Unravelling Genes and Pathways Implicated in Working Memory of Schizophrenia in Han Chinese
Abstract
:1. Introduction
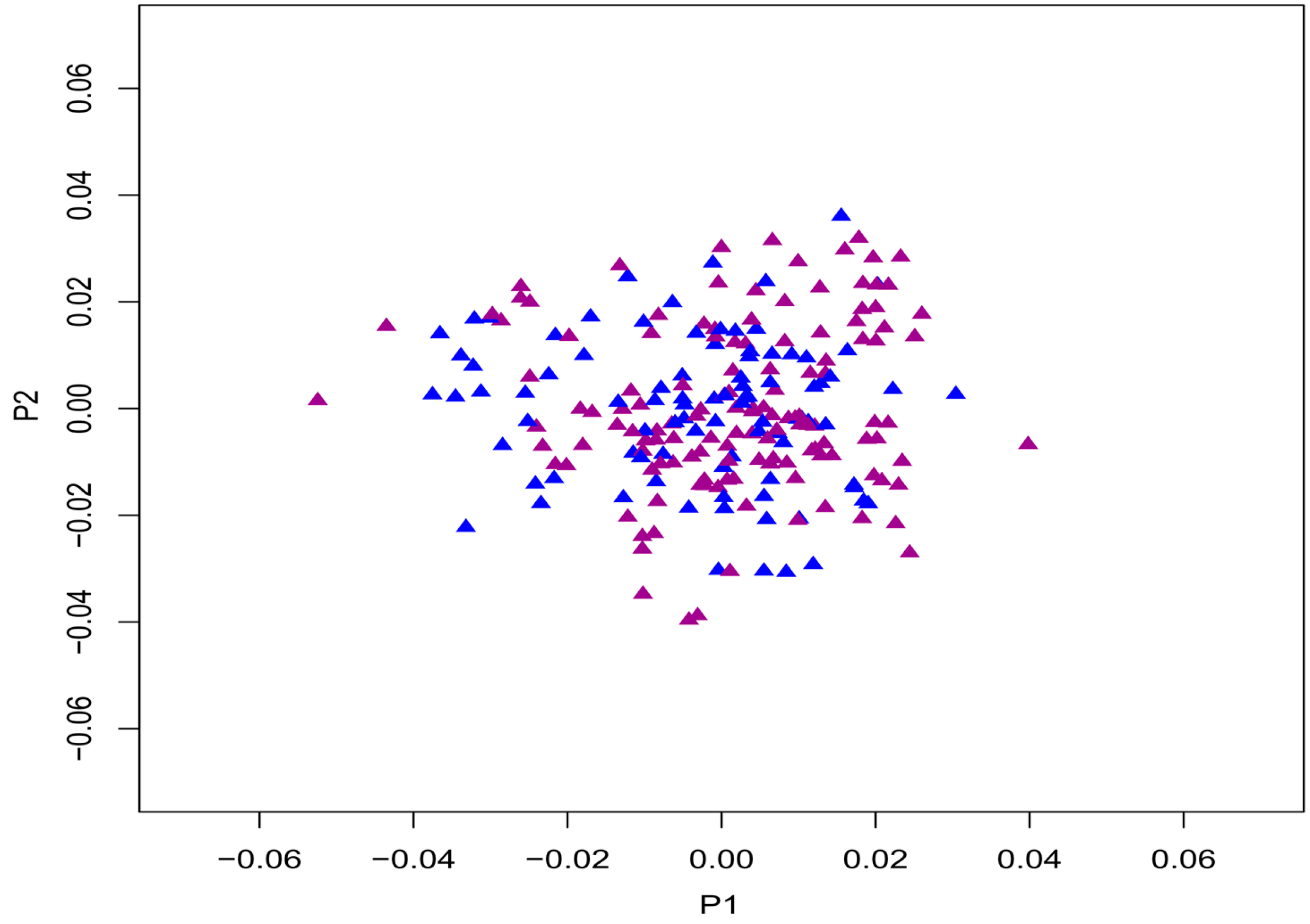
2. Results
2.1. Demographic Characteristics and Delayed-Matching-to-Sample (DMS) Test
| Demographic Characteristics and DMS Measures | Patients | Controls | Statistic Significance |
|---|---|---|---|
| Race (% Chinese) | 100 | 140 | - |
| Gender (% male) | 44.29 | 55.71 | 0.848 |
| Education (year) | 10.60 ± 5.117 | 11.86 ± 5.416 | 0.071 |
| Age | 21.57 ± 12.850 | 21.08 ± 10.776 | 0.755 |
| DMS-TC | 18.31 (58) | 15.593 (67) | 0.049 |
2.2. Analysis of Quantitative Traits (QTs) and Over-Representation Study

| CHR | SNP | Position | Type | Gene | Traits | p |
|---|---|---|---|---|---|---|
| 10 | rs1411832 | 107886255 | Intergenic | Downstream of YWHAZP5 | DMS-TC | 2.02 × 10−7 |
| 20 | rs61131853 | 41749812 | Intron | PTPRT | DMS-TC | 7.10 × 10−7 |
| 18 | rs79589976 | 73305576 | Intron | TADA2L | DMS-TC | 1.51 × 10−6 |
| 13 | rs74108723 | 90358757 | Intergenic | N/A | DMS-TC | 1.64 × 10−6 |
| 10 | rs10999524 | 72525761 | intergenic | Upstream of C10orf27 | DMS-TC | 1.95 × 10−6 |
| 7 | rs4718138 | 64303065 | Intron | ZNF138 | DMS-TC | 2.15 × 10−6 |
| 11 | rs1552511 | 92605986 | Intron | FAT3 | DMS-TC | 2.31 × 10−6 |
| 11 | rs555329 | 95993708 | Intron | FAT3 | DMS-TC | 2.41 × 10−6 |
| 18 | rs2868934 | 10204383 | Intron | TADA2L | DMS-TC | 2.56 × 10−6 |
| 7 | rs60569161 | 98199903 | intergenic | Upstream of NPTX2 | DMS-TC | 3.62 × 10−6 |
| 10 | rs7899885 | 70143774 | Intron | RUFY2 | DMS-TC | 5.02 × 10−6 |
| 12 | rs12811916 | 25550866 | Intron | DSCAML1 | DMS-TC | 5.33 × 10−6 |
| 10 | rs2281698 | 70104320 | Intron | RUFY2 | DMS-TC | 5.42 × 10−6 |
| 16 | rs4780688 | 17567540 | intergenic | Downstream of XYLT1 | DMS-TC | 5.84 × 10−6 |
| 21 | rs76659985 | 26786931 | Intron | LINC00158 | DMS-TC | 6.13 × 10−6 |
| 1 | rs3738516 | 43440201 | Intron | Downstream of SLC2A1 | DMS-TC | 6.19 × 10−6 |
| 3 | rs17609699 | 60487578 | Intron | FHIT | DMS-TC | 6.20 × 10−6 |
| 16 | rs4785000 | 58963145 | intergenic | Upstream of LOC100132798 | DMS-TC | 6.51 × 10−6 |
| 11 | rs630024 | 117534353 | Intron | MAML2 | DMS-TC | 8.09 × 10−6 |
| 10 | rs3781567 | 70105178 | Intron | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs1162753 | 70105560 | synonymous | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs17297439 | 70103461 | Intron | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs3199937 | 70102749 | Intron | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs3781568 | 70105286 | Intron | HNRNPH3 | DMS-TC | 8.11 × 10−6 |
| 10 | rs7897488 | 70179746 | Intron | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs7071140 | 70100250 | Intron | HNRNPH3 | DMS-TC | 8.42 × 10−6 |
