1. Introduction
Endophytes are microorganisms that reside within internal tissues of living plants without visibly harming the host plant [
1]. Endophytic microorganisms have been found in all plant families [
2], including species in many different climate regions of the world [
2,
3,
4,
5]. Endophyte research has increased in recent years because of their taxonomic diversity [
6], their multiple functions including potential uses as genetic vectors [
7], and their influence on host plant growth promotion and fitness [
8,
9,
10]. Recently, there has been a rising interest in the use of eco-friendly biopesticides for control of plant diseases, especially for biological control of plant diseases, and has few quick benefits but can be long lasting, inexpensive, and harmless. Bio-control systems do not eliminate neither pathogen nor disease but bring them into natural balance. Endophytes are a major source of biological control agents these days.
Chinese boxthorn (
Lycium chinense Mill) is also known as matrimony vine, goji berry or wolfberry and belongs to the Solanaceae family. The plant is native to southeastern Europe and Asia. The fruits are well known in China and Korea for their medicinal and high nutritious value [
11]. Consumers of Chinese boxthorn are increasing dramatically, mainly due to the nutrition value (68% of the dry mass exists as carbohydrates, 12% as protein, 10% as fiber and 10% as fat) and there is also high proportion of antioxidants [
12]. The plant may live in association with a number of endophytes. Researchers have attempted to isolate fungi from this plant in Korea.
Colletotrichum causes anthracnose diseases in a wide range of economically important plants, crops, and grasses and is important fungal taxa. More than one species of
Colletotrichum can affect a single plant species [
13]. This phenomenon makes the taxa more important in agriculture. Identification based on morphology is problematic due to the small number of morphological traits that can be used to distinguish species [
14]. Conidial size, shape, appresoria formation, sclerotia, setae, and acervuli are some of their distinguishing characters used to separate species of
