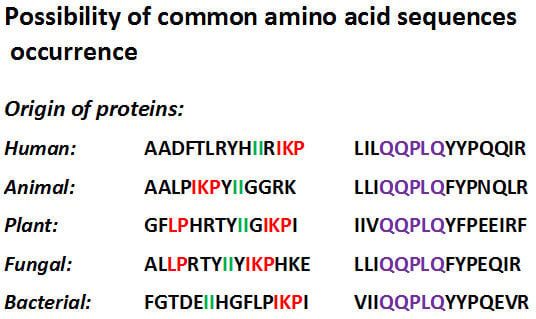
Common Amino Acid Subsequences in a Universal Proteome—Relevance for Food Science
Abstract
:1. Introduction
2. Biologically Active Peptides
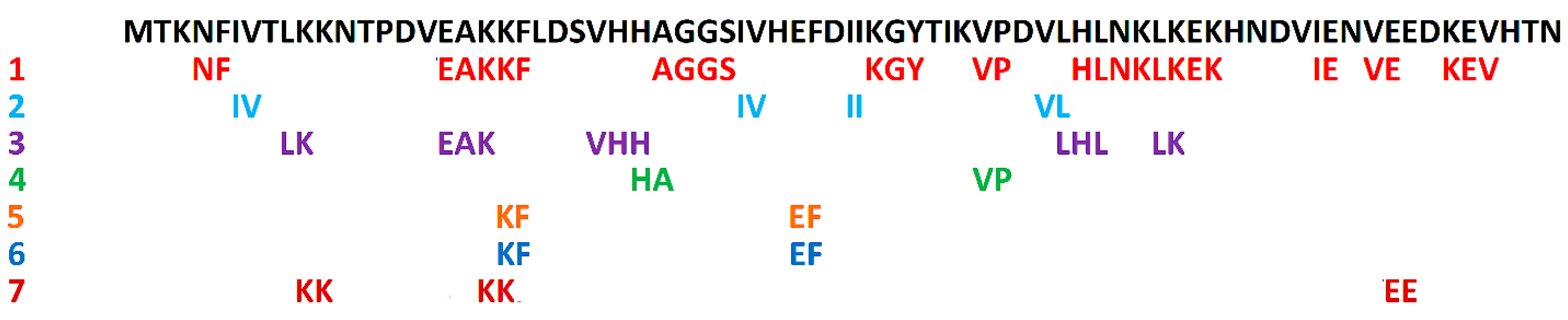
| ID a | Sequence b | Activity | Primary Resource c | Reference |
|---|---|---|---|---|
| 3379 | AKK | ACE inhibitor | Muscle of fish of the genus Sardina d | [25] |
| 3532 | GY | ACE inhibitor | Synthetic | [26] |
| 7587 | VP | ACE inhibitor | Synthetic | [26] |
| 7600 | AG | ACE inhibitor | Synthetic | [26] |
| 7602 | HL | ACE inhibitor | Synthetic | [27] |
| 7604 | KG | ACE inhibitor | Synthetic | [26] |
| 7607 | GS | ACE inhibitor | Synthetic | [26] |
| 7616 | GG | ACE inhibitor | Synthetic | [27] |
| 7623 | EA | ACE inhibitor | Synthetic | [26] |
| 7654 | NKL | ACE inhibitor | Wakame (Undaria pinnatifida) d | [28] |
| 7683 | NF | ACE inhibitor | Garlic (Allium sativum) d | [28] |
| 7692 | KF | ACE inhibitor | Garlic (Allium sativum) d | [28] |
| 7693 | KL | ACE inhibitor | Wakame (Undaria pinnatifida) d | [28] |
| 7698 | NK | ACE inhibitor | Wakame (Undaria pinnatifida) d | [28] |
| 7827 | IE | ACE inhibitor | Bovine (Bos taurus) milk d | [29] |
| 7828 | EV | ACE inhibitor | Bovine (Bos taurus) milk d | [29] |
| 7829 | VE | ACE inhibitor | Bovine (Bos taurus) milk d | [29] |
| 7832 | LN | ACE inhibitor | Bovine (Bos taurus) milk d | [29] |
| 7840 | EK | ACE inhibitor | Bovine (Bos taurus) milk d | [29] |
| 7841 | KE | ACE inhibitor | Bovine (Bos taurus) milk d | [29] |
| 8320 | VL | Glucose uptake stimulating | Bovine (Bos taurus) whey d | [30] |
| 8322 | IV | Glucose uptake stimulating | Bovine (Bos taurus) whey d | [30] |
| 8325 | II | Glucose uptake stimulating | Bovine (Bos taurus) whey d | [30] |
| 8329 | EE | Vasoactive substance release stimulating | Soybean (Glycine max) d | [31] |
| 3305 | LH | Antioxidant | Soybean (Glycine max) d | [32] |
| 3317 | HL | Antioxidant | Soybean (Glycine max) d | [32] |
| 3319 | HH | Antioxidant | Soybean (Glycine max) d | [32] |
| 7794 | VHH | Antioxidant | Chicken (Gallus gallus) egg d | [33] |
| 7995 | LHL | Antioxidant | Synthetic | [34] |
| 8130 | EAK | Antioxidant | Bonito (Katsuwonus pelamis) d | [35] |
| 8217 | LK | Antioxidant | Chicken (Gallus gallus) egg d | [36] |
| 3751 | KK | Bacterial permease ligand | Synthetic | [37] |
| 3181 | VP | Dipeptidyl peptidase IV inhibitor | Rat (Rattus norvegicus) | [38] |
| 3184 | HA | Dipeptidyl peptidase IV inhibitor | Rat (Rattus norvegicus) | [38] |
| 8249 | KF | CaMPDE inhibitor | Pea (Pisum sativum) d | [39] |
| 8250 | EF | CaMPDE inhibitor | Pea (Pisum sativum) d | [39] |
| 8248 | KF | Renin inhibitor | Pea (Pisum sativum) d | [39] |
| 8251 | EF | Renin inhibitor | Pea (Pisum sativum) d | [39] |
| Database Search Application | Reference |
|---|---|
| Location of short, bioactive fragments in sequences of peptides released during hydrolysis of bovine and trout meat proteins in the porcine digestive tract. Peptides used as query sequences were identified by mass spectrometry. | [60] |
| Location of bioactive fragments in sequences of rapeseed proteins. Protein sequences from UniProt were used as queries. | [61] |
| Location of bioactive fragments in sequences of bovine meat proteins. Protein sequences from UniProt were used as queries. | [62] |
| Location of short, bioactive fragments in sequences of peptides released during hydrolysis of fish sarcoplasmic proteins. Peptides used as query sequences were identified by mass spectrometry. | [63] |