| 19 | rs7249563 | 7009813 | intergenic | Downstream of VAPA | DMS-TC | 8.85 × 10−6 |
| 18 | rs652630 | 34899378 | Intron | CELF4 | DMS-TC | 9.70 × 10−6 |
| 2 | rs520102 | 220529636 | intergenic | Downstream of SLC4A3 | DMS-TC | 1.11 × 10−5 |
| 15 | rs2453034 | 101241344 | Intron | NPAS3 | DMS-TC | 1.17 × 10−5 |
| 16 | rs6499996 | 58993725 | intergenic | Downstream of XYLT1 | DMS-TC | 1.17 × 10−5 |
| 3 | rs6773944 | 54516884 | Intron | CACNA2D3 | DMS-TC | 1.23 × 10−5 |
| 17 | rs2322973 | 13699188 | intergenic | Downstream of LOC644649 | DMS-TC | 1.33 × 10−5 |
| 7 | rs58908055 | 83788680 | Intron | SEMA3A | DMS-TC | 1.38 × 10−5 |
| 16 | rs9930442 | 58978493 | intergenic | Downstream of LOC644649 | DMS-TC | 1.45 × 10−5 |
| 5 | rs59017736 | 26250801 | intergenic | Downstream of MSNL1 | DMS-TC | 1.56 × 10−5 |
| 4 | rs4470690 | 188721512 | intergenic | Downstream of LOC644325 | DMS-TC | 1.57 × 10−5 |
| 17 | rs8067120 | 35783565 | Intron | TADA2A | DMS-TC | 1.58 × 10−5 |
| 15 | rs1458888 | 35406129 | Intron | NPAS3 | DMS-TC | 1.60 × 10−5 |
| 1 | rs1286830 | 62270494 | Intron | INADL | DMS-TC | 1.64 × 10−5 |
| 16 | rs8060933 | 65891108 | intergenic | Downstream of LOC644649 | DMS-TC | 1.67 × 10−5 |
| 7 | rs705337 | 98226627 | intergenic | Upstream of NPTX2 | DMS-TC | 1.68 × 10−5 |
| 20 | rs6030661 | 41748131 | Intron | PTPRT | DMS-TC | 1.70 × 10−5 |
| 6 | rs4708759 | 169005721 | Intron | SMOC2 | DMS-TC | 1.76 × 10−5 |
| 16 | rs12373039 | 65894066 | intergenic | Downstream of LOC644649 | DMS-TC | 1.80 × 10−5 |
| 1 | rs1286831 | 62271962 | Intron | INADL | DMS-TC | 1.81 × 10−5 |
| 14 | rs78636353 | 88681552 | Intron | KCNK10 | DMS-TC | 1.84 × 10−5 |
| 3 | rs614673 | 118539108 | intergenic | Upstream of IGSF11 | DMS-TC | 1.87 × 10−5 |
| 15 | rs28436697 | 27624233 | Intron | GABRG3 | DMS-TC | 1.92 × 10−5 |
| 4 | rs77470375 | 83823703 | Intron | THAP9 | DMS-TC | 1.98 × 10−5 |
| 5 | rs2066960 | 131994435 | Intron | IL13 | DMS-TC | 2.08 × 10−5 |
| 20 | rs6132627 | 23444688 | Intron | LGALS4 | DMS-TC | 2.08 × 10−5 |
| 4 | rs62303604 | 82397503 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs16998600 | 82403384 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs62302363 | 82417252 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs17005142 | 82402588 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs17005144 | 82404274 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs6819741 | 82403942 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 15 | rs2575426 | 96405795 | intergenic | Downstream ofLOC441722 | DMS-TC | 2.22 × 10−5 |
| 16 | rs12931857 | 58967112 | intergenic | Upstream of CDH5 | DMS-TC | 2.25× 10−5 |
| 3 | rs80028372 | 118805552 | Intron | IGSF11 | DMS-TC | 2.26 × 10−5 |
| 4 | rs7438406 | 99702148 | intergenic | Downstream of BTF3L3 | DMS-TC | 2.28 × 10−5 |
