Isolation and Characterization of Six AP2/ERF Transcription Factor Genes in Chrysanthemum nankingense
Abstract
:1. Introduction
2. Results and Discussion
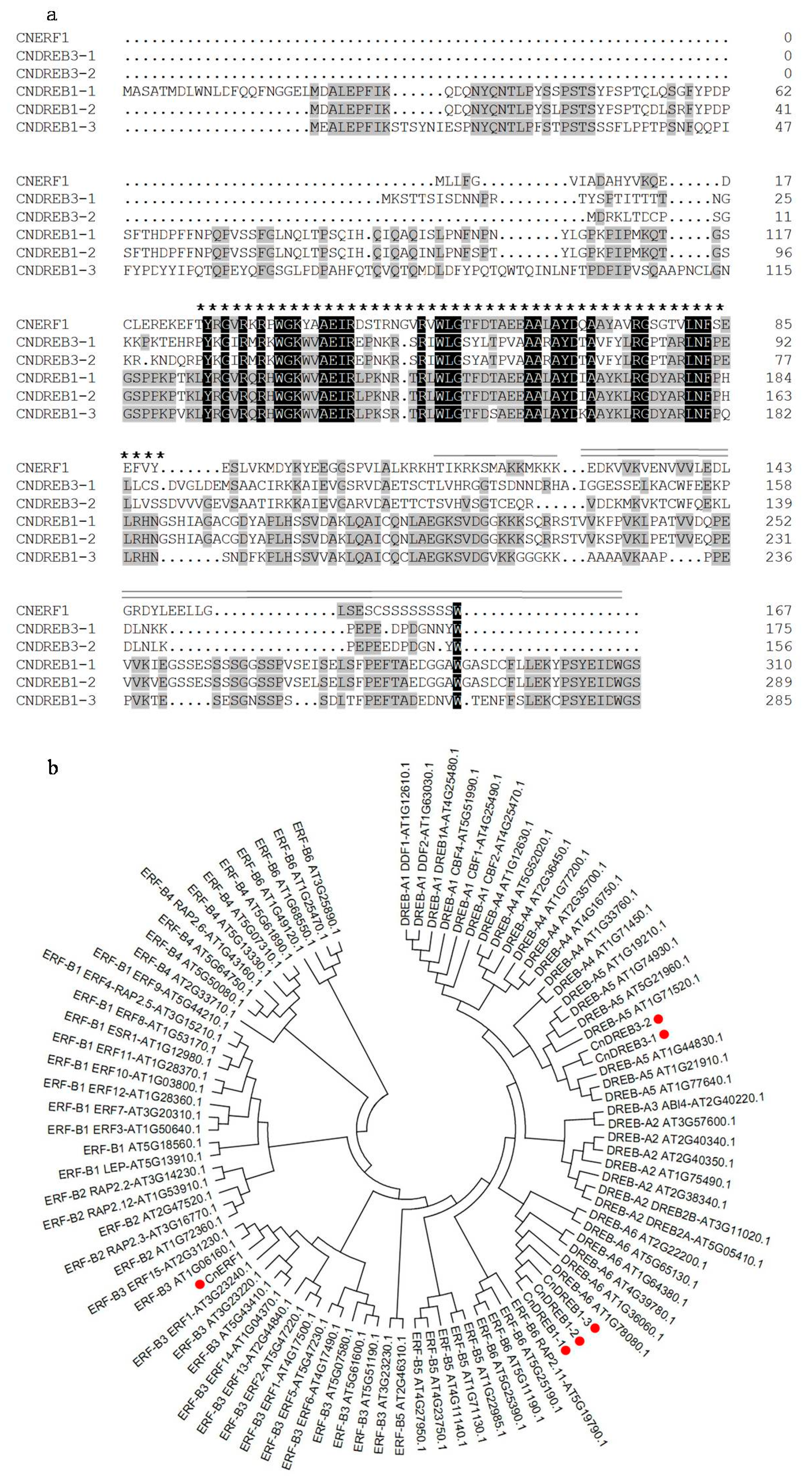
2.1. Identification of Six AP2/ERF TFs in C. nankingense
| Gene | GenBank Accession No. | cDNA Length (bp) | Amino Acids Length (aa) | AtAP2/ERF Orthologs | Locus Name | E-Value |
|---|---|---|---|---|---|---|
| CnERF1 | KF986840 | 683 | 167 | ERF1B | AT3G23240 | 1 × 10−38 |
| CnDREB1-1 | KF986841 | 1241 | 311 | RAP2.4 | AT1G78080.1 | 2 × 10−69 |
| CnDREB1-2 | KF986842 | 1164 | 290 | RAP2.4 | AT1G78080.1 | 3 × 10−69 |
| CnDREB1-3 | KF986843 | 1246 | 286 | ERF055 | AT1G78080.1 | 8 × 10−42 |
| CnDREB3-1 | KF986844 | 881 | 175 | ERF008 | AT2G23340.1 | 4 × 10−45 |
| CnDREB3-2 | KF986845 | 658 | 156 | ERF008 | AT2G23340.1 | 1 × 10−48 |
2.2. Subcellular Localization of Cnap2/Erf Products and the Transcription Activation of the TFs


2.3. Transcription of CnAP2/ERF TFs in Response to Abiotic Stress

3. Experimental Section
3.1. Plant Materials and Stress Treatments
3.2. Isolation and Sequencing of Full-Length CnAP2/ERF cDNAs
| Primer Name | Sequence (5' to 3') | Annotation |
|---|---|---|
| AP | AAGCAGTGGTATCAACGCAGAGTAC | Universal primers for 3' RACE |
| dT-AP | AAGCAGTGGTATCAACGCAGAGTACTTTTTTTTTTTTTTTT | |
| AUAP | GGCCACGCGTCGACTAGTAC | Universal primers for 5' RACE |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | |
| CnERF1-F | ATGCTTCTCTTCGGGGTCATTGCT | ORF of CnAP2/ERFs |
| CnERF1-R | TCACCAACTAGAACTACTGCTGCTGCT | |
| CnDREB1-1-F | ATGGCTTCAGCTACAATGGACTTAT | |
| CnDREB1-1-R | ATATACCCTCATAAACACTGCCACG | |
| CnDREB1-2-F | ATGGATGCACTAGAACCATTCATCAAGC | |
| CnDREB1-2-R | CTAGATAGAACCCCAATCAATCTCGTAC | |
| CnDREB1-3-F | ATGGAAGCACTTGAACCTTTTATCA | |
| CnDREB1-3-R | CTATAATGAACCCCAGTCAATCTCG | |
| CnDREB3-1-F | ATGAAATCCACAACATCCATCAGCG | |
| CnDREB3-1-R | TCACCAATAATTATTTCCATCCGGATC | |
| CnDREB3-2-F | ATGGACAGAAAATTAACAGACTGTCCATC | |
| CnDREB3-2-R | TCACCAATAATTTCCATCCGGATCTTCT | |
| EFIα-F | TTTTGGTATCTGGTCCTGGAG | qRT-PCR for CnEFIα |
| EFIα-R | CCATTCAAGCGACAGACTCA | |
| CnERF1-BamHI-F | CGGGATCCGGATGCTTCTCTTCGGGGTCATTG | Vector construction of CnAP2/ERFs |
| CnERF1-NOTI-R | TTGCGGCCGCGATCACCAACTAGAACTACTGCTGCTG | |
| CnDREB1-1-BamHI-F | CGGGATCCGGATGGCTTCAGCTACAATGGAC | |
| CnDREB1-1-NOTI-R | TTGCGGCCGCGAGATAGAACCCCAATCAATCTCGTA | |
| CnDREB1-2-BamHI-F | CGGGATCCGGATGGATGCACTAGAACCATTCAT | |
| CnDREB1-2-NOTI-R | TTGCGGCCGCGAGATAGAACCCCAATCAATCTCGTA | |
| CnDREB1-3-BamHI-F | CGGGATCCGGATGGAAGCACTTGAACCTTTTATC | |
| CnDREB1-3-NOTI-R | TTGCGGCCGCGATATTGAACCCCAGTCAATCTCGTAC | |
