Differential Salt-Induced Dissociation of the p53 Protein Complexes with Circular and Linear Plasmid DNA Substrates Suggest Involvement of a Sliding Mechanism
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of Salt Concentration on p53-DNA Association
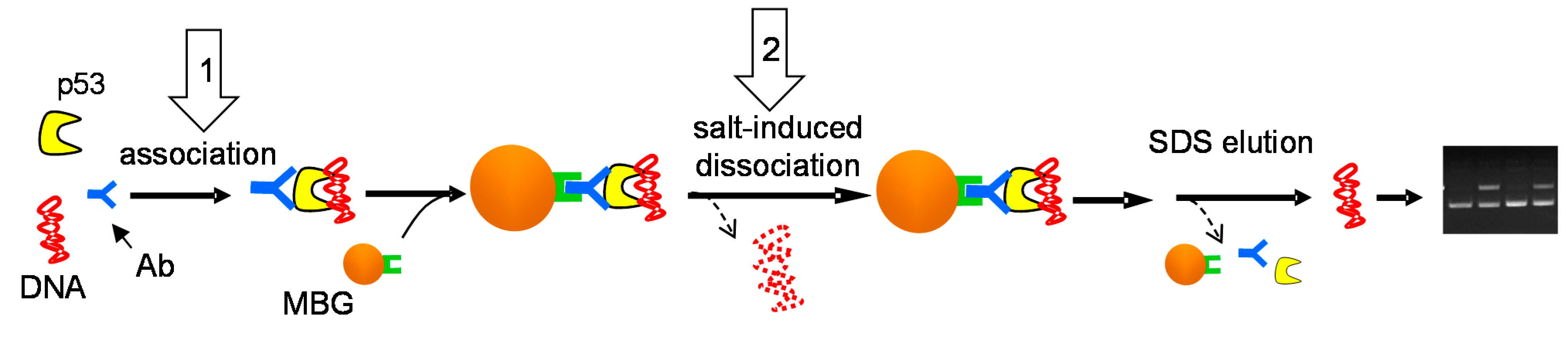

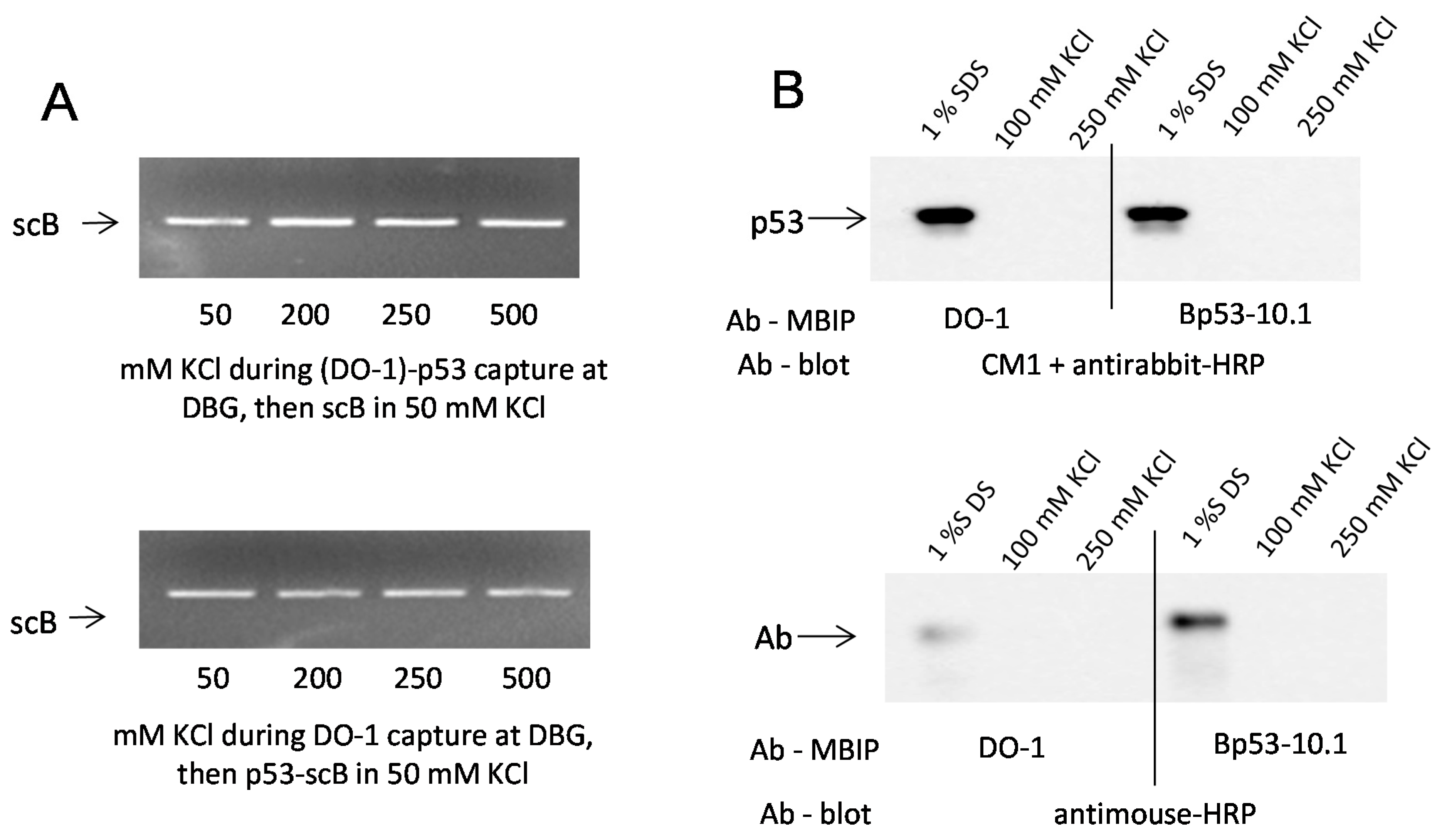
2.2. Effects of DNA Structure, Presence of p53CON and Availability of CTDBS on Salt-Induced Dissociation of the p53-DNA Complexes
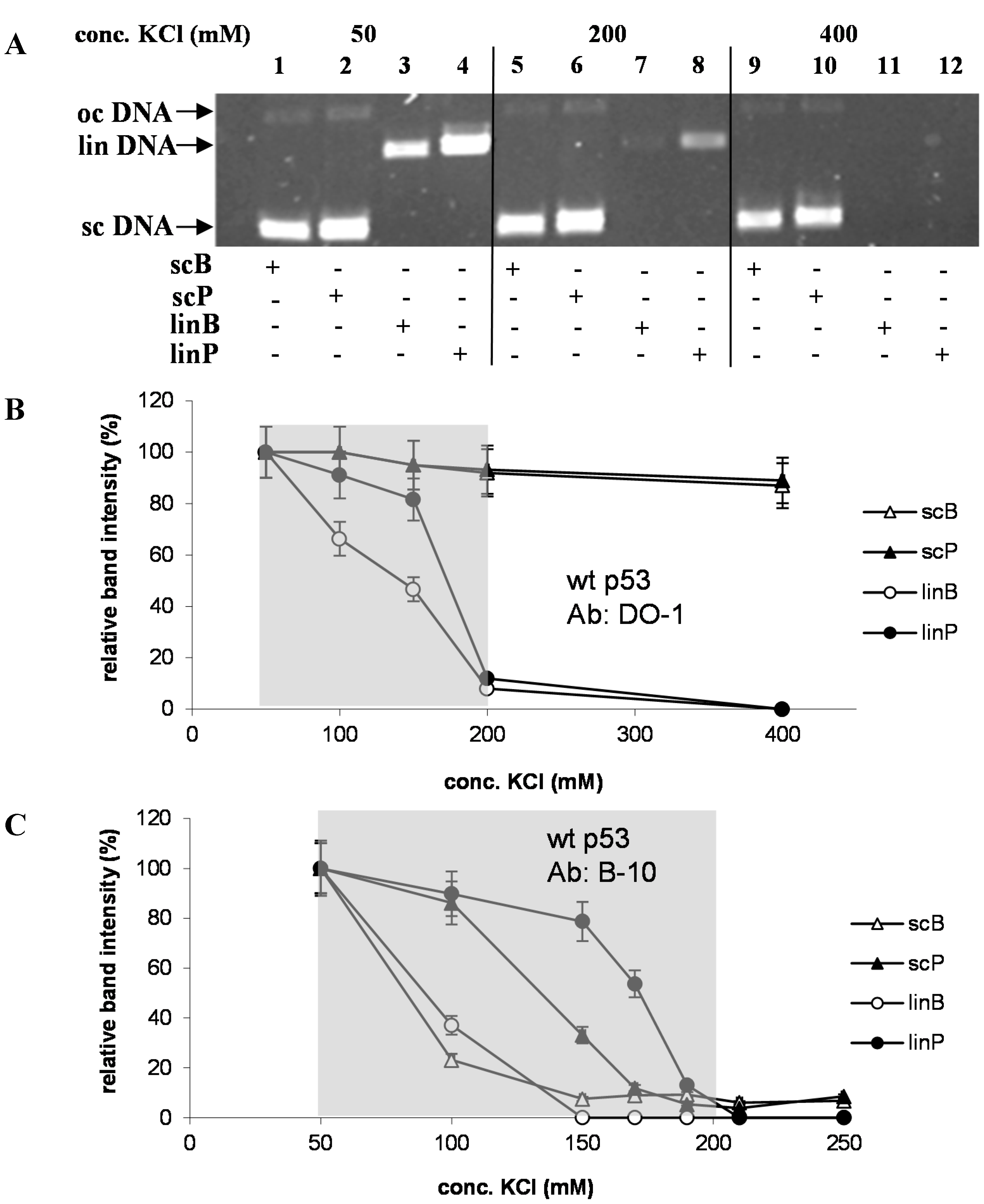
2.3. Circular Shape of p53-Bound DNA Molecule rather than DNA Superhelicity Renders the (DO-1)-p53-DNA Complexes Stability in High Salts
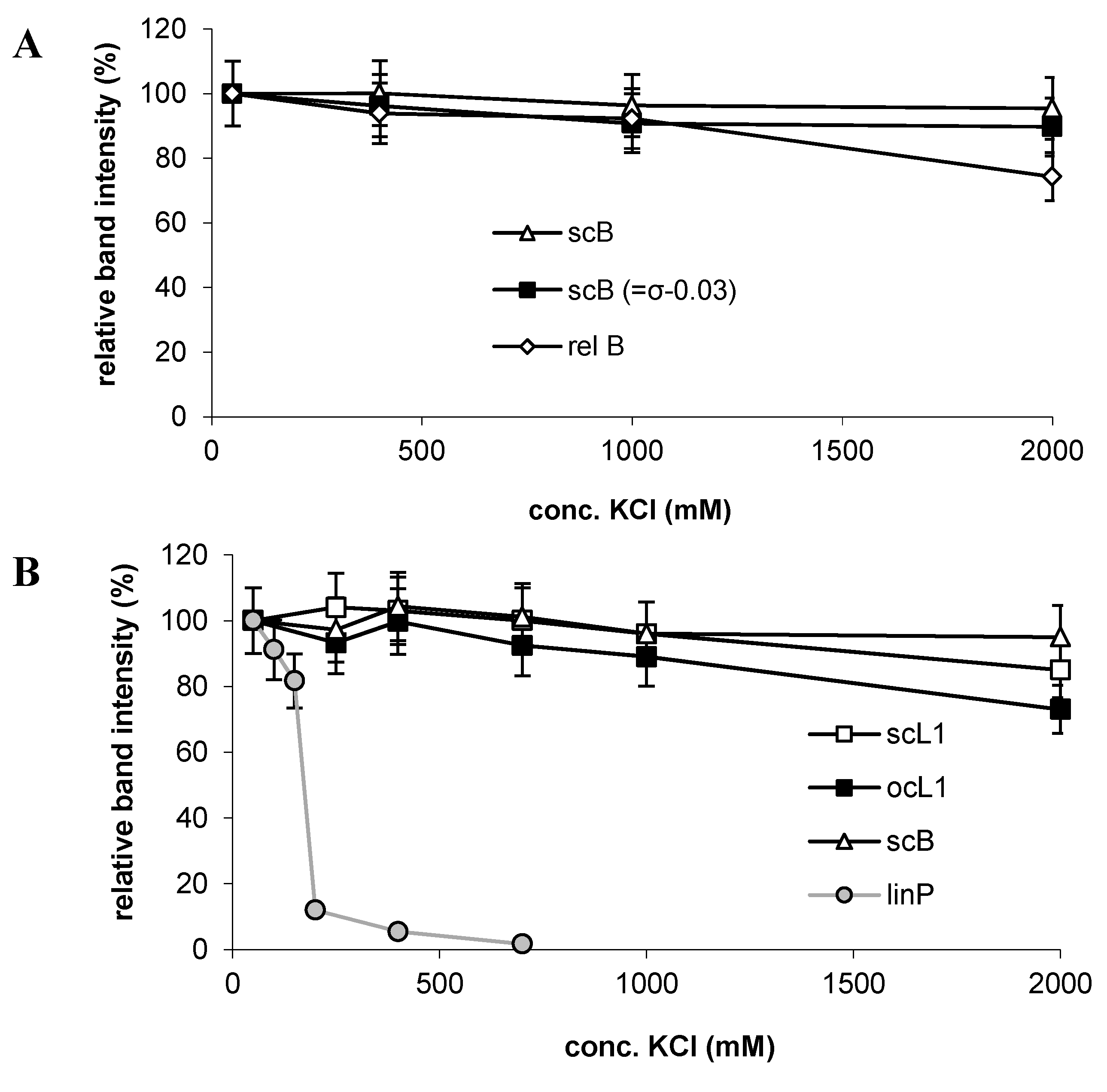
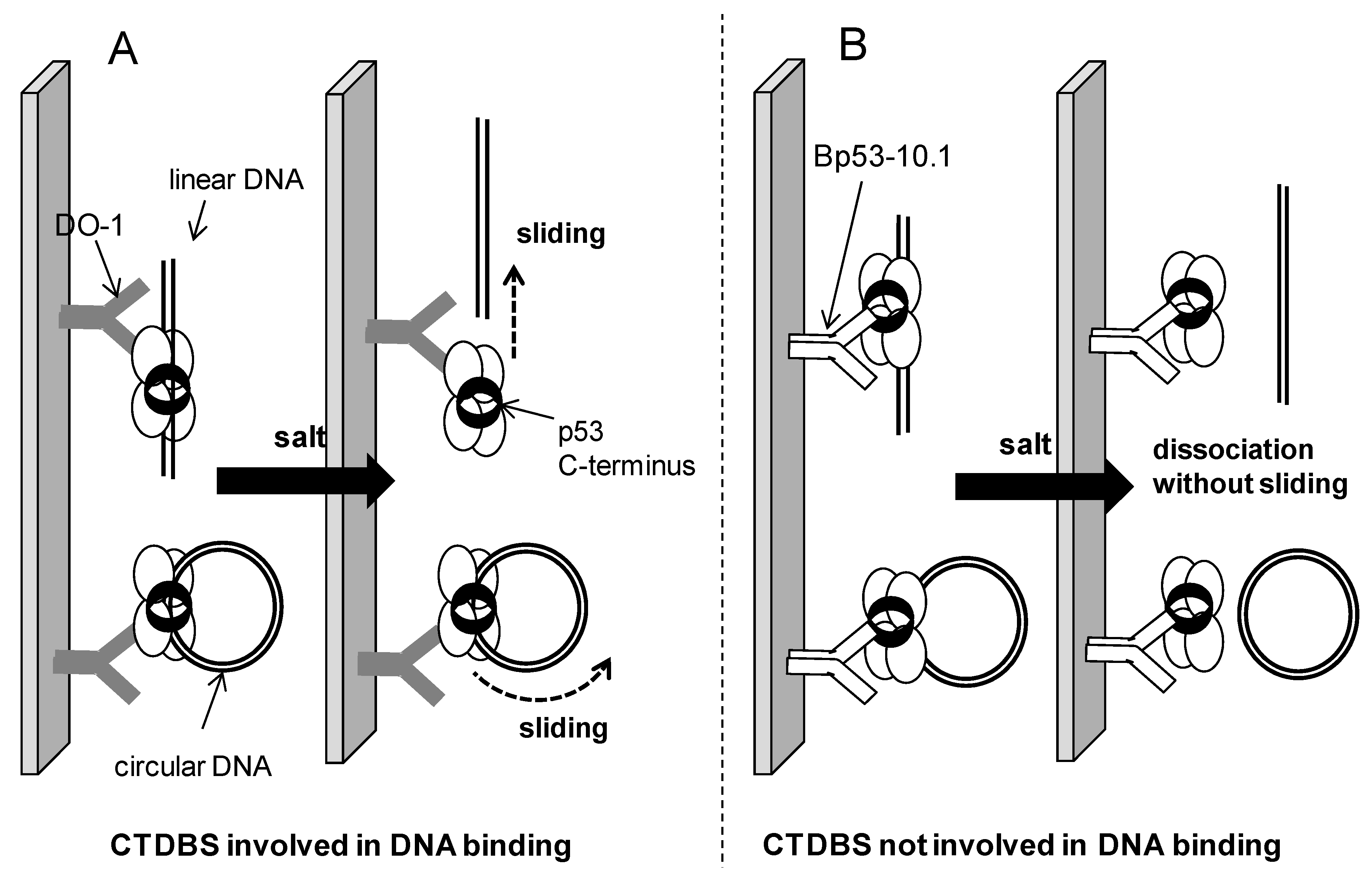
2.4. Possible Mechanism of Salt-Induced Dissociation of the p53-DNA Complexes
3. Experimental Section
3.1. DNA Samples
3.2. Monoclonal Antibodies
3.3. Preparation of Protein p53
3.4. Preparation of the p53 Immune Complexes and p53-DNA Binding
3.5. Immunoprecipitation of the p53-DNA Complexes with Magnetic Beads (MBIP Assay)
3.6. Agarose Gel Electrophoresis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Meek, D.W. Tumour suppression by p53: A role for the DNA damage response? Nat. Rev. Cancer 2009, 9, 714–723. [Google Scholar]
- Menendez, D.; Inga, A.; Resnick, M.A. The expanding universe of p53 targets. Nat. Rev. Cancer 2009, 9, 724–737. [Google Scholar]
