the PLA
1 activity of the
F. oxysporum phospholipase. When used in oil degumming, this enzyme showed no speficity for any of the phospholipids, and reduced the amount of phosphorous to below the 10 mg/kg, with short reaction times [
275]. Lecitase
® Ultra has also been immobilized on different solid supports [
276]. When applied to soybean oil degumming, immobilized PLA
1 reduced the phosphorous content to <10 mg/kg, with more than 99% oil recovery.
Purfine™ is also a recombinant enzyme, and it contains a
B. anthracis-like PLC expressed in
P. pastoris [
9]. Purifine™ catalyzes the splitting off of the phosphate head-group from PC and PE, but has no activity with respect to PI or PA [
277]. The DAGs formed during the degumming process using Purifine™ remain in the oil, thus increasing the oil yield; moreover, this enzyme does not produce FFAs that need to be removed later by any refining process, which would inevitably lead to an oil-yield loss. Interestingly, the two recombinant phospholipases Lecitase Ultra
® and Purifine™ have been used recently in combination with a lipase (Callera Trans L
®, Novozymes) both to perform enzymatic degumming and transesterification in a single step, using crude soybean oil as feedstock, and to convert part of the phospholipids into biodiesel [
271]. After the combined activity of the different enzymes, more than 95% of the methyl esters were produced, and the phosphorous content was reduced to below 5 mg/kg.
In addition to commercial phospholipases, other recombinant enzymes have been developed and tested for their reduction of the phosphorous content in vegetable oils (
Table 6). Very recently, Liu and coworkers [
278] expressed a PLA
2 from
S. violaceoruber in
P. pastoris. The recombinant enzyme hydrolyzed PC at a high rate and, when applied in the degumming of rapeseed oil, this PLA
2 decreased the phosphorous content to 20.74 ± 0.24 mg/kg after a reaction of 6 h, which is still slightly higher than the residual phosphorous content obtained after chemical oil degumming or enzymatic degumming with commercial phospholipases. Nevertheless, the recombinant PLA
2 from
S. violaceoruber is very promising for industrial applications, as it has only phospholipase activity, without concomitant lipase activity, which is crucial to avoid oil-yield loss due to TAG hydrolysis. Another successful application of recombinant phospholipases in oil degumming is represented by a PLB from
P. fluorescens expressed in
P. pastoris (Pf-PLB-P) [
279]. When Pf-PLB-P was used to degum soybean and peanut oils, the phosphorus content decreased to 4.96 and 3.54 mg/kg, respectively, after 5 h of reaction. Similar to the PLA
2 from
S. violaceoruber, Pf-PLB-P did not show lipase activity, thus minimizing loss of TAGs during the degumming process.
Recently, the application to oil degumming of phospholipases recombinantly expressed in
E. coli has also been reported. PLCs from
Clostridium perfringens were shown to decrease the phosphorous content of rapeseed oil to 5.66 mg/kg after a 1.5 h reaction [
280]. The PLC from
Listeria monocytogenes has been applied to degumming of both soybean and rice bran crude oils, and in both cases, it decreased the phosphorous content to lower than 5 mg/kg [
281].
Table 6.
Vegetable oil degumming by recombinant phospholipases.
Table 6.
Vegetable oil degumming by recombinant phospholipases.
| Host Strain | Donor Species | Phospholipase Name | Feedstock | Residual P (mg/kg) | Reaction Time (h) | Refs. |
|---|
| Escherichia coli | Clostridium perfringens | PLC | Rapeseed oil | 5.66 | 1.5 | [280] |
| Listeria monocytogenes | PLC | Soybean rice bran oil | 3.70, 4.50 | - | [281] |
| Pichia pastoris | Streptomyces violaceoruber | PLA2 | Rapeseed oil | 20.74 | 6 | [278] |
| Pseudomonas fluorescens | PLB | Soybean and peanut oil | 4.96, 3.54 | 5 | [279] |
5.4. The Detergent Industry
Certain enzymes, such as lipase, protease and amylase, have been used as catalysts in detergents since the 1960s to efficiently remove certain stains. In particular, the addition of lipases in detergent formulations is needed to remove fat-containing stains, thus allowing the content of undesirable chemicals to be reduced; moreover, they are biodegradable, leave no harmful residues, have no negative impact on sewage treatment processes, and pose no risk to aquatic life. Lipases used in detergent formulations are chosen on the basis of their broad substrate specificity, and their resistance to the harsh washing conditions of alkaline pHs and high temperatures, to oxidizing and chelating agents, and to damage induced by surfactants and other enzymes, such as proteases, which are found in many detergent formulations [
131]. In recent years, the trend toward low washing temperatures has made the removal of fatty stains more difficult. The use of cold-active lipases in the formulation of detergents has solved this problem, with the additional advantages of a reduction in energy consumption and wear and tear of the textiles [
282].