| Location of bioactive fragments in sequences of cereal proteins. Protein sequences from UniProt were used as queries. | [64] |
| The BIOPEP database was used to determine the profiles of potential biological activity of salmon proteins. Some of the predicted peptides were identified in protein hydrolysates by liquid chromatography and mass spectrometry. | [58] |
| Location of bioactive fragments in sequences of proteins from the human digestive tract, followed by proteolysis simulation by digestive proteolytic enzymes. Protein sequences from UniProt were used as queries. | [55] |
| Location of bioactive fragments in sequences of amaranthus proteins. Protein sequences from UniProt were used as queries. | [65] |
| No | Protein Name | Entry Name in UniProtKB | Organism a |
|---|---|---|---|
| 1. | Uncharacterized protein | TR:W4ZV89_WHEAT | Triticum aestivum (4565) |
| 2. | Glyceraldehyde-3-phosphate dehydrogenase | SP:G3P3_YEAST | Saccharomyces cerevisiae (strain ATCC 204508/S288c) (559292) |
| 3. | Glyceraldehyde-3-phosphate dehydrogenase | TR:A6ZUK2_YEAS7 | Saccharomyces cerevisiae (strain YJM789) (307796) |
| 4. | Glyceraldehyde-3-phosphate dehydrogenase | TR:B3LI45_YEAS1 | Saccharomyces cerevisiae (strain RM11-1a) (285006) |
| 5. | Glyceraldehyde-3-phosphate dehydrogenase | TR:B5VJD4_YEAS6 | Saccharomyces cerevisiae (strain AWRI1631) (545124) |
| 6. | Glyceraldehyde-3-phosphate dehydrogenase | TR:C8Z985_YEAS8 | Saccharomyces cerevisiae (strain Lalvin EC1118 / Prise de mousse) (643680) |
| 7. | Glyceraldehyde-3-phosphate dehydrogenase | TR:E7KD02_YEASA | Saccharomyces cerevisiae (strain AWRI796) (764097) |
| 8. | Glyceraldehyde-3-phosphate dehydrogenase | TR:E7KP33_YEASL | Saccharomyces cerevisiae (strain Lalvin QA23) (764098) |
| 9. | Glyceraldehyde-3-phosphate dehydrogenase | TR:E7LUX3_YEASV | Saccharomyces cerevisiae (strain VIN 13) (764099) |
| 10. | Glyceraldehyde-3-phosphate dehydrogenase | TR:E7NI37_YEASO | Saccharomyces cerevisiae (strain FostersO) (764101) |
| 11. | Glyceraldehyde-3-phosphate dehydrogenase | TR:E7Q4A2_YEASB | Saccharomyces cerevisiae (strain FostersB) (764102) |
| 12. | Glyceraldehyde-3-phosphate dehydrogenase | TR:E7QF80_YEASZ | Saccharomyces cerevisiae (strain Zymaflore VL3) (764100) |
| 13. | Glyceraldehyde-3-phosphate dehydrogenase | TR:G2WES0_YEASK | Saccharomyces cerevisiae (strain Kyokai no. 7/NBRC 101557) (721032) |
| 14. | Tdh3p | TR:H0GGT7_9SACH | Saccharomyces cerevisiae x Saccharomyces kudriavzevii VIN7 (1095631) |
| 15. | Uncharacterized protein | TR:J7S7S3_KAZNA | Kazachstania naganishii (strain ATCC MYA-139/BCRC 22969/CBS 8797/CCRC 22969/KCTC 17520/NBRC 10181/NCYC 3082) (1071383) |
| 16. | Tdh3p | TR:N1P2H7_YEASC | Saccharomyces cerevisiae (strain CEN.PK113-7D) (889517) |
| 17. | Tdh3p | TR:W7PUI3_YEASX | Saccharomyces cerevisiae R008 (1182966) |
| 18. | Tdh3p | TR:W7RBG4_YEASX | Saccharomyces cerevisiae P283 (1177187) |
3. Linear Epitopes
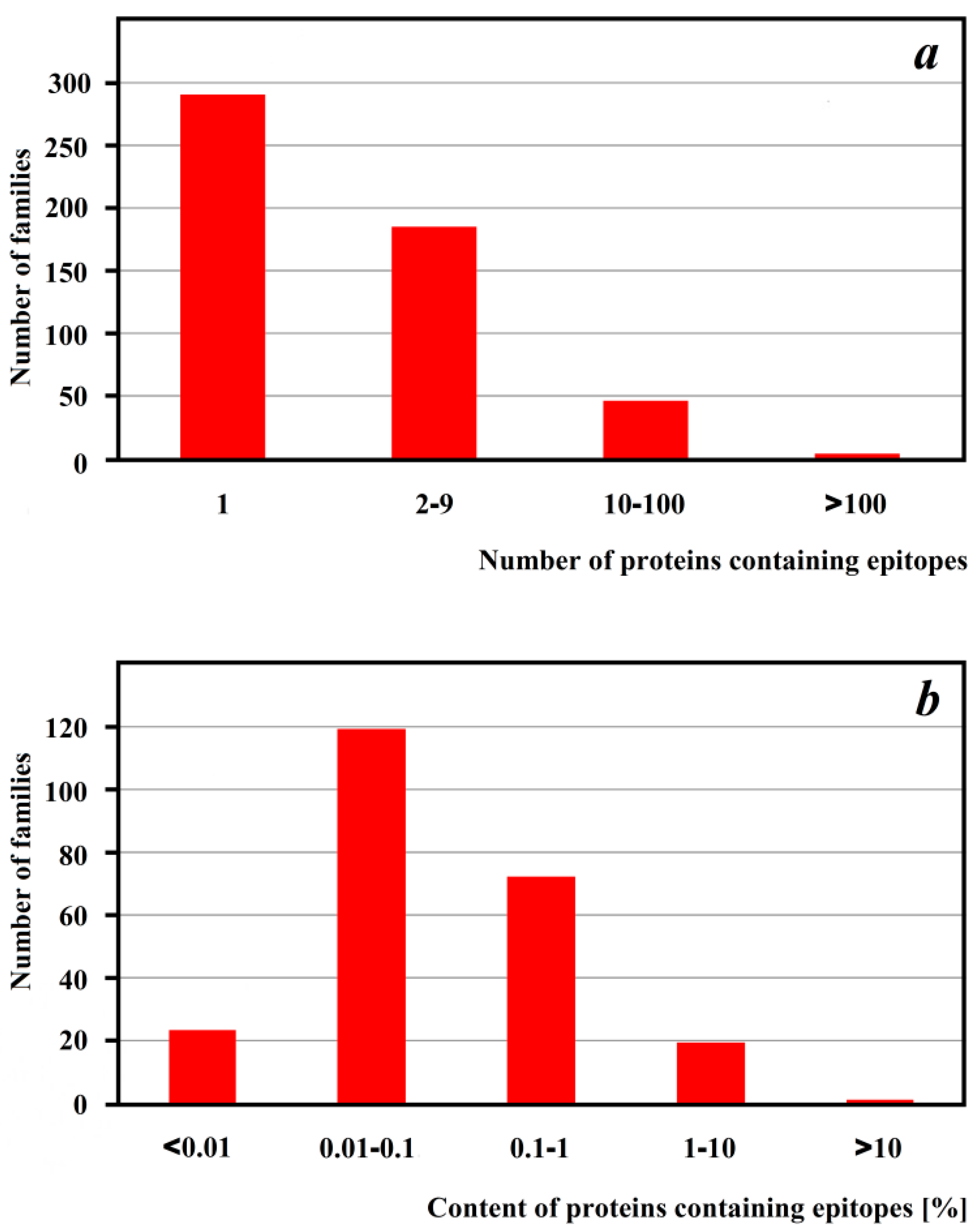
4. Peptides Relevant as Allergen Markers
| No | Entry Name in UniProtKB | Allergome Annotation | Organism a |
|---|---|---|---|
| Peptide (R)FFVAPFPEVFGK b—marker of αs1-casein | |||
| 1. | CASA1_BOVIN | Bos d 9.0101; Code 10197 | Bos taurus (9913) |
| 2. | CASA1_BUBBU | Bub b 8; Code 1259 | Bubalus bubalis (89462) |
| 3. | G3C8Y4_BUBBU | Bubalus bubalis (89462) | |
