Validation of Tuba1a as Appropriate Internal Control for Normalization of Gene Expression Analysis during Mouse Lung Development
Abstract
:1. Introduction
2. Results
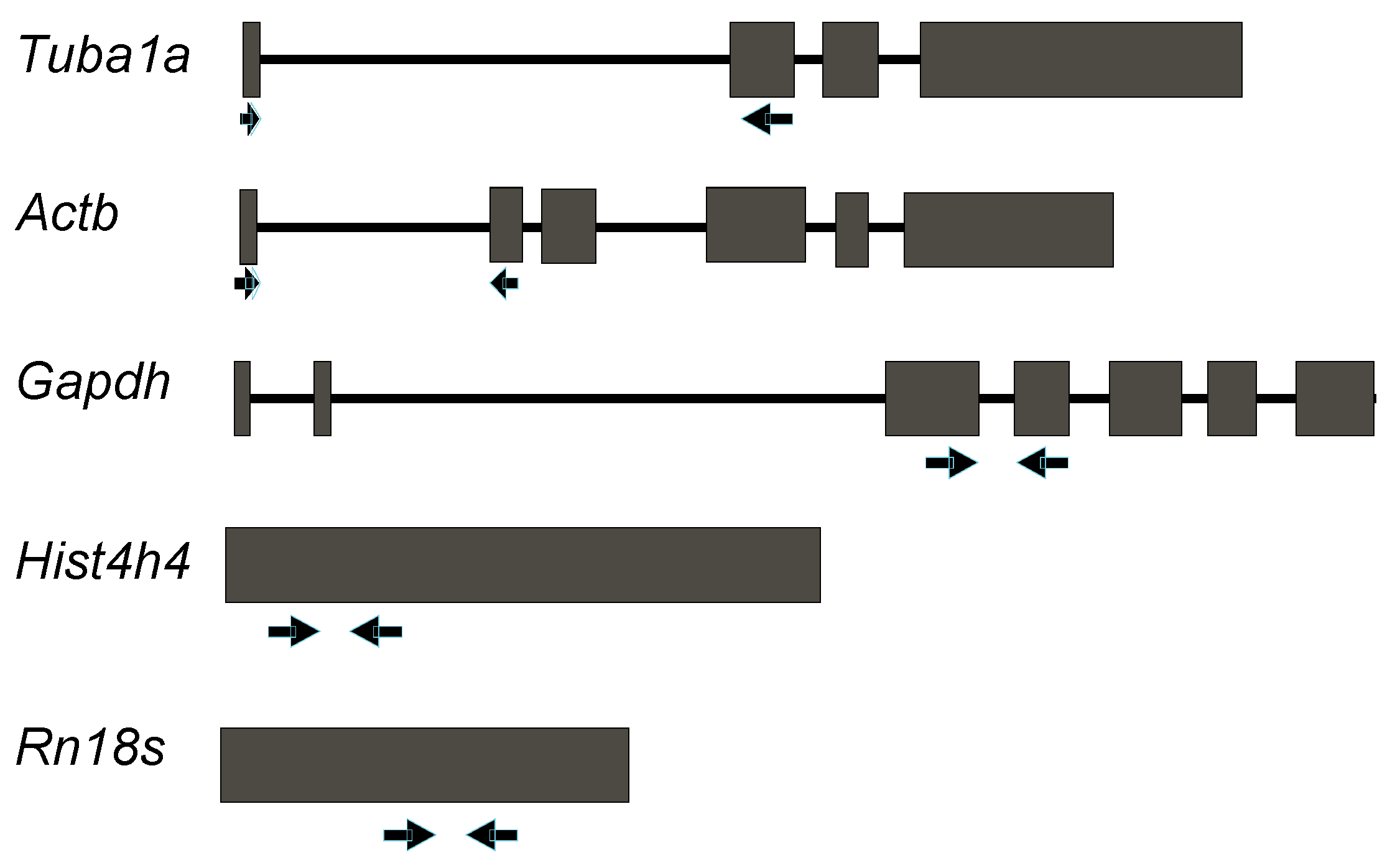
2.1. Design of Primer Pairs for qRT-PCR Expression Analysis of Reference Genes that Are Widely Used in Mouse Lung Development
| Gene Symbol | Accession | Name | Function | Localization | Pseudogenes † |
|---|---|---|---|---|---|
| Tuba1a | NM_011653 | tubulin, alpha 1A | Essential for structure and kinetics of cytoskeleton | Chr 15 (15 F1; 15 55.29 cM) | 3 |
| Actb | NM_007393 | actin, beta | Essential for structure and kinetics of cytoskeleton | Chr 5 (5 G2; 5 81.8 cM) | 39 |
| Gapdh | NM_008084 | glyceraldehyde-3-phosphate dehydrogenase | Important enzyme of glycolytic pathway | Chr 6 (6 F2; 6 59.32 cM) | 309 |
| Hist4h4 | NM_175652 | histone cluster 4, H4 | Structural component of nucleosome | Chr 6 (6 G1; 6) | 0 |
| Rn18S | NR_003278 | 18S ribosomal RNA | Ribosomal subunits | Chr 6 (6; 17) | 1 |
| Gene Symbol | Forward Primer (5'–3') | Tm (°C) | E Value | Reverse Primer (5'–3') | Tm (°C) | E Value | Primer Dimer | Secondary Structure • | Intron Spanning | Amplicon Size | Slope (R > 0.99) † | Amplification Efficiency (%) ‡ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tuba1a | CCGCGAAGCAGCAACCAT | 61.43 | 0.012 | CCAGGTCTACGAACACTGCC | 60.39 | 0.001 | No | None | Yes | 227 | −3.3711 | 97.36 |
| Actb | ACACCCGCCACCAGTTC | 64.78 | 0.032 | TACAGCCCGGGGAGCAT | 62.21 | 0.032 | No | Very weak | Yes | 110 | −3.3797 | 97.64 |
| Gapdh | TGAGTATGTCGTGGAGTCTAC | 56.33 | 4 × 10−4 | TGGACTGTGGTCATGAGCC | 59.32 | 0.004 | No | None | Yes | 261 | −3.3904 | 97.22 |