| CnDREB3-1-BamHI-F | CGGGATCCGGATGAAATCCACAACATCCATCAGC | |
| CnDREB3-1-NOTI-R | TTGCGGCCGCGATCACCAATAATTATTTCCATCCGG | |
| CnDREB3-2-BamHI-F | CGGGATCCGGATGGACAGAAAATTAACAGACTGTC | |
| CnDREB3-2-NOTI-R | TTGCGGCCGCGATCACCAATAATTTCCATCCGG |
3.3. Sequence Alignment and Phylogenetic Analysis
3.4. Subcellular Localization of CnAP2/ERF
3.5. Trans-Activation Activity Assay of CnAP2/ERF
3.6. Transcription Profiling of the CnAP2/ERF TFs
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M. Inducing drought tolerance in plants: Recent advances. Biotechnol. Adv. 2010, 28, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; van Montagu, M.; Jofuku, K.D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Cai, B.; Peng, R.; Zhu, B.; Jin, X.; Xue, Y.; Gao, F.; Fu, X.; Tian, Y.; Zhao, W. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Peng, R.; Cheng, Z.M.; Zhang, J.; Cai, B.; Zhang, Z.; Gao, F.; Zhu, B.; Fu, X.; Jin, X. Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Sci. Hortic. 2009, 123, 73–81. [Google Scholar] [CrossRef]
- Qin, Q.; Liu, J.; Zhang, Z.; Peng, R.; Xiong, A.; Yao, Q.; Chen, J. Isolation, optimization, and functional analysis of the cDNA encoding transcription factor OsDREB1B in Oryza Sativa L. Mol. Breed. 2007, 19, 329–340. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; van Lammeren, A.A.; Miki, B.L. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Sohn, K.H.; Lee, S.C.; Jung, H.W.; Hong, J.K.; Hwang, B.K. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006, 61, 897–915. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Xu, Z.; Xia, L.; Chen, M.; Cheng, X.; Zhang, R.; Li, L.; Zhao, Y.; Lu, Y.; Ni, Z.; Liu, L. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 2007, 65, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Riaño-Pachón, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Chen, X.; Xu, Z.; Guan, S.; Li, L.; Li, A.; Guo, J.; Mao, L.; Ma, Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 2008, 59, 4095–4107. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Chen, J.; Yao, Q.; Xiong, F.; Sun, C.; Zhou, X.; Zhang, J.; Xiong, A. Discovery and expression profile analysis of AP2/ERF family genes from Triticum aestivum. Mol. Biol. Rep. 2011, 38, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, Z.; Hu, X.; Yin, J.; Li, W.; Rao, G.; Zhang, X.; Huang, C.; Anderson, N.; Zhang, Q. Chrysanthemum genetic resources and related genera of Chrysanthemum collected in China. Genet. Res. Crop. Evol. 2009, 56, 937–946. [Google Scholar] [CrossRef]