| 4. | B5B3R8_BOVIN | Bos d 9; Code 2734 | Bos taurus (9913) |
| 5. | L8I5S0_9CETA | Bos mutus (Bos grunniens) (72004) | |
| 6. | G3C8Y5_BUBBU | Bubalus bubalis (89462) | |
| 7. | Q4F6X6_BUBBU | Bubalus bubalis (89462) | |
| Peptide (K)FESNFNTQATNR c—marker of lysozyme C | |||
| 1. | LYSC_CHICK | Gal d 4.0101; Code 3294 | Gallus gallus (9031) |
| 2. | LYSC_COTJA | Coturnix coturnix japonica (93934) | |
| 3. | B8YK77_GALLA | Gal la 4; Code 9143 | Gallus lafayetii (9032) |
| 4. | B8YK75_GALSO | Gal so 4; Code 9144 | Gallus sonneratii (9033) |
| 5. | B8YK79_CHICK | Gal d 4; Code 362 | Gallus gallus (9031) |
| 6. | B8YJP1_CHICK | Gal d 4; Code 362 | Gallus gallus (9031) |
| 7. | B8YJN9_CHICK | Gal d 4; Code 362 | Gallus gallus (9031) |
| 8. | B8YJT7_CHICK | Gal d 4; Code 362 | Gallus gallus (9031) |
5. Mass Spectrometry as a Tool for Experimental Identification of Common Subsequences
| Aim of the Experiment | Mass Spectrometry Technique | Separation Method | Reference |
|---|---|---|---|
| Identification of Angiotensin I-converting enzyme (ACE) inhibitory peptides released during simulated gastrointestinal digestion of salmon (Salmo salar) muscles | ESI-IT-MS/MS, SRM | RP-HPLC, low TFA concentration in mobile phase | [58] |
| Detection and quantitative determination of peptides that are markers of bovine (Bos taurus) casein and chicken (Gallus gallus) egg proteins | ESI-MS/MS, SRM | RP-HPLC | [126] |
| Detection and quantitative determination of peptides that are markers of mustard allergen Sin a 1 in foods | ESI-MS/MS, SRM | RP-HPLC | [128] |
| Identification of peptides from peanut (Arachis hypogaea) allergens | nano-ESI Q-TOF MS/MS | capillary RP-HPLC | [129] |
| Identification of peptides from soybean (Glycine max) allergens | MALDI-TOF and MALDI-TOF-TOF | RP-HPLC | [130] |
6. Final Remarks
Supplementary Information
Acknowledgments
Conflict of Interest
References
- Kusalik, A.; Trost, B.; Bickis, M.; Fasano, C.; Capone, G.; Kanduc, D. Codon number shapes peptide redundancy in the universal proteome composition. Peptides 2009, 30, 1940–1944. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Valcu, C.-M.; Jungueira, M. Tools for exploiting proteomosphere. J. Proteom. 2009, 72, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Levitt, M. Nature of the protein universe. Proc. Natl. Acad. Sci. USA 2009, 106, 11079–11084. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.P.; Wu, L.Y.; Wang, Y.; Zhang, X.S.; Chen, L. Bridging protein local structures and protein functions. Amino Acids 2008, 35, 627–650. [Google Scholar] [CrossRef] [PubMed]
- Lucchese, G.; Stufano, A.; Trost, B.; Kusalik, A.; Kanduc, D. Peptidology: Short amino acid modules in cell biology and immunology. Amino Acids 2007, 33, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Schlimme, E.; Meisel, H. Bioactive peptides derived from milk proteins. Structural, physiological and analytical aspects. Nahrung 1995, 39, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Dziuba, J.; Darewicz, M.; Iwaniak, A.; Dziuba, M.; Nałęcz, D. Food peptidomics. Food Technol. Biotechnol. 2008, 46, 1–10. [Google Scholar]
- Wang, L.; Wang, Q.; Qian, J.; Liang, Q.; Wang, Z.; Xu, J.; He, S.; Ma, H. Bioavailability and bioavailable forms of collagen after oral administration to rats. J. Agric. Food Chem. 2015, 63, 3752–3756. [Google Scholar] [CrossRef] [PubMed]
- ChEMBL Database. Available online: https://www.ebi.ac.uk/chembldb/ (accessed on 1 May 2015).