| HistH4 | ATGTCAGGACGAGGAAAAGGC | 60.34 | 4 × 10−4 | TTAGCCGCCGAAGCCGTAC | 62.70 | 0.004 | No | None | No | 312 | −3.4822 | 99.17 |
| Rn18S | GTAACCCGTTGAACCCCATT | 58.1 | 0.013 | CCATCCAATCGGTAGTAGCG | 57.93 | 0.013 | None | No | 151 | −3.4239 | 95.91 |
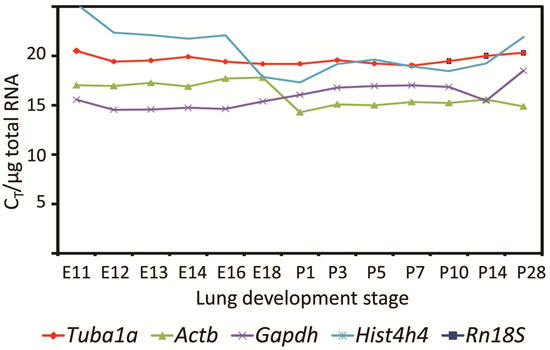
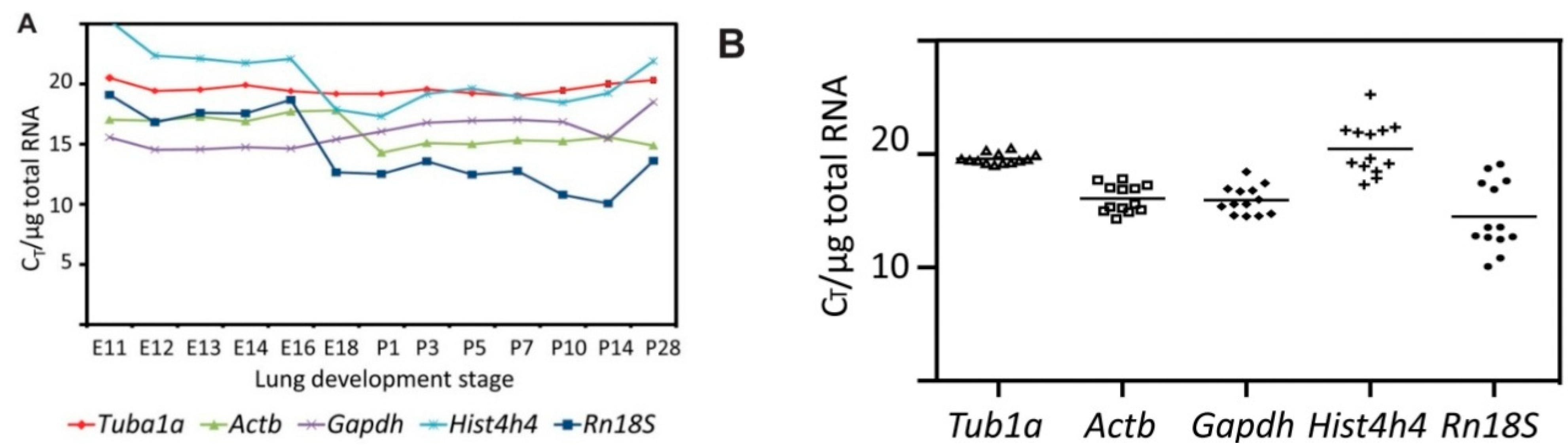
2.2. Characterization of Primer Pairs by Determining the Amplification Efficiency and through Dissociation Curve Analysis


2.3. Tuba1a Is the Least Variable Endogenous Reference for the Normalization of qRT-PCR-Based Expression Analysis during Mouse Lung Development
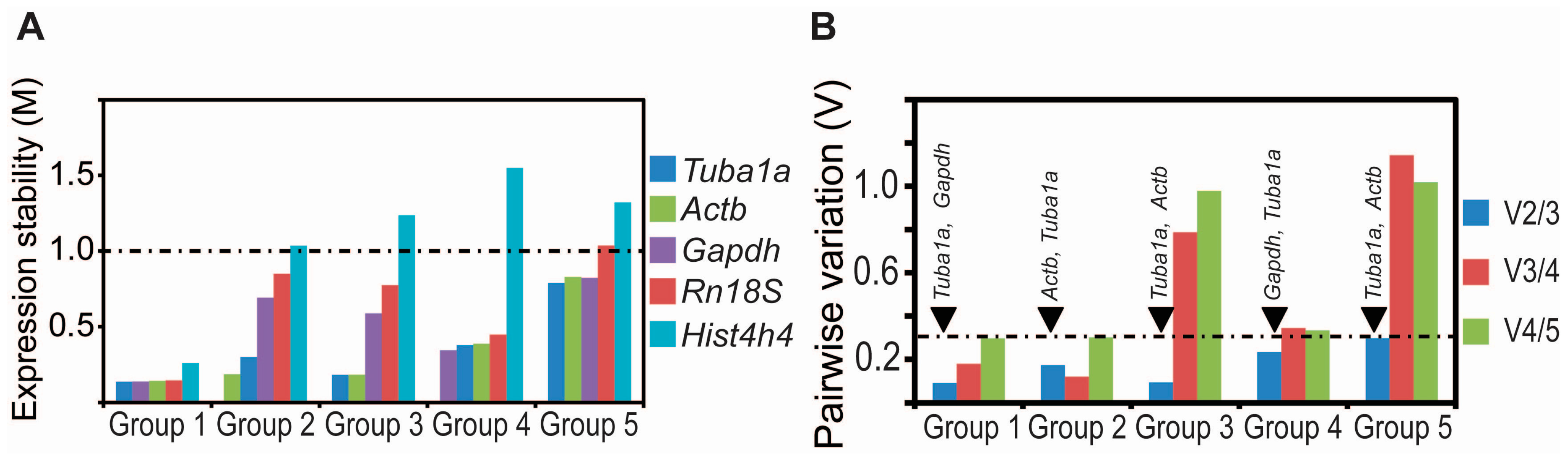
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Ranking | Stability Value (M) | Ranking | Stability Value (M) | Ranking | Stability Value (M) | Ranking | Stability Value (M) | Ranking | Stability Value (M) |
| Tuba1a | 0.136 | Actb | 0.185 | Tuba1a | 0.182 | Gapdh | 0.343 | Tuba1a | 0.788 |
| Gapdh | 0.137 | Tuba1a | 0.299 | Actb | 0.183 | Tuba1a | 0.376 | Actb | 0.822 |
| Actb | 0.142 | Gapdh | 0.691 | Gapdh | 0.586 | Actb | 0.386 | Gapdh | 0.829 |
| Rn18S | 0.145 | Rn18S | 0.849 | Rn18S | 0.773 | Rn18S | 0.447 | Rn18S | 1.035 |
| Hist4h4 | 0.258 | Hist4h4 | 1.035 | Hist4h4 | 1.236 | Hist4h4 | 1.55 | Hist4h4 | 1.321 |
| Group | V2/3 | V3/4 | V4/5 |
|---|---|---|---|
| Group 1 | Tuba1a, Gapdh | Tuba1a, Gapdh, Actb | Tuba1a, Gapdh, Rn18S |
| 0.091462 | 0.179303 | 0.296517 | |
| Group 2 | Tuba1a, Actb | Tuba1a, Gapdh, Actb | Tuba1a, Gapdh, Rn18S |
| 0.173682 | 0.12009 | 0.30073 | |
| Group 3 | Tuba1a, Actb | Tuba1a, Gapdh, Actb | Tuba1a, Gapdh, Rn18S |
| 0.094428 | 0.786888 | 0.978725 | |
| Group 4 | Tuba1a, Gapdh | Tuba1a, Gapdh, Actb | Tuba1a, Gapdh, Rn18S |