- El-Deiry, W.S.; Kern, S.E.; Pietenpol, J.A.; Kinzler, K.W.; Vogelstein, B. Definition of a consensus binding site for p53. Nat. Genet. 1992, 1, 45–49. [Google Scholar]
- Weinberg, R.L.; Veprintsev, D.B.; Fersht, A.R. Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 2004, 341, 1145–1159. [Google Scholar]
- Brazdova, M.; Navratilova, L.; Tichy, V.; Nemcova, K.; Lexa, M.; Hrstka, R.; Pecinka, P.; Adamik, M.; Vojtesek, B.; Palecek, E.; et al. Preferential binding of hot spot mutant p53 proteins to supercoiled DNA in vitro and in cells. PLoS One 2013, 8, e59567. [Google Scholar]
- Joerger, A.C.; Fersht, A.R. Structure–function–rescue: The diverse nature of common p53 cancer mutants. Oncogene 2007, 26, 2226–2242. [Google Scholar]
- Brazdova, M.; Palecek, J.; Cherny, D.I.; Billova, S.; Fojta, M.; Pecinka, P.; Vojtesek, B.; Jovin, T.M.; Palecek, E. Role of tumor suppressor p53 domains in selective binding to supercoiled DNA. Nucleic Acids Res. 2002, 30, 4966–4974. [Google Scholar]
- Fojta, M.; Pivonkova, H.; Brazdova, M.; Nemcova, K.; Palecek, J.; Vojtesek, B. Investigations of the supercoil-selective DNA binding of wild type p53 suggest a novel mechanism for controlling p53 function. Eur. J. Biochem. 2004, 271, 3865–3876. [Google Scholar]
- Palecek, E.; Vlk, D.; Stankova, V.; Brazda, V.; Vojtesek, B.; Hupp, T.R.; Schaper, A.; Jovin, T.M. Tumor suppressor protein p53 binds preferentially to supercoiled DNA. Oncogene 1997, 15, 2201–2209. [Google Scholar]