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Krüger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL bioactivity database: An update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Reymond, J.-L.; Awale, M. Exploring chemical space for drug discovery using the chemical universe database. ACS Chem. Neurosci. 2012, 3, 649–657. [Google Scholar] [CrossRef] [PubMed]
- PubChem Database. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 1 May 2015).
- Wang, Y.; Suzek, T.; Zhang, J.; Wang, J.; He, S.; Cheng, T.; Shoemaker, B.A.; Gindulyte, A.; Bryant, S.H. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014, 42, D1075–D1082. [Google Scholar] [CrossRef] [PubMed]
- ChemSpider Database. Available online: http://www.chemspider.com/Default.aspx (accessed on 1 May 2015).
- Pence, H.E.; Williams, A. Chemspider: An online chemical information resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Karelin, A.A.; Blischenko, E.Y.; Ivanov, V.T. A novel system of peptidergic regulation. FEBS Lett. 1998, 428, 7–12. [Google Scholar] [CrossRef]
- Shevchenko, A.; Sunyaev, S.; Loboda, A.; Shevchenko, A.; Bork, P.; Ens, W.; Standing, K.G. Charting the proteomes of organism with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 2001, 73, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Dallas, D.C.; Guerrero, A.; Parker, E.A.; Robinson, R.C.; Gan, J.; German, J.B.; Barile, D.; Lebrilla, C.B. Current peptidomics: Applications, purification, identification, quantification, and functional analysis. Proteomics 2015, 15, 1026–1038. [Google Scholar] [CrossRef] [PubMed]
- Catherman, A.D.; Skinner, O.S.; Kelleher, N.L. Top down proteomics: Facts and perspectives. Biochem. Biophys. Res. Commun. 2014, 445, 683–693. [Google Scholar] [CrossRef] [PubMed]
- UniProtKB Website. Available online: http://www.uniprot.org/help/uniprotkb (accessed on 1 May 2015).
- UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar]
- BIOPEP Database. Available online: http://www.uwm.edu.pl/biochemia/index.php/pl/biopep (accessed on 1 May 2015).
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP database and other programs for processing bioactive peptide sequences. J. AOAC Int. 2008, 91, 965–980. [Google Scholar] [PubMed]
- Matsufuji, H.; Matsui, T.; Seki, E.; Osajima, K.; Nakashima, M.; Osajima, Y. Angiotensin I-converting enzyme inhibitory peptides in an alkaline proteinase hydrolysate derived from sardine muscle. Biosci. Biotechnol. Biochem. 1994, 58, 2244–2245. [Google Scholar] [CrossRef] [PubMed]
- Cheung, H.-S.; Wang, F.-L.; Ondetti, M.A.; Sabo, E.F.; Cushman, D.W. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. J. Biol. Chem. 1980, 255, 401–407. [Google Scholar] [PubMed]
- Cushman, D.W. Angiotensin converting enzyme inhibitors: Evolution of a new class of antihypertensive drugs. In Mechanisms of Action and Clinical Implications; Horovitz, Z.P., Ed.; Urban & Schwarzenberg: Munich, Germany, 1981; p. 19. [Google Scholar]
- Meisel, H.; Walsh, D.J.; Murray, B.; FitzGerald, R.J. ACE inhibitory peptides. In Nutraceutical Proteins and Peptides in Health and Disease; Mine, Y., Shahidi, F., Eds.; In CRC Taylor & Francis Group: Boca Raton, FL, USA; London, UK; New York, NY, USA, 2006; pp. 269–315. [Google Scholar]
- Van Platerink, C.J.; Janssen, H.-G.M.; Haverkamp, J. Application of at-line two-dimensional liquid chromatography-mass spectrometry for identification of small hydrophilic angiotensin I-inhibiting peptides in milk hydrolysates. Anal. Bioanal. Chem. 2008, 391, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Morifuji, M.; Koga, J.; Kawanaka, K.; Higuchi, M. Branched-chain amino acid-containing dipeptides, identified from whey protein hydrolysates, stimulate glucose uptake in L6 myotubes and isolated skeletal muscles. J. Nutr. Sci. Vitaminol. 2009, 55, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Ringseis, R.; Motthes, B.; Lehmann, V.; Becker, K.; Schöps, R.; Ulbrich-Hofmann, R.; Eder, K. Peptides and hydrolysates from casein and soy protein modulate the release of vasoactive substances from human aortic endothelial cells. Biochim. Biophys. Acta Gen. Subj. 2005, 1721, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-M.; Muramoto, K.; Yamauchi, F.; Nokihara, K. Antioxidant activity of designed peptides based on the antioxidant peptide isolated from digests of a soybean protein. J. Agric. Food Chem. 1996, 44, 2619–2623. [Google Scholar] [CrossRef]
- Mine, Y. Egg proteins and peptides in human health-chemistry, bioactivity and production. Curr. Pharm. Des. 2007, 13, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Jin, D.-H.; Ogawa, T.; Muramoto, K.; Hatakeyama, E.; Yasuhara, T.; Nokihara, K. Antioxidative properties of tripeptide libraries prepared by the combinatorial chemistry. J. Agric. Food Chem. 2003, 51, 3668–3674. [Google Scholar] [CrossRef] [PubMed]
- Suetsuna, K. Separation and identification of antioxidant peptides from proteolytic digest of dried bonito. Nippon Suisan Gakkaishi 1999, 65, 92–96, (In Japanese, Abstract in English). [Google Scholar] [CrossRef]
- Huang, W.-Y.; Majumder, K.; Wu, J. Oxygen radical absorbance capacity of peptides from egg white protein ovotransferrin and their interactions with phytochemicals. Food Chem. 2010, 123, 635–641. [Google Scholar] [CrossRef]
- Sleigh, S.H.; Tame, J.R.H.; Dodson, E.J.; Wilkinson, A.J. Peptide binding in OppA, the crystal structure of the periplasmic oligopeptide-binding protein in the unliganded form and in complex with lysillysine. Biochemistry 1997, 36, 9747–9758. [Google Scholar] [CrossRef] [PubMed]
- Bella, A.M.; Erickson, R.H., Jr.; Kim, Y.S. Rat intestinal brush border membrane dipeptidyl-aminopeptidase IV: Kinetic properties and substrate specificities of the purified enzyme. Arch. Biochem. Biophys. 1982, 218, 156–162. [Google Scholar] [CrossRef]
- Li, H.; Aluko, R.E. Identification and inhibitory properties of multifunctional peptides from pea protein hydrolysate. J. Agric. Food Chem. 2010, 58, 11471–11476. [Google Scholar] [CrossRef] [PubMed]
- BRENDA Database. Available online: http://www.brenda-enzymes.org/ (accessed on 1 May 2015).