| 0.234143 | 0.343936 | 0.333048 | |
| Group 5 | Tuba1a, Actb | Tuba1a, Gapdh, Actb | Tuba1a, Gapdh, Rn18S |
| 0.29673 | 1.143188 | 1.018239 |
2.4. Expression Analysis of Two Cell Lineage Markers Using Different Normalizing Genes Suggesting Tuba1a Is the Most Suitable Endogenous Reference during Mouse Lung Development
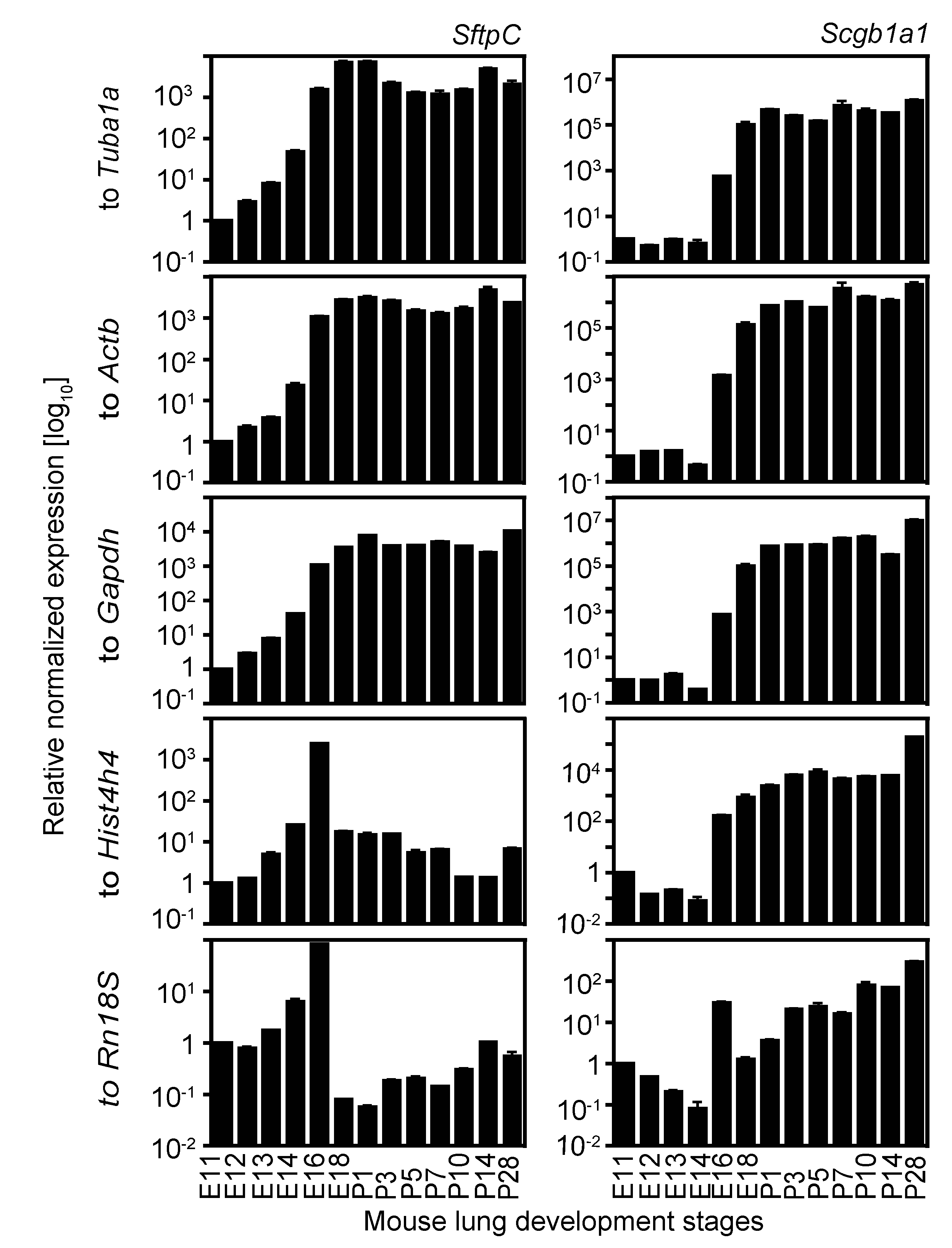
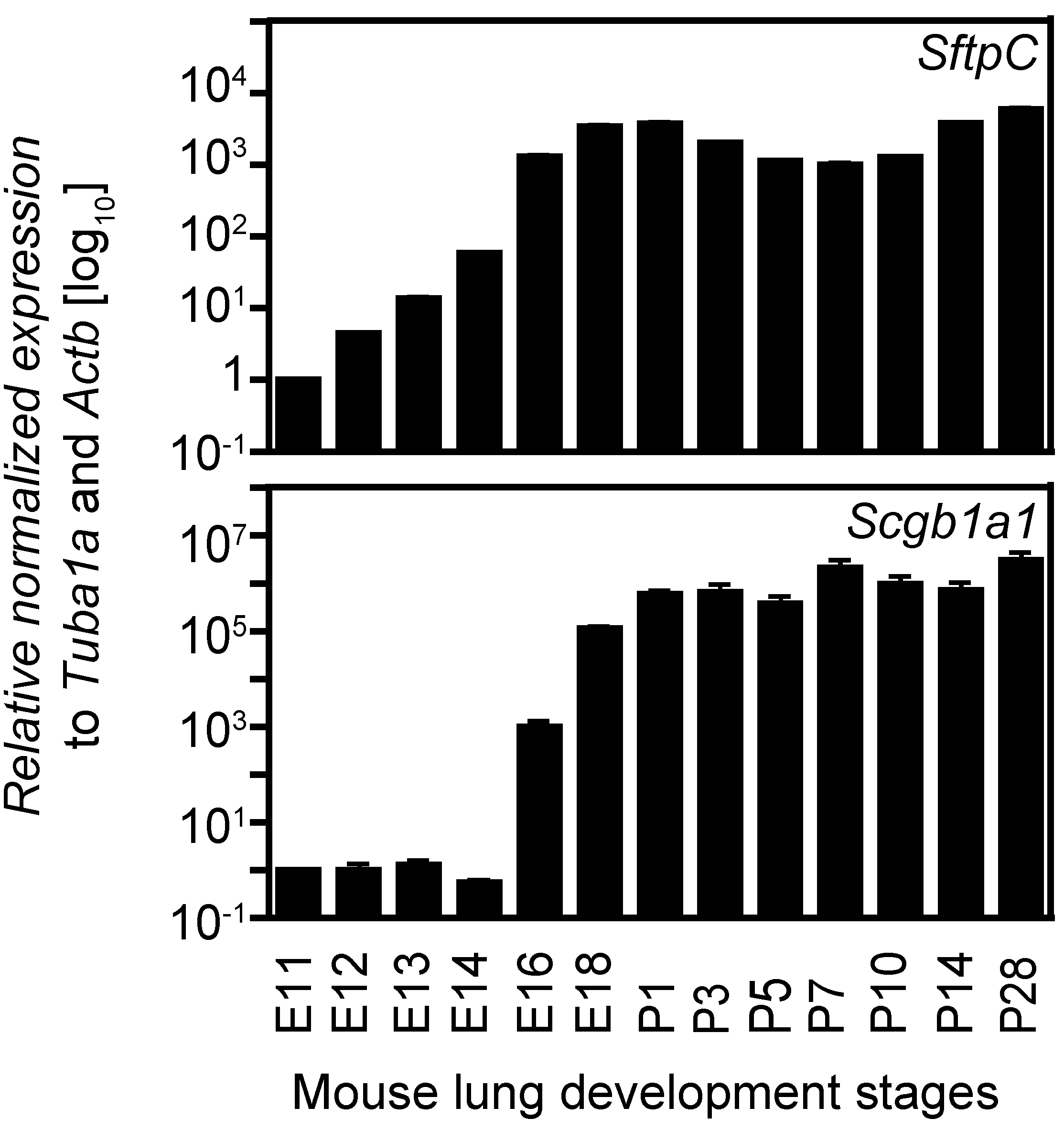
3. Discussion
4. Experimental Section
4.1. Ethics Statement
4.2. Work with Animals
4.3. RNA Isolation, cDNA Synthesis and qRT-PCR
4.4. Sequencing of PCR Products
4.5. Stability Ranking of Candidate Reference Genes
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Citri, A.; Pang, Z.P.; Sudhof, T.C.; Wernig, M.; Malenka, R.C. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat. Protoc. 2012, 7, 118–127. [Google Scholar] [CrossRef]
- Kralj, J.G.; Salit, M.L. Characterization of in vitro transcription amplification linearity and variability in the low copy number regime using External RNA Control Consortium (ERCC) spike-ins. Anal. Bioanal. Chem. 2013, 405, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Fort, P.; Marty, L.; Piechaczyk, M.; el Sabrouty, S.; Dani, C.; Jeanteur, P.; Blanchard, J.M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985, 13, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.H.; Kalinichenko, V.V.; Lim, L. Transcription factors in mouse lung development and function. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001, 280, L823–L838. [Google Scholar] [PubMed]