- Pivonkova, H.; Sebest, P.; Pecinka, P.; Ticha, O.; Nemcova, K.; Brazdova, M.; Jagelska, E.B.; Brazda, V.; Fojta, M. Selective binding of tumor suppressor p53 protein to topologically constrained DNA: Modulation by intercalative drugs. Biochem. Biophys. Res. Commun. 2010, 393, 894–899. [Google Scholar]
- Coufal, J.; Jagelska, E.B.; Liao, J.C.C.; Brazda, V. Preferential binding of p53 tumor suppressor to p21 promoter sites that contain inverted repeats capable of forming cruciform structure. Biochem. Biophys. Res. Commun. 2013, 441, 83–88. [Google Scholar]
- Jagelska, E.B.; Brazda, V.; Pecinka, P.; Palecek, E.; Fojta, M. DNA topology influences p53 sequence-specific DNA binding through structural transitions within the target sites. Biochem. J. 2008, 412, 57–63. [Google Scholar]
- Jagelska, E.B.; Pivonkova, H.; Fojta, M.; Brazda, V. The potential of the cruciform structure formation as an important factor influencing p53 sequence-specific binding to natural DNA targets. Biochem. Biophys. Res. Commun. 2010, 391, 1409–1414. [Google Scholar]
- Pivonkova, H.; Brazdova, M.; Kasparkova, J.; Brabec, V.; Fojta, M. Recognition of cisplatin-damaged DNA by p53 protein: Critical role of the p53 C-terminal domain. Biochem. Biophys. Res. Commun. 2006, 339, 477–484. [Google Scholar]
- Wetzel, C.C.; Berberich, S.J. P53 binds to cisplatin-damaged DNA. Biochim. Biophys. Acta 2001, 1517, 392–397. [Google Scholar]
- Brazda, V.; Jagelska, E.B.; Fojta, M.; Palecek, E. Searching for target sequences by p53 protein is influenced by DNA length. Biochem. Biophys. Res. Commun. 2006, 341, 470–477. [Google Scholar]