- Chang, A.; Schomburg, I.; Placzek, P.; Jeske, L.; Ulbrich, M.; Xiao, M.; Sensen, C.W.; Schomburg, D. BRENDA in 2015: Exciting developments in its 25th year of existence. Nucleic Acids Res. 2015, 43, D439–D446. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-originating ACE inhibitors, including antihypertensive peptides, as preventive food components in blood pressure reduction. Compr. Rev. Food Sci. Food Saf. 2014, 13, 114–134. [Google Scholar] [CrossRef]
- AHTPDB Database. Available online: http://crdd.osdd.net/raghava/ahtpdb/ (accessed on 1 May 2015).
- Kumar, R.; Chaudhary, K.; Sharma, M.; Nagpal, G.; Chauhan, J.S.; Singh, S.; Gautam, A.; Raghava, G.P.S. AHTPDB: A comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Res. 2015, 43, D956–D962. [Google Scholar] [CrossRef] [PubMed]
- Azizi, M.; Ménard, J. Renin inhibitors and cardiovascular and renal protection: An endless quest? Cardiovasc. Drugs Ther. 2013, 27, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Gotoh, T.; Hata, K.; Tokiwano, T.; Yoshizawa, Y.; Hiwatashi, K.; Ogasawara, H.; Hori, K. Renin inhibitors in foodstuffs: Structure-function relationship. J. Biol. Macromol. 2014, 14, 71–84. [Google Scholar]
- Udenigwe, C.C.; Mohan, A. Mechanisms of food protein-derived antihypertensive peptides other than ACE inhibition. J. Funct. Foods 2014, 8C, 45–52. [Google Scholar] [CrossRef]
- Juillerat-Jeanneret, L. Dipeptidyl peptidase IV and its inhibitors: Therapeutics for type 2 diabetes and what else. J. Med. Chem. 2014, 57, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, M.J.; Cereto-Massagué, A.; Valls, C.; Pujadas, G. DPP-IV, an important target for antidiabetic functional food design. In Foodinformatics. Applications of Chemical Information to Food Chemistry; Martinez-Mayorga, K., Medina-Franco, J.L., Eds.; Springer International Publishing AG: Cham, Switzerland, 2014; pp. 177–212. [Google Scholar]
- Fajardo, A.M.; Piazza, G.A.; Tinsley, H.N. The role of cyclic nucleotide signaling pathways in cancer: Targets for prevention and treatment. Cancers 2014, 6, 436–458. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Gil, C. cAMP-specific phosphodiesterase inhibitors: Promising drugs for inflammatory and neurological diseases. Expert Opin. Ther. Pat. 2014, 24, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.S. Phosphodiesterase inhibition in the treatment of autoimmune and inflammatory diseases: Current status and potential. J. Recept. Ligand Channel Res. 2015, 8, 19–30. [Google Scholar] [CrossRef]
- Freitas, A.C.; Andrade, J.C.; Silva, F.M.; Rocha-Santos, T.A.P.; Duarte, A.C.; Gomes, A.M. Antioxidative peptides: Trends and perspectives for future research. Curr. Med. Chem. 2013, 20, 4575–4594. [Google Scholar] [CrossRef] [PubMed]
- Ormsbee, M.J.; Bach, C.W.; Baur, D.A. Pre-exercise nutrition: The role of macronutrients, modified starches and supplements on metabolism and endurance performance. Nutrients 2014, 6, 1782–1808. [Google Scholar] [CrossRef] [PubMed]
- Dave, L.A.; Montoya, C.A.; Rutherfurd, S.M.; Moughan, P.J. Gastrointestinal endogenous proteins as a source of bioactive peptides—An in silico study. PLoS ONE 2014, 9, e98922. [Google Scholar] [CrossRef] [PubMed]
- Barba de la Rosa, A.P.; Barba Montoya, A.; Martínez-Cuevas, P.; Hernández-Ledesma, B.; León-Galván, M.F.; de León-Rodríguez, A.; González, C. Tryptic amaranth glutelin digests induce endothelial nitric oxide production through inhibition of ACE: Antihypertensive role of amaranth peptides. Nitric Oxide 2010, 23, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Kanawjia, S.K.; Khetra, Y.; Saini, P. Discordance between in silico & in vitro analyses of ACE inhibitory & antioxidative peptides from mixed milk tryptic whey protein hydrolysate. J. Food Sci. Technol. 2015. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska, J.; Vegarud, G.E.; Minkiewicz, P.; Iwaniak, A. Angiotensin I-converting enzyme (ACE) inhibitory activity and ACE inhibitory peptides of salmon (Salmo salar) protein hydrolysates obtained by human and porcine gastrointestinal enzymes. Int. J. Mol. Sci. 2014, 15, 14077–14101. [Google Scholar] [CrossRef] [PubMed]
- Guinane, C.M.; Kent, R.M.; Norberg, S.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Generation of the antimicrobial peptide caseicin A from casein by hydrolysis with thermolysin enzymes. Int. Dairy J. 2015, 49, 1–7. [Google Scholar] [CrossRef]
- Bauchart, C.; Morzel, M.; Chambon, C.; Mirand, P.P.; Reynès, C.; Buffière, C.; Rémond, D. Peptides reproducibly released by in vivo digestion of beef meat and trout flesh in pigs. Br. J. Nutr. 2007, 98, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Wanasundara, J.P.D. Proteins of Brassicaceae oilseeds and their potential as a plant protein source. Crit. Rev. Food Sci. Nutr. 2011, 51, 635–677. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Dziuba, J.; Michalska, J. Bovine meat proteins as potential precursors of biologically active peptides—A computational study based on the BIOPEP database. Food Sci. Technol. Int. 2011, 17, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.; Cañas, B.; Gallardo, J.M. The sarcoplasmic fish proteome: Pathways, metabolic networks and potential bioactive peptides for nutritional inferences. J. Proteom. 2013, 78, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Cavazos, A.; González de Mejía, E. Identification of bioactive peptides from cereal storage proteins and their potential role in prevention of chronic diseases. Compr. Rev. Food Sci. Food Saf. 2013, 12, 364–380. [Google Scholar] [CrossRef]