Colletotrichum [
15,
16]. Recently, multilocus molecular characteristics have become increasingly important in the identification of
Colletotrichum species [
17].
The objectives of this study were (1) to investigate the occurrence, isolation and sequence based identification of endophytic fungi from symptomless tissues of Chinese boxthorn plant in Korea and (2) to isolate, identify, and characterize two new Colletotrichum species by molecular and morphological data analysis.
2. Results
A total of 10 endophytic fungal morphospecies obtained from
L. chinense in Korea were selected from 14 isolates for identification (
Table 1). Endophytic fungi were identified by analysis of the ITS region of the rDNA gene.
Table 1.
Closest relatives of endophytic fungi isolated from Lycium chinense with BLAST search analyses based on ITS gene sequence.
Table 1.
Closest relatives of endophytic fungi isolated from Lycium chinense with BLAST search analyses based on ITS gene sequence.
| Isolate No. | GenBank Closest Hit (Accession Number) | Similarity (%) | Sequence Based Identification | Host Tissue | Accssion Number |
|---|
| CNU122031 | C. fructicola C1263.3 (JX010164) | 100 | C. fructicola | Fruit | KJ651254 |
| CNU122032 | C. brevisporum LC0600 (KC790943) | 99 | C. brevisporum | Leaf | KJ651255 |
| | Glomerella magna L2.5 (DQ003103) | 99 | | | |
| CNU122033 | Acremonium sp. r116 (HQ649797) | 100 | Acremonium sp. | Fruit | KJ651256 |
| | Acremonium strictum F21 (EU497953) | 99 | | | |
| CNU122034 | Cochliobolus lunatus Cs-1C (JN107740) | 99 | Cochliobolus lunatus | Leaf | KJ651257 |
| | Cochliobolus sp. P2E4 (JN207244) | 99 | | | |
| CNU122035 | Fusarium cf. equiseti AM-48 (JN038489) | 99 | Fusarium equiseti | Leaf | KJ651258 |
| | Fusarium equiseti ATT040 (HQ607811) | 99 | | | |
| CNU122036 | C. truncatum tc-1 (KC460308) | 99 | Colletotrichum sp. | Leaf | KJ651259 |
| | Colletotrichum sp. ITCC 2041 (JN390888) | 99 | | | |
| CNU122037 | Hypocrea citrina GJS 91-61 (DQ000630) | 99 | Hypocrea citrina | Fruit | KJ651260 |
| CNU122038 | Nemania sp. NDJL-2009a (GU166482) | 99 | Nemania sp. | Leaf | KJ651261 |