- Leith, J.S.; Tafvizi, A.; Huang, F.; Uspal, W.E.; Doyle, P.S.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. Sequence-dependent sliding kinetics of p53. Proc. Natl. Acad. Sci. USA 2012, 109, 16552–16557. [Google Scholar]
- McKinney, K.; Mattia, M.; Gottifredi, V.; Prives, C. p53 Linear diffusion along DNA requires its C-terminus. Mol. Cell 2004, 16, 413–424. [Google Scholar]
- Tafvizi, A.; Huang, F.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 563–568. [Google Scholar]
- Tafvizi, A.; Huang, F.; Leith, J.S.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys. J. 2008, 95, L1–L3. [Google Scholar]
- Terakawa, T.; Kenzaki, H.; Takada, S. P53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains. J. Am. Chem. Soc. 2012, 134, 14555–14562. [Google Scholar]
- Butcher, S.; Hainaut, P.; Milner, J. Increased salt concentration reversibly destabilizes p53 quaternary structure and sequence-specific DNA-binding. Biochem. J. 1994, 298, 513–516. [Google Scholar]
- Cobb, A.M.; Jackson, B.R.; Kim, E.; Bond, P.L.; Bowater, R.P. Sequence-specific and DNA structure-dependent interactions of Escherichia coli MutS and human p53 with DNA. Anal. Biochem. 2013, 442, 51–61. [Google Scholar]
- Arbely, E.; Natan, E.; Brandt, T.; Allen, M.D.; Veprintsev, D.B.; Robinson, C.V.; Chin, J.W.; Joerger, A.C.; Fersht, A.R. Acetylation of lysine 120 of p53 endows DNA-binding specificity at effective physiological salt concentration. Proc. Natl. Acad. Sci. USA 2011, 108, 8251–8256. [Google Scholar]