- Montoya-Rodríguez, A.; Gómez-Favela, M.A.; Reyes-Moreno, C.; González de Mejía, E.; Milán-Carrillo, J. Identification of bioactive peptide sequences from amaranth (Amaranthus hypochondriacus) seed proteins and their potential role in the prevention of chronic diseases. Compr. Rev. Food Sci. Food Saf. 2015, 14, 139–158. [Google Scholar] [CrossRef]
- Kohama, Y.; Nagase, Y.; Oka, H.; Nakagawa, T.; Teramoto, T.; Murayama, N.; Tsujibo, H.; Inamori, Y.; Mimura, T. Production of angiotensin-converting enzyme inhibitors from baker’s yeast glyceraldehyde-3-phosphate dehydrogenase. J. Pharmacobiodyn. 1990, 13, 766–771. [Google Scholar] [CrossRef] [PubMed]
- PepBank Database. Available online: http://pepbank.mgh.harvard.edu/ (accessed on 1 May 2015).
- Duchrow, T.; Shtatland, T.; Guettler, D.; Pivovarov, M.; Kramer, S.; Weissleder, R. Enhancing navigation in biomedical databases by community voting and database-driven text classification. BMC Bioinform. 2009, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- BLAST Program. Available online: http://www.ebi.ac.uk/Tools/sss/wublast/ (accessed on 1 May 2015).
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Bucholska, J.; Darewicz, M.; Borawska, J. Epitopic hexapeptide sequences from Baltic cod parvalbumin beta (allergen Gad c 1) are common in the universal proteome. Peptides 2012, 38, 105–109. [Google Scholar] [CrossRef] [PubMed]
- NCBI Taxonomy Website. Available online: http://www.ncbi.nlm.nih.gov/taxonomy (accessed on 1 May 2015).
- Federhen, S. Type material in the NCBI Taxonomy Database. Nucleic Acids Res. 2015, 43, D1086–D1098. [Google Scholar] [CrossRef] [PubMed]
- InterPro Website. Available online: http://www.ebi.ac.uk/interpro/ (accessed on 1 May 2015).
- Mitchell, A.; Chang, H.-Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Gift, N.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- Zambrowicz, A.; Eckert, E.; Pokora, M.; Bobak, Ł.; Dąbrowska, A.; Szołtysik, M.; Trziszka, T.; Chrzanowska, J. Antioxidant and antidiabetic activities of peptides isolated from a hydrolysate of an egg-yolk protein by-product prepared with a proteinase from Asian pumpkin (Cucurbita ficifolia). RSC Adv. 2015, 5, 10460–10467. [Google Scholar] [CrossRef]
- Eckert, E.; Zambrowicz, A.; Pokora, M.; Polanowski, A.; Chrzanowska, J.; Szołtysik, M.; Dąbrowska, A.; Różański, H.; Trziszka, T. Biologically active peptides derived from egg proteins. World’s Poultry Sci. J. 2013, 69, 375–386. [Google Scholar] [CrossRef]
- Albrecht, M.; Kühne, Y.; Ballmer-Weber, B.K.; Becker, W.-M.; Holzhauser, T.; Lauer, I.; Reuter, A.; Randow, S.; Falk, S.; Wangorsch, A.; et al. Relevance of IgE binding to short peptides for the allergenic activity of food allergens. J. Allergy Clin. Immunol. 2009, 124, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Dall’Antonia, F.; Pavkov-Keller, T.; Zangger, K.; Keller, W. Structure of allergens and structure based epitope predictions. Methods 2014, 66, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.; Hawranek, T.; Gruber, P.; Wopfner, N.; Mari, A. Allergic cross-reactivity: From gene to the clinic. Allergy 2004, 59, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Bartuzi, Z. The molecular traits of food allergens (Molekularne cechy alergenów pokarmowych). Post. Dermatol. Alergol. 2009, 26, 310–312. (In Polish) [Google Scholar]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breiteneder, H.; Chapman, M.D. Allergen nomenclature. In Allergens and Allergen Immunotherapy, 5th ed.; Lockey, R.F., Ledford, D.K., Eds.; CRC Press: Boca Raton, FL, USA, 2014; pp. 37–49. [Google Scholar]
- Bindslev-Jensen, C.; Sten, E.; Earl, L.K.; Crevel, R.W.R.; Bindslev-Jensen, U.; Hansen, T.K.; Skov, P.S.; Poulsen, L.K. Assessment of the potential allergenicity of ice structuring protein type III HPLC 12 using the FAO/WHO 2001 decision tree for novel foods. Food Chem. Toxicol. 2003, 41, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.E. Practical and predictive bioinformatic methods for the identification of potentially cross-reactive protein matches. Mol. Nutr. Food Res. 2006, 50, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Schein, C.H.; Ivanciuc, O.; Braun, W. Bioinformatic approaches to classifying allergens and predicting cross-reactivity. Immunol. Allergy Clin. N. Am. 2007, 27, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Kleter, G.A.; Peijnenburg, A.A.C.M. Screening of transgenic proteins expressed in transgenic food crops for the presence of short amino acid sequences identical to potential, IgE-binding linear epitopes of allergens. BMC Struct. Biol. 2002, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Dziuba, J.; Gładkowska-Balewicz, I. Update of the list of allergenic proteins from milk based on local amino acid sequence identity with known epitopes from bovine milk proteins—A short report. Pol. J. Food Nutr. Sci. 2011, 61, 153–158. [Google Scholar] [CrossRef]
- Dessailly, B.H.; Redfern, O.C.; Cuff, A.; Orengo, C.A. Exploiting structural classifications for function prediction: Towards a domain grammar for protein function. Curr. Opin. Struct. Biol. 2009, 19, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Pfam Database. Available online: http://pfam.xfam.org/ (accessed on 1 May 2015).