- Warburton, D.; Schwarz, M.; Tefft, D.; Flores-Delgado, G.; Anderson, K.D.; Cardoso, W.V. The molecular basis of lung morphogenesis. Mech. Dev. 2000, 92, 55–81. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, W.V.; Lu, J. Regulation of early lung morphogenesis: Questions, facts and controversies. Development 2006, 133, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Shibai, Y.; Mitsushita, J.; Shang, W.H.; Hirose, K.; Kamata, T. Oncogenic Ras upregulates NADPH oxidase 1 gene expression through MEK-ERK-dependent phosphorylation of GATA-6. Oncogene 2008, 27, 4921–4932. [Google Scholar] [CrossRef] [PubMed]
- Erickson, H.P. Evolution of the cytoskeleton. Bioessays 2007, 29, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, G.S.; Dmitriev, A.A.; Snezhkina, A.V.; Kudryavtseva, A.V. Deregulation of glycolysis in cancer: Glyceraldehyde-3-phosphate dehydrogenase as a therapeutic target. Expert Opin. Ther. Targets 2013, 17, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. Structure of chromatin. Annu. Rev. Biochem. 1977, 46, 931–954. [Google Scholar] [CrossRef] [PubMed]
- Elsevier, S.M.; Ruddle, F.H. Location of genes coding for 18S and 28S ribosomal RNA within the genome of Mus musculus. Chromosoma 1975, 52, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Lee, D.; Schulman, A.H. Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. Genomics 2011, 98, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-Time PCR; Meuer, S., Wittwer, C., Nakagawara, K.-I., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar]
- Bookout, A.L.; Mangelsdorf, D.J. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal. 2003, 1, e012. [Google Scholar] [CrossRef] [PubMed]