- Nemcova, K.; Sebest, P.; Havran, L.; Orsag, P.; Fojta, M.; Pivonkova, H. Electrochemical detection of DNA binding by tumor suppressor p53 protein using osmium-labeled oligonucleotide probes and catalytic hydrogen evolution at the mercury electrode. Anal. Bioanal. Chem. 2014, 406, 5843–5852. [Google Scholar]
- Nemcova, K.; Havran, L.; Sebest, P.; Brazdova, M.; Pivonkova, H.; Fojta, M. A label-free electrochemical test for DNA-binding activities of tumor suppressor protein p53 using immunoprecipitation at magnetic beads. Anal. Chim. Acta 2010, 668, 166–170. [Google Scholar]
- Palecek, E.; Brazdova, M.; Brazda, V.; Palecek, J.; Billova, S.; Subramaniam, V.; Jovin, T.M. Binding of p53 and its core domain to supercoiled DNA. Eur. J. Biochem. 2001, 268, 573–581. [Google Scholar]
- Kim, E.; Albrechtsen, N.; Deppert, W. DNA-conformation is an important determinant of sequence-specific DNA binding by tumor suppressor p53. Oncogene 1997, 15, 857–869. [Google Scholar]
- Palecek, E.; Brazda, V.; Jagelska, E.; Pecinka, P.; Karlovska, L.; Brazdova, M. Enhancement of p53 sequence-specific binding by DNA supercoiling. Oncogene 2004, 23, 2119–2127. [Google Scholar]
- Gohler, T.; Jager, S.; Warnecke, G.; Yasuda, H.; Kim, E.; Deppert, W. Mutant p53 proteins bind DNA in a DNA structure-selective mode. Nucleic Acids Res. 2005, 33, 1087–1100. [Google Scholar]