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- AllFam Database. Available online: http://www.meduniwien.ac.at/allergens/allfam/ (accessed on 1 June 2015).
- Radauer, C.; Bublin, M.; Wagner, S.; Mari, A.; Breiteneder, H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 2008, 121, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Kanduc, D. Pentapeptides as minimal functional units in cell biology and immunology. Curr. Protein Pept. Sci. 2013, 14, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kanduc, D. Correlating low-similarity peptide sequences and allergenic epitopes. Curr. Pharm. Des. 2008, 14, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Kanduc, D. Homology, similarity, and identity in peptide epitope immunodefinition. J. Pept. Sci. 2012, 18, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Tachyon Program. Available online: http://tachyon.bii.a-star.edu.sg/index.action (accessed on 1 May 2015).
- Tan, J.; Kuchibhatla, D.; Sirota, F.L.; Sherman, W.A.; Gattermayer, T.; Kwoh, C.Y.; Eisenhaber, F.; Schneider, G.; Maurer-Stroh, S. Tachyon search speeds up retrieval of similar sequences by several orders of magnitude. Bioinformatics 2012, 28, 1645–1646. [Google Scholar] [CrossRef] [PubMed]
- MimicMe Program. Available online: http://mimicme.uwaterloo.ca/ (accessed on 1 May 2015).
- Petrenko, P.; Doxey, A.C. MimicMe: A web server for prediction and analysis of host-like proteins in microbial pathogens. Bioinformatics 2015, 31, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, H.; Morita, E.; Tatham, A.S.; Morimoto, K.; Horikawa, T.; Osuna, H.; Ikezawa, Z.; Kaneko, S.; Kohno, K.; Dekio, S. Identification of the IgE-binding epitope in omega-5 gliadin, a major wheat allergen in wheat-dependent exercise-induced anaphylaxis. J. Biol. Chem. 2004, 279, 12135–12140. [Google Scholar] [CrossRef] [PubMed]
- Abe, R.; Shimizu, S.; Yasuda, K.; Sugai, M.; Okada, Y.; Chiba, K.; Akao, M.; Kumagai, H.; Kumagai, H. Evaluation of reduced allergenicity of deamidated gliadin in a mouse model of wheat-gliadin allergy using an antibody prepared by a peptide containing three epitopes. J. Agric. Food Chem. 2014, 62, 2845–2852. [Google Scholar] [CrossRef] [PubMed]
- Immune Epitope Database. Available online: http://www.iedb.org/ (accessed on 1 May 2015).
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef] [PubMed]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Heller, S.; McNaught, A.; Stein, S.; Tchekhovskoi, D.; Pletnev, I. InChI—The worldwide chemical structure identifier standard. J. Cheminform. 2013, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Protasiewicz, M.; Mogut, D. Chemometrics and cheminformatics in the analysis of biologically active peptides from food sources. J. Funct. Foods 2015, 16, 334–351. [Google Scholar] [CrossRef]
- MetaComBio Website. Available online: http://www.uwm.edu.pl/metachemibio/index.php/about-metacombio (accessed on 1 July 2015).
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. Using internet databases for food science organic chemistry students to discover chemical compound information. J. Chem. Educ. 2015, 92, 874–876. [Google Scholar] [CrossRef]
- Southan, C. InChI in the wild: An assessment of InChIKey searching in Google. J. Cheminform. 2013, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Open Babel Program. Available online: http://openbabel.org/wiki/Main_Page (accessed on 1 May 2015).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Sokołowska, J.; Darewicz, M. The occurrence of sequences identical with epitopes from the allergen Pen a 1.0102 among food and non-food proteins. Pol. J. Food Nutr. Sci. 2015, 65, 21–29. [Google Scholar] [CrossRef]
- Custovic, A. To what extent is allergen exposure a risk factor for the development of allergic disease? Clin. Exp. Allergy 2015, 45, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Allergome Database. Available online: http://www.allergome.org/ (accessed on 1 May 2015).