- Ririe, K.M.; Rasmussen, R.P.; Wittwer, C.T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997, 245, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Jiang, G.; Asmann, Y.W.; Tomaszek, S.; Jen, J.; Kislinger, T.; Wigle, D.A. MicroRNA networks in mouse lung organogenesis. PLoS One 2010, 5, e10854. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.K.; Wert, S.E.; Nagy, A.; Lobe, C.G.; Whitsett, J.A. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc. Natl. Acad. Sci. USA 2002, 99, 10482–10487. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.K.; Wert, S.E.; Loudy, D.E.; Shan, Z.; Blair, P.A.; Whitsett, J.A. Conditional recombination reveals distinct subsets of epithelial cells in trachea, bronchi, and alveoli. Am. J. Respir. Cell Mol. Biol. 2005, 33, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Guckenberger, M.; Andratschke, N.; Alheit, H.; Holy, R.; Moustakis, C.; Nestle, U.; Sauer, O. Definition of stereotactic body radiotherapy Principles and practice for the treatment of stage I non-small cell lung cancer. Strahlenther Onkol. 2014, 190, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Wert, S.E.; Glasser, S.W.; Korfhagen, T.R.; Whitsett, J.A. Transcriptional elements from the human SP-C gene direct expression in the primordial respiratory epithelium of transgenic mice. Dev. Biol. 1993, 156, 426–443. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.; Mehta, A.; Contreras, A.; Boettger, T.; Carraro, G.; Wheeler, M.; Cabrera-Fuentes, H.A.; Bellusci, S.; Seeger, W.; Braun, T.; et al. Hmga2 is required for canonical WNT signaling during lung development. BMC Biol. 2014, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. BioTechniques 2000, 29, 332–337. [Google Scholar] [PubMed]