| 20 | rs2325606 | 41738666 | Intron | PTPRT | DMS-TC | 2.33 × 10−5 |
| 3 | rs17659192 | 3108711 | Intron | IL5RA | DMS-TC | 2.41 × 10−5 |
| 3 | rs12630657 | 118842442 | Intron | IGSF11 | DMS-TC | 2.43 × 10−5 |
| 4 | rs10034975 | 122175499 | intergenic | Upstream of GPR103 | DMS-TC | 2.46 × 10−5 |
| 9 | rs7041922 | 34938198 | intergenic | Upstream of KIAA1045 | DMS-TC | 2.49 × 10−5 |
| 2 | rs6544074 | 37634473 | intergenic | Downstream of QPCT | DMS-TC | 2.52 × 10−5 |
| 4 | rs10015146 | 82401332 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.65 × 10−5 |
| 6 | rs2744229 | 25341580 | Intron | LRRC16A | DMS-TC | 2.71 × 10−5 |
| 12 | rs12318900 | 66044284 | intergenic | Upstream of KRAS | DMS-TC | 2.72 × 10−5 |
| 15 | rs9708085 | 27619217 | Intron | GABRG3 | DMS-TC | 2.83 × 10−5 |
| 5 | rs6894424 | 34732456 | Intron | RAI14 | DMS-TC | 2.85 × 10−5 |
| 10 | rs2503870 | 43796180 | Intron | HNRNPH3 | DMS-TC | 2.86 × 10−5 |
| 3 | rs6808187 | 175861931 | intergenic | Upstream of LOC730168 | DMS-TC | 2.91 × 10−5 |
| 2 | rs755300 | 241652703 | Intron | KIF1A | DMS-TC | 2.94 × 10−5 |
| 3 | rs4687154 | 190304172 | Intron | IL1RAP | DMS-TC | 2.98 × 10−5 |
| 19 | rs1353166 | 6992943 | intergenic | BRUNOL4 | DMS-TC | 3.02 × 10−5 |
| 1 | rs347272 | 162318498 | Intron | NOS1AP | DMS-TC | 3.04 × 10−5 |
| 1 | rs11577628 | 162319524 | Intron | NOS1AP | DMS-TC | 3.04 × 10−5 |
| 1 | rs347273 | 162317513 | Intron | NOS1AP | DMS-TC | 3.04 × 10−5 |
| 17 | rs8065154 | 17614947 | Intron | RAI1 | DMS-TC | 3.11 × 10−5 |
| 17 | rs6502615 | 17612023 | Intron | TADA2L | DMS-TC | 3.11 × 10−5 |
| 10 | rs12268934 | 13581758 | intergenic | Downstream of RASGEF1A | DMS-TC | 3.21 × 10−5 |
| 17 | rs11263747 | 35742069 | Intron | RAI1 | DMS-TC | 3.30 × 10−5 |
| 17 | rs11263750 | 35816826 | Intron | RAI1 | DMS-TC | 3.30 × 10−5 |
| 17 | rs11868171 | 35816330 | Intron | C17orf78 | DMS-TC | 3.30 × 10−5 |
| 17 | rs2898656 | 35806418 | Intron | ACACA | DMS-TC | 3.30 × 10−5 |
| 20 | rs2024886 | 5700696 | Intron | PTPRT | DMS-TC | 3.31 × 10−5 |
| 11 | rs583983 | 117525125 | Intron | DSCAML1 | DMS-TC | 3.32 × 10−5 |
| 18 | rs72899323 | 39845104 | Intron | LINC00907 | DMS-TC | 3.34 × 10−5 |
| 19 | rs1035525 | 39299362 | Intron | LGALS4 | DMS-TC | 3.50 × 10−5 |
| 12 | rs10846743 | 125310305 | Intron | SCARB1 | DMS-TC | 3.51 × 10−5 |
| 21 | rs7283946 | 42143503 | Intron | DSCAM | DMS-TC | 3.51 × 10−5 |
| 14 | rs10131813 | 23745533 | Intron | HOMEZ | DMS-TC | 3.58 × 10−5 |
| 14 | rs10144278 | 23749595 | Intron | HOMEZ | DMS-TC | 3.58 × 10−5 |
| 15 | rs72633609 | 96389640 | intergenic | Upstream of LOC100132798 | DMS-TC | 3.66 × 10−5 |
| 15 | rs11858405 | 96331346 | Intron | GABRG3 | DMS-TC | 3.66 × 10−5 |
| 2 | rs6544072 | 99112745 | Intron | INPP4A | DMS-TC | 3.68 × 10−5 |