- Mari, A.; Rasi, C.; Palazzo, P.; Scala, E. Allergen databases: Current status and perspectives. Curr. Allergy Asthma Rep. 2009, 9, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Darewicz, M.; Dziuba, J.; Minkiewicz, P. Computational characterisation and identification of peptides for in silico detection of potentially celiac-toxic proteins. Food Sci. Technol. Int. 2007, 13, 125–133. [Google Scholar] [CrossRef]
- Vojdani, A.; Tarash, I. Cross-reaction between gliadin and different food and tissue antigens. Food Nutr. Sci. 2013, 4, 20–32. [Google Scholar] [CrossRef]
- Yapar, N. Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 2014, 10, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Mirza, Z.K.; Sastri, B.; Lin, J.J.C.; Amenta, P.S.; Das, K.M. Autoimmunity against human tropomyosin isoforms in ulcerative colitis—Localization of specific human tropomyosin isoforms in the intestine and extraintestinal organs. Inflamm. Bowel Dis. 2006, 12, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Fæste, C.K.; Rønning, H.T.; Christians, U.; Granum, P.E. Liquid chromatography and mass spectrometry in food allergen detection. J. Food Protect. 2011, 74, 316–345. [Google Scholar] [CrossRef] [PubMed]
- Cunsolo, V.; Muccilli, V.; Saletti, R.; Foti, S. Mass spectrometry in food proteomics: A tutorial. J. Mass Spectrom. 2014, 49, 768–784. [Google Scholar] [CrossRef] [PubMed]
- Koeberl, M.; Clarke, D.; Lopata, A.L. Next generation of food allergen quantification using mass spectrometric systems. J. Proteome Res. 2014, 13, 3499–3509. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, S.; Mullen, W.; Cristobal, S. High-throughput proteomics: A new tool for quality and safety in fishery products. Curr. Protein Pept. Sci. 2014, 15, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Pilolli, R.; de Angelis, E.; Godula, M.; Visconti, A.; Monaci, L. Orbitrap™ monostage MS versus hybrid linear ion trap MS: Application to multi-allergen screening in wine. J. Mass Spectrom. 2014, 49, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, A.; Boye, J. Simultaneous detection of multi-allergens in an incurred food matrix using ELISA, multiplex flow cytometry and liquid chromatography mass spectrometry (LC–MS). Food Chem. 2015, 175, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Posada-Ayala, M.; Alvarez-Llamas, G.; Maroto, A.S.; Maes, X.; Muñoz-Garcia, E.; Villalba, M.; Rodríguez, R.; Perez-Gordo, M.; Vivanco, F.; Pastor-Vargas, C.; et al. Novel liquid chromatography-mass spectrometry method for sensitive determination of the mustard allergen Sin a 1 in food. Food Chem. 2015, 183, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Chassaigne, H.; Nørgaard, J.V.; van Hengel, A.J. Proteomics-based approach to detect and identify major allergens in processed peanuts by capillary LC-Q-TOF (MS/MS). J. Agric. Food Chem. 2007, 55, 4461–4473. [Google Scholar] [CrossRef] [PubMed]
- Cucu, T.; de Meulenaer, B.; Devreese, B. MALDI based identification of soybean protein markers—Possible analytical targets for allergen detection in processed foods. Peptides 2012, 33, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.E.; Baumgartner, S.; Aldick, T.; Bessant, C.; Giosafatto, V.; Heick, J.; Mamone, G.; O’Connor, G.; Poms, R.; Popping, B.; et al. Current perspectives and recommendations for the development of mass spectrometry methods for the determination of allergens in foods. J. AOAC Int. 2011, 94, 1026–1033. [Google Scholar] [PubMed]
- NCBI Proteins Database. Available online: http://www.ncbi.nlm.nih.gov/protein (accessed on 1 May 2015).
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015, 43, D6–D17. [Google Scholar]
- Dziuba, M.; Minkiewicz, P.; Dąbek, M. Peptides, specific proteolysis products as molecular markers of allergenic proteins—In silico studies. Acta Sci. Polon. Technol. Aliment. 2013, 12, 101–112. [Google Scholar]
- Belluco, S.; Losasso, C.; Maggioletti, M.; Alonzi, C.C.; Paoletti, M.G.; Ricci, A. Edible insects in a food safety and nutritional perspective: A critical review. Compr. Rev. Food Sci. Food Saf. 2013, 12, 296–313. [Google Scholar] [CrossRef]
- Mlcek, J.; Rop, O.; Borkovcova, M.; Bednarova, M. A comprehensive look at the possibilities of edible insects as food in Europe—A review. Pol. J. Food Nutr. Sci. 2014, 64, 147–157. [Google Scholar] [CrossRef]
- Carrasco-Castilla, J.; Hernández-Álvarez, A.J.; Jiménez-Martínez, C.; Gutiérrez-López, G.F.; Dávila-Ortiz, G. Use of proteomics and peptidomics methods in food bioactive peptide science and engineering. Food Eng. Rev. 2012, 4, 224–243. [Google Scholar] [CrossRef]
- Sauer, S.; Luge, T. Nutriproteomics: Facts, concepts, and perspectives. Proteomics 2015, 15, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, C.; Simó, C.; García-Cañas, V.; Cifuentes, A.; Castro-Puyana, M. Metabolomics, peptidomics and proteomics applications of capillary electrophoresis-mass spectrometry in foodomics: A review. Anal. Chim. Acta 2013, 802, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rivera, L.; Martínez-Maqueda, D.; Cruz-Huerta, E.; Miralles, B.; Recio, I. Peptidomics for discovery, bioavailability and monitoring of dairy bioactive peptides. Food Res. Int. 2014, 63, 170–181. [Google Scholar]
- Lan, V.T.T.; Ito, K.; Ohno, M.; Motoyama, T.; Ito, S.; Kawarasaki, Y. Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor. Food Chem. 2015, 175, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Chanput, W.; Nakai, S.; Theerakulkait, C. Introduction of new computer softwares for classification and prediction purposes of bioactive peptides: Case study in antioxidative tripeptides. Int. J. Food Prop. 2010, 13, 947–959. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Sokołowska, J.; Starowicz, P.; Bucholska, J.; Hrynkiewicz, M. Common Amino Acid Subsequences in a Universal Proteome—Relevance for Food Science. Int. J. Mol. Sci. 2015, 16, 20748-20773. https://doi.org/10.3390/ijms160920748
Minkiewicz P, Darewicz M, Iwaniak A, Sokołowska J, Starowicz P, Bucholska J, Hrynkiewicz M. Common Amino Acid Subsequences in a Universal Proteome—Relevance for Food Science. International Journal of Molecular Sciences. 2015; 16(9):20748-20773. https://doi.org/10.3390/ijms160920748
Chicago/Turabian StyleMinkiewicz, Piotr, Małgorzata Darewicz, Anna Iwaniak, Jolanta Sokołowska, Piotr Starowicz, Justyna Bucholska, and Monika Hrynkiewicz. 2015. "Common Amino Acid Subsequences in a Universal Proteome—Relevance for Food Science" International Journal of Molecular Sciences 16, no. 9: 20748-20773. https://doi.org/10.3390/ijms160920748