- Piechaczyk, M.; Blanchard, J.M.; Marty, L.; Dani, C.; Panabieres, F.; el Sabouty, S.; Fort, P.; Jeanteur, P. Post-transcriptional regulation of glyceraldehyde-3-phosphate-dehydrogenase gene expression in rat tissues. Nucleic Acids Res. 1984, 12, 6951–6963. [Google Scholar] [CrossRef] [PubMed]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genomics 2005, 21, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Dighiero, G.; Intrator, L.; Cordonnier, C.; Tortevoye, P.; Vernant, J.P. High levels of anti-cytoskeleton autoantibodies are frequently associated with chronic GVHD. Br. J. Haematol. 1987, 67, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Solanas, M.; Moral, R.; Escrich, E. Unsuitability of using ribosomal RNA as loading control for Northern blot analyses related to the imbalance between messenger and ribosomal RNA content in rat mammary tumors. Anal. Biochem. 2001, 288, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Bilodeau-Goeseels, S.; Schultz, G.A. Changes in ribosomal ribonucleic acid content within in vitro-produced bovine embryos. Biol. Reprod. 1997, 56, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Wisnieski, F.; Calcagno, D.Q.; Leal, M.F.; dos Santos, L.C.; Gigek Cde, O.; Chen, E.S.; Pontes, T.B.; Assumpcao, P.P.; de Assumpcao, M.B.; Demachki, S.; et al. Reference genes for quantitative RT-PCR data in gastric tissues and cell lines. World J. Gastroenterol. 2013, 19, 7121–7128. [Google Scholar] [CrossRef] [PubMed]
- Thorrez, L.; van Deun, K.; Tranchevent, L.-C.; van Lommel, L.; Engelen, K.; Marchal, K.; Moreau, Y.; van Mechelen, I.; Schuit, F. Using Ribosomal Protein Genes as Reference: A Tale of Caution. PLoS One 2008, 3, e1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, S.; Furukawa, T.; Plotkin, S.A. Human cytomegalovirus stimulates host cell RNA synthesis. J. Virol. 1975, 15, 297–304. [Google Scholar] [PubMed]