- Bates, A.D.; Maxwell, A. DNA Topology; IRL Press at Oxford University Press: Oxford, UK, 1993. [Google Scholar]
- Bowater, R.P. Supercoiled DNA: Structure. In Encyclopedia of Life Sciences; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef] [Green Version]
- Gowers, D.M.; Wilson, G.G.; Halford, S.E. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc. Natl. Acad. Sci. USA 2005, 102, 15883–15888. [Google Scholar]
- Vacek, J.; Cahova, K.; Palecek, E.; Bullard, D.R.; Lavesa-Curto, M.; Bowater, R.P.; Fojta, M. Label-free electrochemical monitoring of DNA ligase activity. Anal. Chem. 2008, 80, 7609–7613. [Google Scholar]
- Bellamy, S.R.; Milsom, S.E.; Scott, D.J.; Daniels, L.E.; Wilson, G.G.; Halford, S.E. Cleavage of individual DNA strands by the different subunits of the heterodimeric restriction endonuclease BbvCI. J. Mol. Biol. 2005, 384, 641–653. [Google Scholar]
- Petty, T.J.; Emamzadah, S.; Costantino, L.; Petkova, I.; Stavridi, E.S.; Saven, J.G.; Vauthey, E.; Halazonetis, T.D. An induced fit mechanism regulates p53 DNA binding kinetics to confer sequence specificity. EMBO J. 2011, 30, 2167–2176. [Google Scholar]
- Bhattacherjee, A.; Levy, Y. Search by proteins for their DNA target site: 1. The effect of DNA conformation on protein sliding. Nucleic Acids Res. 2014, 42, 12404–12414. [Google Scholar]
- Fojta, M.; Palecek, E. Supercoiled DNA-modified mercury electrode: A highly sensitive tool for the detection of DNA damage. Anal. Chim. Acta 1997, 342, 1–12. [Google Scholar]