The first lipase introduced into detergent formulations was commercialized in 1988 by the company Novo Nordisk. This enzyme is known as Lipolase
®, and it was naturally produced by a selected strain of
T. lanuginosus, although at concentrations too low for industrial applications. So, to enhance the production yield, the gene that encodes this lipase was cloned and inserted into the host
A. oryzae. This heterologous system allowed its expression to reach commercially significant levels of production. In the 1990s, other recombinant lipases were introduced onto the market for detergent applications. In particular, two lipases from
P. mendocina and
P. alcaligenes, which are known as Lumafast
® and Lipomax
®, respectively, were commercialized by Genencor (now Du Pont), while a lipase from
P. glumae was commercialized by Unilever. All these three enzymes were recombinantly expressed in
B. subtilis [
91].
In the subsequent years, these enzymes have been subjected to protein engineering to improve their stabilities and catalytic properties under washing conditions [
283]. For instance, different variants of Lipolase
® were obtained by SDM approaches. In particular, this enzyme was engineered to improve its binding to fatty stains. To achieve this goal, the negatively charged aspartate residue in the lipid contact zone was exchanged for an uncharged leucine residue. In this way, a new enzyme was obtained, then called Lipolase
® Ultra, with improved washing performance at low temperature. Moreover, the T231R and N233R mutations generated new Lipolase
® variants, known as Lipex
®, which showed better performances in the first wash at temperatures as low as 20 °C and showed beneficial synergistic results when combined with bleach catalysts. Another engineering variant of Lipolase
® that shows better first washing performance is LipoPrime
®, which carries the mutation S216W. For the
P. alcaligenes lipase, Lipomax
® is an engineered variant of the WT enzyme in which the substitution of the methionine at position 21 with leucine resulted in increased washing performance of this lipase. This enzyme was also engineered for improved stability with anionic surfactants, which are responsible for the inactivation of the enzyme in liquid detergents. To block the entrance of the anionic surfactant into the enzyme, and to prevent denaturation of the enzyme, the positively charged amino acids on the protein surface were substituted with negatively charged amino acids, and a new more stable variant was obtained. Also, a lipase from
P. glumae was engineered to improve its resistance to proteases. To achieve this goal, mutations were introduced in the cleavage sites, to introduce amino-acid residues that are not preferred by proteases. Moreover, the M254I mutation was also introduced to improve the stability of this enzyme in the presence of oxidizing agents.
Very recently, Kranen and coworkers [
284] applied a whole-cell biocatalyst in the real-life laundry process for the first time. A lipase from
B. cepacia and its chaperone, the so-called lipase-specific foldase (Lif) that is responsible for correct folding of the lipase, were co-expressed on the surface of
E. coli via autodisplay. The whole-cell biocatalyst and the outer-membrane preparations were used in a standardized laundry tests that imitated conventional machine washing. Both of these preparations appeared to be stable enough to endure the standard European laundry test, while they showed the same lipolytic activity as the purified lipase from
B. cepacia and the commercial Lipex
®. These results clearly show that autodisplay represents a more convenient alternative to obtain a functional biocatalyst without the need for prior laborious purification steps, and that successful removal of fat or grease spots can be achieved by simply working with a cell-free preparation containing
E. coli outer membrane preparations that contain the active lipase.