- Bonefeld, B.E.; Elfving, B.; Wegener, G. Reference genes for normalization: A study of rat brain tissue. Synapse 2008, 62, 302–309. [Google Scholar] [CrossRef] [PubMed]
- McCurley, A.T.; Callard, G.V. Characterization of housekeeping genes in zebrafish: Male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Boil. 2008, 9, 102. [Google Scholar] [CrossRef]
- Olsvik, P.A.; Lie, K.K.; Jordal, A.E.; Nilsen, T.O.; Hordvik, I. Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Mol. Biol. 2005, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Dummler, B.; Tschopp, O.; Hynx, D.; Yang, Z.Z.; Dirnhofer, S.; Hemmings, B.A. Life with a single isoform of Akt: Mice lacking Akt2 and Akt3 are viable but display impaired glucose homeostasis and growth deficiencies. Mol. Cell. Biol. 2006, 26, 8042–8051. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Villanueva, D.; Deng, W.; Lopez-Camacho, C.; Shore, P. The Runx transcriptional co-activator, CBFbeta, is essential for invasion of breast cancer cells. Mol. Cancer 2010, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, Y.; Luo, D.Z.; Liao, D.J. Pseudogenes as weaknesses of ACTB (Actb) and GAPDH (Gapdh) used as reference genes in reverse transcription and polymerase chain reactions. PLoS One 2012, 7, e41659. [Google Scholar] [CrossRef] [PubMed]
- Del Moral, P.M.; Warburton, D. Explant culture of mouse embryonic whole lung, isolated epithelium, or mesenchyme under chemically defined conditions as a system to evaluate the molecular mechanism of branching morphogenesis and cellular differentiation. Methods Mol. Biol. 2010, 633, 71–79. [Google Scholar] [PubMed]
- Geneious. Available online: http://www.geneious.com (accessed on 10 November 2013).