- Bowater, R.; Aboul-Ela, F.; Lilley, D.M. Two-dimensional gel electrophoresis of circular DNA topoisomers. Methods Enzymol. 1992, 212, 105–120. [Google Scholar]
- Vojtesek, B.; Bartek, J.; Midgley, C.A.; Lane, D.P. An immunochemical analysis of the human nuclear phosphoprotein p53: New monoclonal antibodies and epitope mapping using recombinant p53. J. Immunol. Methods 1992, 151, 237–244. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šebest, P.; Brázdová, M.; Fojta, M.; Pivoňková, H. Differential Salt-Induced Dissociation of the p53 Protein Complexes with Circular and Linear Plasmid DNA Substrates Suggest Involvement of a Sliding Mechanism. Int. J. Mol. Sci. 2015, 16, 3163-3177. https://doi.org/10.3390/ijms16023163
Šebest P, Brázdová M, Fojta M, Pivoňková H. Differential Salt-Induced Dissociation of the p53 Protein Complexes with Circular and Linear Plasmid DNA Substrates Suggest Involvement of a Sliding Mechanism. International Journal of Molecular Sciences. 2015; 16(2):3163-3177. https://doi.org/10.3390/ijms16023163
Chicago/Turabian StyleŠebest, Peter, Marie Brázdová, Miroslav Fojta, and Hana Pivoňková. 2015. "Differential Salt-Induced Dissociation of the p53 Protein Complexes with Circular and Linear Plasmid DNA Substrates Suggest Involvement of a Sliding Mechanism" International Journal of Molecular Sciences 16, no. 2: 3163-3177. https://doi.org/10.3390/ijms16023163