| 8 | rs10111291 | 121266654 | Intron | COL14A1 | DMS-TC | 3.68 × 10−5 |
| 17 | rs58509949 | 35016090 | Intron | TADA2L | DMS-TC | 3.70 × 10−5 |
| 12 | rs7137152 | 66063249 | Intron | SCARB1 | DMS-TC | 3.75 × 10−5 |
| 12 | rs17120580 | 66018826 | intergenic | Upstream of LOC204010 | DMS-TC | 3.75 × 10−5 |
| 10 | rs2349764 | 85675348 | intergenic | At upstream of PRPF18 | DMS-TC | 3.76 × 10−5 |
| 14 | rs8005082 | 28802074 | Intron | HOMEZ | DMS-TC | 3.78 × 10−5 |
| 12 | rs7968661 | 99137384 | Intron | ANKS1B | DMS-TC | 3.81 × 10−5 |
| 13 | rs9315422 | 37055511 | intergenic | Downstream of CCNA1 | DMS-TC | 3.86 × 10−5 |
| 1 | rs4660239 | 43431528 | Intron | SLC2A1-AS1 | DMS-TC | 3.95 × 10−5 |
| 14 | rs1958005 | 33646022 | Intron | HOMEZ | DMS-TC | 3.97 × 10−5 |
| 2 | rs6544072 | 37619430 | intergenic | Downstream of QPCT | DMS-TC | 4.01 × 10−5 |
| 8 | rs7829966 | 53081817 | Intron | ST18 | DMS-TC | 4.01 × 10−5 |
| 7 | rs7794560 | 143029983 | Intron | CLCN1 | DMS-TC | 4.06 × 10−5 |
| 3 | rs17659353 | 3111436 | Intron | IL5RA | DMS-TC | 4.09 × 10−5 |
| 5 | rs10059239 | 18879807 | intergenic | Downstream of LOC646241 | DMS-TC | 4.15 × 10−5 |
| 7 | rs3298 | 154685873 | Intron | DPP6 | DMS-TC | 4.17 × 10−5 |
| 16 | rs9924423 | 64526807 | intergenic | Upstream of CDH5 | DMS-TC | 4.18 × 10−5 |
| 9 | rs12237468 | 125491634 | Intergenic | Downstream of OR1L4 | DMS-TC | 4.19 × 10−5 |
| 5 | rs56196053 | 115759247 | intergenic | Upstream of SEMA6A | DMS-TC | 4.20 × 10−5 |
| 5 | rs1480583 | 105022790 | intergenic | Downstream of RAB9P1 | DMS-TC | 4.25 × 10−5 |
| 14 | rs912857 | 33657980 | Intron | NPAS3 | DMS-TC | 4.85 × 10−5 |
| 1 | rs12024045 | 14297220 | Intron | KAZN | DMS-TC | 4.86 × 10−5 |
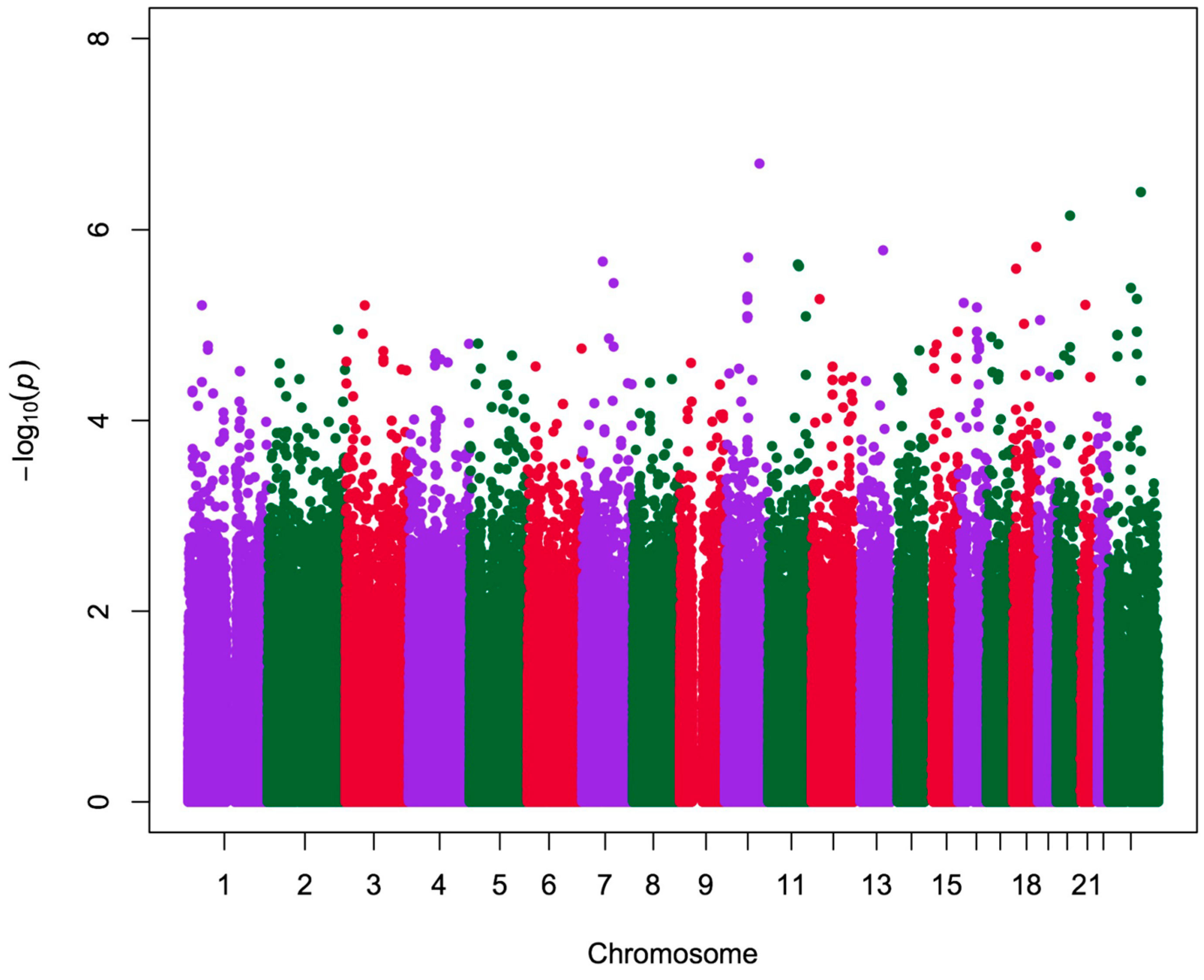
3. Discussion
3.1. Association Study of Working Memory Deficit as QT
3.2. Over-Representation of Genes in Single Pathway
3.3. Functional Characterization of Genes Post GWAS
4. Experimental Section
4.1. Participants
4.2. DMS Test Paradigm
4.3. Statistical Analysis
4.3.1. DMS Test
4.3.2. Genotyping and Quality Control
4.3.3. Correction for Population Stratification
4.3.4. Linear Regression Analysis
4.4. Over-Representation Study
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Os, J.; Kapur, S. Schizophrenia. Lancet 2009, 374, 635–645. [Google Scholar]
- McGuffin, P.; Farmer, A.E.; Gottesman, II; Murray, R.M.; Reveley, A.M. Twin concordance for operationally defined schizophrenia. Confirmation of familiality and heritability. Arch. Gen. Psychiatry 1984, 41, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Flint, J.; Munafo, M. Schizophrenia: Genesis of a complex disease. Nature 2014, 511, 412–413. [Google Scholar] [CrossRef] [PubMed]
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS). Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011, 43, 969–976. [Google Scholar]