| | Nemania serpens BF330 (EF155504) | 95 | | | |
| CNU122039 | Nemania sp. AX48 (KC507255) | 100 | Nemania sp. | Leaf | KJ651262 |
| | Nemania diffusa Z26 (JN198514) | 100 | | | |
| CNU122040 | Nemania sp. AX48 (KC507255) | 99 | Nemania sp. | Leaf | KJ651263 |
Six distinctive fungal taxa were detected at a
>90% sequence similarity threshold (
Table 1) representing six fungal taxa (
Figure 1) and they were
Acremonium,
Cochliobolus,
Colletotrichum,
Fusarium,
Hypocrea and
Nemania. The species of fungi isolated from Chinese boxthorn were
Acremonium sp. (CNU122033),
Cochliobolus lunatus (CNU122034),
Colletotrichum brevisporum (CNU122032),
Colletotrichum fructicola (CNU122031),
Colletotrichum sp. (CNU122036),
Fusarium equiseti (CNU122035),
Hypocrea citrina (CNU122037), and
Nemania sp. (CNU122038, CNU122039, & CNU122040).
The sequence of CNU122031 completely matched with C. fructicola C1263.3 when retrieved from GenBank with maximum bootstrap value shown in the phylogenetic tree. The isolate CNU122032 showed 99% sequence similarity with two similar isolates C. brevisporum LC0600 and Glomerella magna L2.5 with 100% bootstrap support. Therefore, we used Actin and GAPDH primers to confirm the identification, and sequence similarity with C. brevisporum was 99%–100%. The isolate CNU122033, CNU122034, CNU122035, CNU122036 and CNU122037 showed 99%–100% sequence similarity with Acremonium sp. r116, Cochliobolus lunatus Cs-1C, Fusarium cf. equiseti AM-48, Colletotrichum sp. ITCC 2041 and Hypocrea citrina GJS 91–61, and a high bootstrap value. CNU122038, CNU122039 and CNU122040 showed 99%–100% sequence similarity with Nemania species isolates from GenBank.
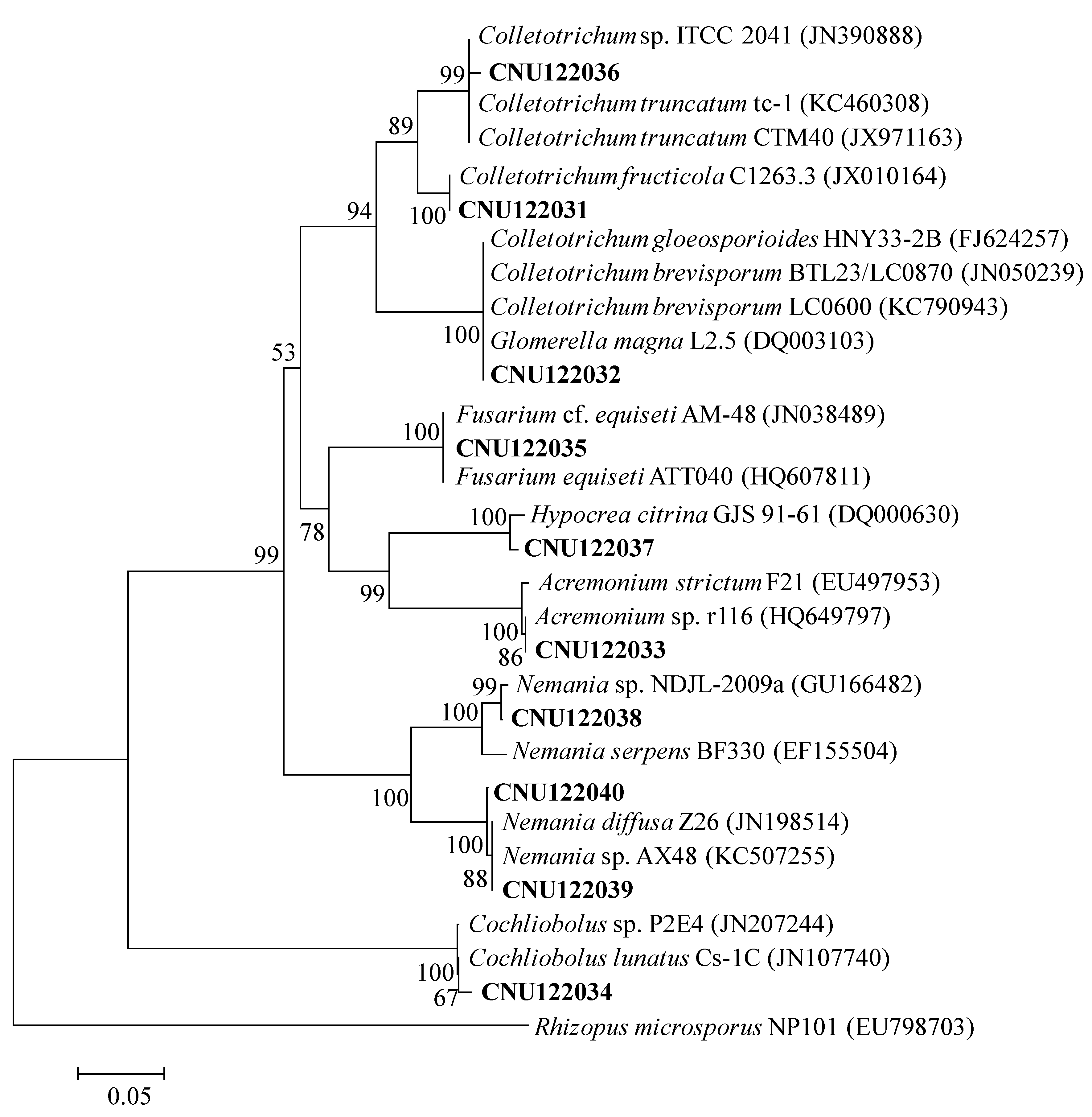
Figure 1.
Neighbor-joining phylogenetic tree showing the placement of the representative endophytic isolates based on the sequences of the ITS region. The Kimura two-parameter model is used for pairwise distance measurement. The tree is rooted with Rhizopus microsporus (EU798703). Only bootstrap values >50% (1000 replications) are shown in at the internal nodes.
Figure 1.