- OligoEvaluator™. Available online: http://www.sigmaaldrich.com/life-science/custom-oligos/custom-dna/learning-center/calculator.html (accessed on 10 November 2013).
- Primer-BLAST. Available online: http://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 10 November 2013).
- Hellemans, J.; Mortier, G.; de Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Loureiro, A.; Batista, D.; Monteiro, F.; Varzea, V.; Pais, M.S.; Gichuru, E.K.; Silva, M.C. Validation of reference genes for normalization of qPCR gene expression data from Coffea spp. hypocotyls inoculated with Colletotrichum kahawae. BMC Res. Notes 2013, 6, 388. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehta, A.; Dobersch, S.; Dammann, R.H.; Bellusci, S.; Ilinskaya, O.N.; Braun, T.; Barreto, G. Validation of Tuba1a as Appropriate Internal Control for Normalization of Gene Expression Analysis during Mouse Lung Development. Int. J. Mol. Sci. 2015, 16, 4492-4511. https://doi.org/10.3390/ijms16034492
Mehta A, Dobersch S, Dammann RH, Bellusci S, Ilinskaya ON, Braun T, Barreto G. Validation of Tuba1a as Appropriate Internal Control for Normalization of Gene Expression Analysis during Mouse Lung Development. International Journal of Molecular Sciences. 2015; 16(3):4492-4511. https://doi.org/10.3390/ijms16034492
Chicago/Turabian StyleMehta, Aditi, Stephanie Dobersch, Reinhard H. Dammann, Saverio Bellusci, Olga N. Ilinskaya, Thomas Braun, and Guillermo Barreto. 2015. "Validation of Tuba1a as Appropriate Internal Control for Normalization of Gene Expression Analysis during Mouse Lung Development" International Journal of Molecular Sciences 16, no. 3: 4492-4511. https://doi.org/10.3390/ijms16034492