- Yue, W.-H.; Wang, H.-F.; Sun, L.-D.; Tang, F.-L.; Liu, Z.-H.; Zhang, H.-X.; Li, W.-Q.; Zhang, Y.-L.; Zhang, Y.; Ma, C.-C. Genome-wide association study identifies a susceptibility locus for schizophrenia in han chinese at 11p11. 2. Nat. Genet. 2011, 43, 1228–1231. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (GWAS). Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [Green Version]
- Potkin, S.G.; Turner, J.; Fallon, J.; Lakatos, A.; Keator, D.; Guffanti, G.; Macciardi, F. Gene discovery through imaging genetics: Identification of two novel genes associated with schizophrenia. Mol. Psychiatry 2008, 14, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Potkin, S.G.; Guffanti, G.; Lakatos, A.; Turner, J.A.; Kruggel, F.; Fallon, J.H.; Saykin, A.J.; Orro, A.; Lupoli, S.; Salvi, E. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for alzheimerʼs disease. PLoS One 2009, 4, e6501. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.H.; Schultz, L.E.; Long, B.C.; Pletcher, M.T. Quantitative trait locus analysis identifies gabra3 as a regulator of behavioral despair in mice. Mamm. Genome 2010, 21, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Andrew, T.; Nyholt, D.R. Genome-wide association studies of quantitative traits with related individuals: Little (power) lost but much to be gained. Eur. J. Hum. Genet. 2008, 16, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A. Cohort studies and the genetics of complex disease. Nat. Genet. 2009, 41, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.; Straub, R.E.; Trampush, J.W.; Gao, Y.; Feng, N.; Xie, B.; Shin, J.H.; Lim, H.K.; Ursini, G.; Bigos, K.L. Differential effects of common variants in SCN2A on general cognitive ability, brain physiology, and messenger rna expression in schizophrenia cases and control individuals. JAMA Psychiatry 2014, 71, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Potkin, S.G.; Turner, J.A.; Guffanti, G.; Lakatos, A.; Fallon, J.H.; Nguyen, D.D.; Mathalon, D.; Ford, J.; Lauriello, J.; Macciardi, F. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr. Bull. 2009, 35, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Silver, H.; Feldman, P.; Bilker, W.; Gur, R.C. Working memory deficit as a core neuropsychological dysfunction in schizophrenia. Am. J. Psychiatry 2003, 160, 1809–1816. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Holzman, P.S.; Goldman-Rakic, P.S. Spatial working memory deficits in the relatives of schizophrenic patients. Arch. Gen. Psychiatry 1995, 52, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Conklin, H.M.; Curtis, C.E.; Katsanis, J.; Iacono, W.G. Verbal working memory impairment in schizophrenia patients and their first-degree relatives: Evidence from the digit span task. Am. J. Psychiatry 2000, 157, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Heath, S.C.; Sobin, C.; Roos, J.L.; Galke, B.L.; Blundell, M.L.; Lenane, M.; Robertson, B.; Wijsman, E.M.; Rapoport, J.L. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc. Natl. Acad. Sci. USA 2002, 99, 3717–3722. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yu, X.; Zhang, B.; Yuan, Y.; Xu, Q.; Shen, Y.; Shen, Y. An association study between polymorphisms in three genes of 14-3-3 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein) family and paranoid schizophrenia in northern chinese population. Eur. Psychiatry 2004, 19, 377–379. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Gao, R.; Xing, Q.; Du, J.; Liu, Z.; Chen, Q.; Wang, H.; Feng, G.; He, L. A family-based association study of schizophrenia with polymorphisms at three candidate genes. Neurosci. Lett. 2005, 379, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Silberberg, G.; Baruch, K.; Navon, R. Detection of stable reference genes for real-time PCR analysis in schizophrenia and bipolar disorder. Anal. Biochem. 2009, 391, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Subramanian, R.R.; Masters, S.C. 14-3-3 proteins: Structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 617–647. [Google Scholar] [CrossRef] [PubMed]
- Berg, D.; Holzmann, C.; Riess, O. 14-3-3 proteins in the nervous system. Nat. Rev. Neurosci. 2003, 4, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Dixon, A.L.; Liang, L.; Moffatt, M.F.; Chen, W.; Heath, S.; Wong, K.C.; Taylor, J.; Burnett, E.; Gut, I.; Farrall, M. A genome-wide association study of global gene expression. Nat. Genet. 2007, 39, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Sadee, W. Measuring cis-acting regulatory variants genome-wide: New insights into expression genetics and disease susceptibility. Genome Med. 2009, 1. [Google Scholar] [CrossRef]
- Pennington, B.F.; Moon, J.; Edgin, J.; Stedron, J.; Nadel, L. The neuropsychology of down syndrome: Evidence for hippocampal dysfunction. Child. Dev. 2003, 74, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.L.; Magnol, L.; Sahún, I.; Brault, V.; Duchon, A.; Prandini, P.; Gruart, A.; Bizot, J.-C.; Chadefaux-Vekemans, B.; Deutsch, S. A new mouse model for the trisomy of the ABCG1-U2AF1 region reveals the complexity of the combinatorial genetic code of down syndrome. Hum. Mol. Genet. 2009, 18, 4756–4769. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Garg, M.L.; Smith, D.W. Altered expression of histone and synaptic plasticity associated genes in the hippocampus of streptozotocin-induced diabetic mice. Metab. Brain Dis. 2013, 28, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Jarrold, C.; Baddeley, A.; Hewes, A. Genetically dissociated components of working memory: Evidence from downs and williams syndrome. Neuropsychologia 1999, 37, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; McClellan, J.M.; McCarthy, S.E.; Addington, A.M.; Pierce, S.B.; Cooper, G.M.; Nord, A.S.; Kusenda, M.; Malhotra, D.; Bhandari, A. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 2008, 320, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Gulsuner, S.; Walsh, T.; Watts, A.C.; Lee, M.K.; Thornton, A.M.; Casadei, S.; Rippey, C.; Shahin, H.; Nimgaonkar, V.L.; Go, R.C. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 2013, 154, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Besco, J.A.; Hooft van Huijsduijnen, R.; Frostholm, A.; Rotter, A. Intracellular substrates of brain-enriched receptor protein tyrosine phosphatase RHO (RPTPP/PTPRT). Brain Res. 2006, 1116, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Peng, G.; Zhu, Y.; Dong, H.; Amos, C.I.; Xiong, M. Genome-wide gene and pathway analysis. Eur. J. Hum. Genet. 2010, 18, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Luo, L.; Siu, H.; Zhu, Y.; Hu, P.; Hong, S.; Zhao, J.; Zhou, X.; Reveille, J.D.; Jin, L. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur. J. Hum. Genet. 2009, 18, 111–117. [Google Scholar] [CrossRef]
- Ramanan, V.K.; Shen, L.; Moore, J.H.; Saykin, A.J. Pathway analysis of genomic data: Concepts, methods, and prospects for future development. TRENDS Genet. 2012, 28, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Eleftherohorinou, H.; Wright, V.; Hoggart, C.; Hartikainen, A.-L.; Jarvelin, M.-R.; Balding, D.; Coin, L.; Levin, M. Pathway analysis of gwas provides new insights into genetic susceptibility to 3 inflammatory diseases. PLoS One 2009, 4, e8068. [Google Scholar] [CrossRef] [PubMed]
- Pharoah, P.D.; Tyrer, J.; Dunning, A.M.; Easton, D.F.; Ponder, B.A.; Investigators, S. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007, 3, e42. [Google Scholar] [CrossRef] [PubMed]
- OʼDushlaine, C.; Kenny, E.; Heron, E.A.; Segurado, R.; Gill, M.; Morris, D.W.; Corvin, A. The SNP ratio test: Pathway analysis of genome-wide association datasets. Bioinformatics 2009, 25, 2762–2763. [Google Scholar] [CrossRef] [PubMed]
- Lesnick, T.G.; Papapetropoulos, S.; Mash, D.C.; Ffrench-Mullen, J.; Shehadeh, L.; de Andrade, M.; Henley, J.R.; Rocca, W.A.; Ahlskog, J.E.; Maraganore, D.M. A genomic pathway approach to a complex disease: Axon guidance and parkinson disease. PLoS Genet. 2007, 3, e98. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Bucan, M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007, 81, 1278–1283. [Google Scholar] [CrossRef] [PubMed]