- Cheng, X.; Chen, S.; Chen, F.; Fang, W.; Deng, Y.; She, L. Interspecific hybrids between Dendranthema morifolium (Ramat.) Kitamura and D. nankingense (Nakai) Tzvel. achieved using ovary rescue and their cold tolerance characteristics. Euphytica 2010, 172, 101–108. [Google Scholar] [CrossRef]
- Gao, H.; Song, A.; Zhu, X.; Chen, F.; Jiang, J.; Chen, Y.; Sun, Y.; Shan, H.; Gu, C.; Li, P. The heterologous expression in Arabidopsis of a chrysanthemum Cys2/His2 zinc finger protein gene confers salinity and drought tolerance. Planta 2012, 235, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; An, J.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 2014, 80, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; He, K.; Liu, D.; Bai, S.; Gu, X.; Wei, L.; Luo, J. DATF: A database of Arabidopsis transcription factors. Bioinformatics 2005, 21, 2568–2569. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration-and Cold-Inducible Gene Expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Khong, G.N.; Richaud, F.; Coudert, Y.; Pati, P.K.; Santi, C.; Périn, C.; Breitler, J.C.; Meynard, D.; Vinh, D.N.; Guiderdoni, E. Modulating rice stress tolerance by transcription factors. Biotechnol. Genet. Eng. 2008, 25, 381–404. [Google Scholar] [CrossRef]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Ohta, M.; Matsui, K.; Hiratsu, K.; Shinshi, H.; Ohme-Takagi, M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 2001, 13, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Q.-Y.; Cheng, X.-G.; Xu, Z.-S.; Li, L.-C.; Ye, X.-G.; Xia, L.-Q.; Ma, Y.-Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007, 353, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Nat. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi‐Shinozaki, K. Identification of cold‐inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004, 38, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, T.; Kato, W.; Asada, Y.; Sako, K.; Sato, T.; Sonoda, Y.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Tamaoki, M.; Arakawa, K. DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J. Plant Res. 2009, 122, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-J.; Liu, J.-Y. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar]
- Tournier, B.; Sanchez-Ballesta, M.T.; Jones, B.; Pesquet, E.; Regad, F.; Latché, A.; Pech, J.; Bouzayen, M. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett. 2003, 550, 149–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhai, Q.; Wei, J.; Li, S.; Wang, B.; Huang, T.; Du, M.; Sun, J.; Kang, L.; Li, C. Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores. PLoS Genet. 2013, 9, e1003964. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Liao, P.; Kuo, W.; Lin, T. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Ni, Z.; Liu, L.; Nie, L.; Li, L.; Chen, M.; Ma, Y. Characterization of the TaAIDFa gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol. Genet. Genomics 2008, 280, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Moran, P.J.; Thompson, G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001, 125, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiang, J.; Chen, S.; Qi, X.; Peng, H.; Li, P.; Song, A.; Guan, Z.; Fang, W.; Liao, Y. Next-generation sequencing of the Chrysanthemum nankingense (Asteraceae) transcriptome permits large-scale unigene assembly and SSR marker discovery. PLoS One 2013, 8, e62293. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Zhu, X.; Chen, F.; Gao, H.; Jiang, J.; Chen, S. A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int. J. Mol. Sci. 2014, 15, 5063–5078. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Lu, J.; Jiang, J.; Chen, S.; Guan, Z.; Fang, W.; Chen, F. Isolation and characterisation of Chrysanthemum crassum SOS1, encoding a putative plasma membrane Na+/H+ antiporter. Plant Biol. 2012, 14, 706–713. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, J.; Song, A.; Chen, S.; Shan, H.; Luo, H.; Gu, C.; Sun, J.; Zhu, L.; Fang, W.; Chen, F. Ambient temperature enhanced freezing tolerance of Chrysanthemum dichrum CdICE1 Arabidopsis via miR398. BMC Biol. 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Li, P.; Jiang, J.; Chen, S.; Li, H.; Zeng, J.; Shao, Y.; Zhu, L.; Zhang, Z.; Chen, F. Phylogenetic and transcription analysis of chrysanthemum WRKY transcription factors. Int. J. Mol. Sci. 2014, 15, 14442–14455. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- de Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, C.; Li, P.; Song, A.; Wang, H.; Wang, Y.; Ren, L.; Qi, X.; Chen, F.; Jiang, J.; Chen, S. Isolation and Characterization of Six AP2/ERF Transcription Factor Genes in Chrysanthemum nankingense. Int. J. Mol. Sci. 2015, 16, 2052-2065. https://doi.org/10.3390/ijms16012052
Gao C, Li P, Song A, Wang H, Wang Y, Ren L, Qi X, Chen F, Jiang J, Chen S. Isolation and Characterization of Six AP2/ERF Transcription Factor Genes in Chrysanthemum nankingense. International Journal of Molecular Sciences. 2015; 16(1):2052-2065. https://doi.org/10.3390/ijms16012052
Chicago/Turabian StyleGao, Chunyan, Peiling Li, Aiping Song, Haibin Wang, Yinjie Wang, Liping Ren, Xiangyu Qi, Fadi Chen, Jiafu Jiang, and Sumei Chen. 2015. "Isolation and Characterization of Six AP2/ERF Transcription Factor Genes in Chrysanthemum nankingense" International Journal of Molecular Sciences 16, no. 1: 2052-2065. https://doi.org/10.3390/ijms16012052