- Poltorak, M.; Hemperly, J.J.; Williams, J.R.; El-Mallakh, R.; Freed, W.J. Disturbances in cell recognition molecules (N-CAM and L1 antigen) in the CSF of patients with schizophrenia. Exp. Neurol. 1995, 131, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Conant, K.; Herman, M.M.; van Kammen, D.P.; Sedvall, G. Characterization of human cleaved N-Cam and association with schizophrenia. Exp. Neurol. 2001, 172, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Brennaman, L.H.; Maness, P.F. NCAM in neuropsychiatric and neurodegenerative disorders. In Structure and Function of the Neural Cell Adhesion Molecule Ncam; Springer: NY, USA, 2010; pp. 299–317. [Google Scholar]
- Vance, A.; Hall, N.; Casey, M.; Karsz, F.; Bellgrove, M.A. Visuospatial memory deficits in adolescent onset schizophrenia. Schizophr. Res. 2007, 93, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P.; Tarone, R.; McLaughlin, J.K. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology 2011, 22, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Wang, L.; Meltzer, H.Y.; Zhao, Z. Common variants conferring risk of schizophrenia: A pathway analysis of gwas data. Schizophr. Res. 2010, 122, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Yang, X.; Kaplan, L.M.; Molony, C.; Schadt, E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet. 2010, 86, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Menashe, I.; Maeder, D.; Garcia-Closas, M.; Figueroa, J.D.; Bhattacharjee, S.; Rotunno, M.; Kraft, P.; Hunter, D.J.; Chanock, S.J.; Rosenberg, P.S. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 2010, 70, 4453–4459. [Google Scholar] [CrossRef] [PubMed]
- Nicolae, D.L.; Gamazon, E.; Zhang, W.; Duan, S.; Dolan, M.E.; Cox, N.J. Trait-associated SNPs are more likely to be eqtls: Annotation to enhance discovery from gwas. PLoS Genet. 2010, 6, e1000888. [Google Scholar] [CrossRef] [PubMed]
- Spencer, C.C.; Su, Z.; Donnelly, P.; Marchini, J. Designing genome-wide association studies: Sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009, 5, e1000477. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, H.; Zhang, C.; Huang, C.; Li, N.; Li, M.; Li, Y.; Deng, W.; Ma, X.; Xiang, B.; Wang, Q.; et al. Unravelling Genes and Pathways Implicated in Working Memory of Schizophrenia in Han Chinese. Int. J. Mol. Sci. 2015, 16, 2145-2161. https://doi.org/10.3390/ijms16012145
Ren H, Zhang C, Huang C, Li N, Li M, Li Y, Deng W, Ma X, Xiang B, Wang Q, et al. Unravelling Genes and Pathways Implicated in Working Memory of Schizophrenia in Han Chinese. International Journal of Molecular Sciences. 2015; 16(1):2145-2161. https://doi.org/10.3390/ijms16012145
Chicago/Turabian StyleRen, Hongyan, Chengcheng Zhang, Chaohua Huang, Na Li, Mingli Li, Yinfei Li, Wei Deng, Xiaohong Ma, Bo Xiang, Qiang Wang, and et al. 2015. "Unravelling Genes and Pathways Implicated in Working Memory of Schizophrenia in Han Chinese" International Journal of Molecular Sciences 16, no. 1: 2145-2161. https://doi.org/10.3390/ijms16012145