Neighbor-joining phylogenetic tree showing the placement of the representative endophytic isolates based on the sequences of the ITS region. The Kimura two-parameter model is used for pairwise distance measurement. The tree is rooted with Rhizopus microsporus (EU798703). Only bootstrap values >50% (1000 replications) are shown in at the internal nodes.
2.1. Taxonomy of Two Colletotrichum Species
2.1.1. Molecular Phylogeny
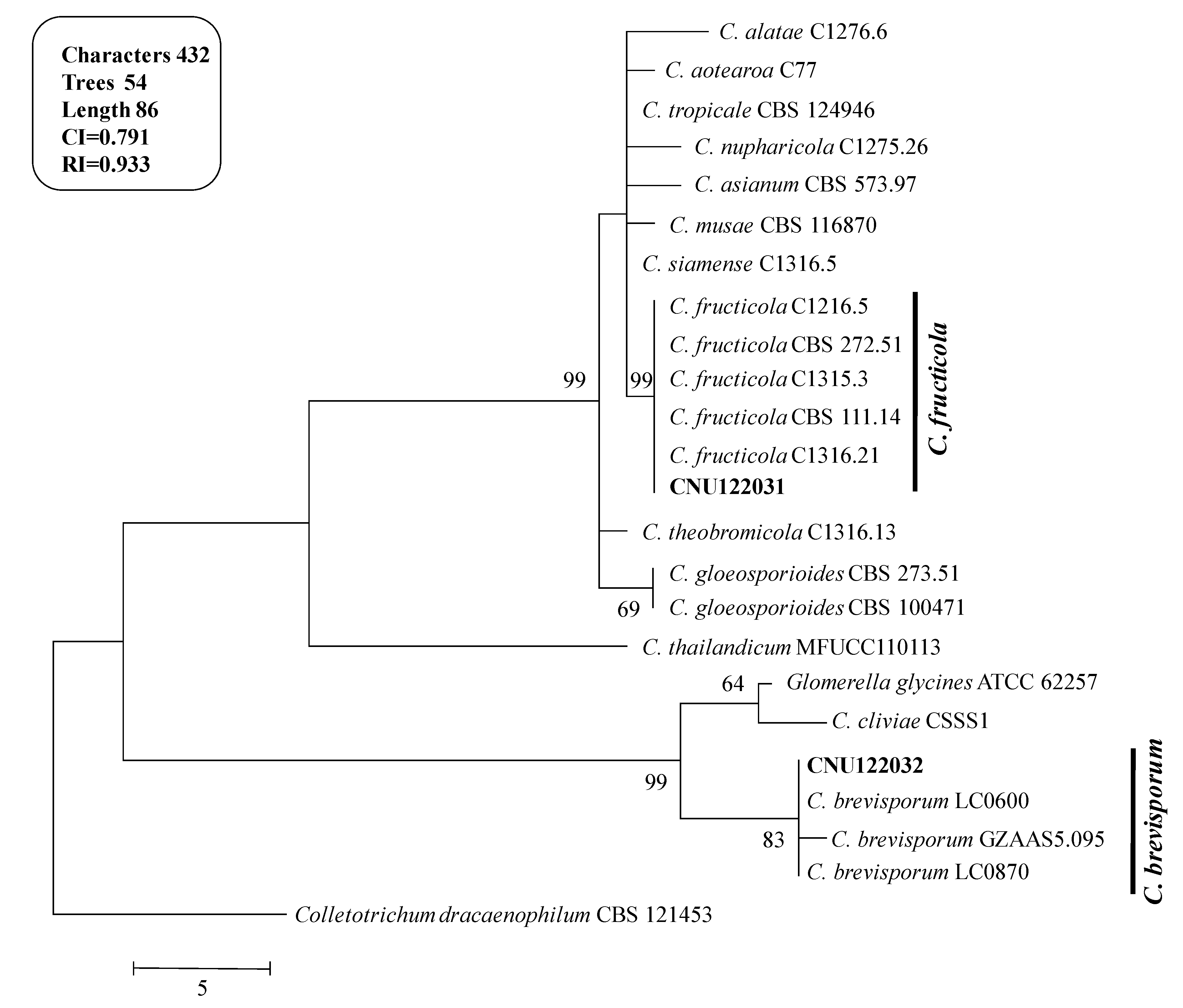
A molecular phylogenetic analysis was generated using the multilocus molecular dataset (
Figure 2). The combined dataset of
ITS, ACT and GAPDH comprised 24 sequences and produced 54 most parsinimonious trees (TL = 86, CI = 0.791 and RI = 0.933). One of the most parsimonious trees is shown in
Figure 2.
Figure 2.
Phylogenetic tree showing the placement of the Colletotrichum isolates from the present study and their related species generated using the maximum parsimony analysis of combined dataset of ITS, ACT, and GAPDH gene sequences. Numbers at the nodes indicate bootstrap values (>50%) from 1000 replications. The bar indicates the number of substitutions per position. The tree is rooted with C. dracaenophilum CBS121453.
Figure 2.
Phylogenetic tree showing the placement of the Colletotrichum isolates from the present study and their related species generated using the maximum parsimony analysis of combined dataset of ITS, ACT, and GAPDH gene sequences. Numbers at the nodes indicate bootstrap values (>50%) from 1000 replications. The bar indicates the number of substitutions per position. The tree is rooted with C. dracaenophilum CBS121453.
The isolates CNU122031 and CNU122032 formed a monophyletic clade with reliable reference relatives from GenBank isolates. The phylogenetic tree constructed from the combined dataset of ITS, GAPDH, and ACT gene sequences clearly showed that the isolates CNU122031 and CNU122032 are
C. fructicola and
C. brevisporum, respectively with high bootstrap support (
Figure 2).
2.1.2. Morphological Characterization
Taxonomic descriptions and micromorphographs of the morphological structures for the two species (C. fructicola CNU122031 and C. brevisporum CNU122032) are shown in details below.
2.1.3. CNU122031-Colletotrichum fructicola
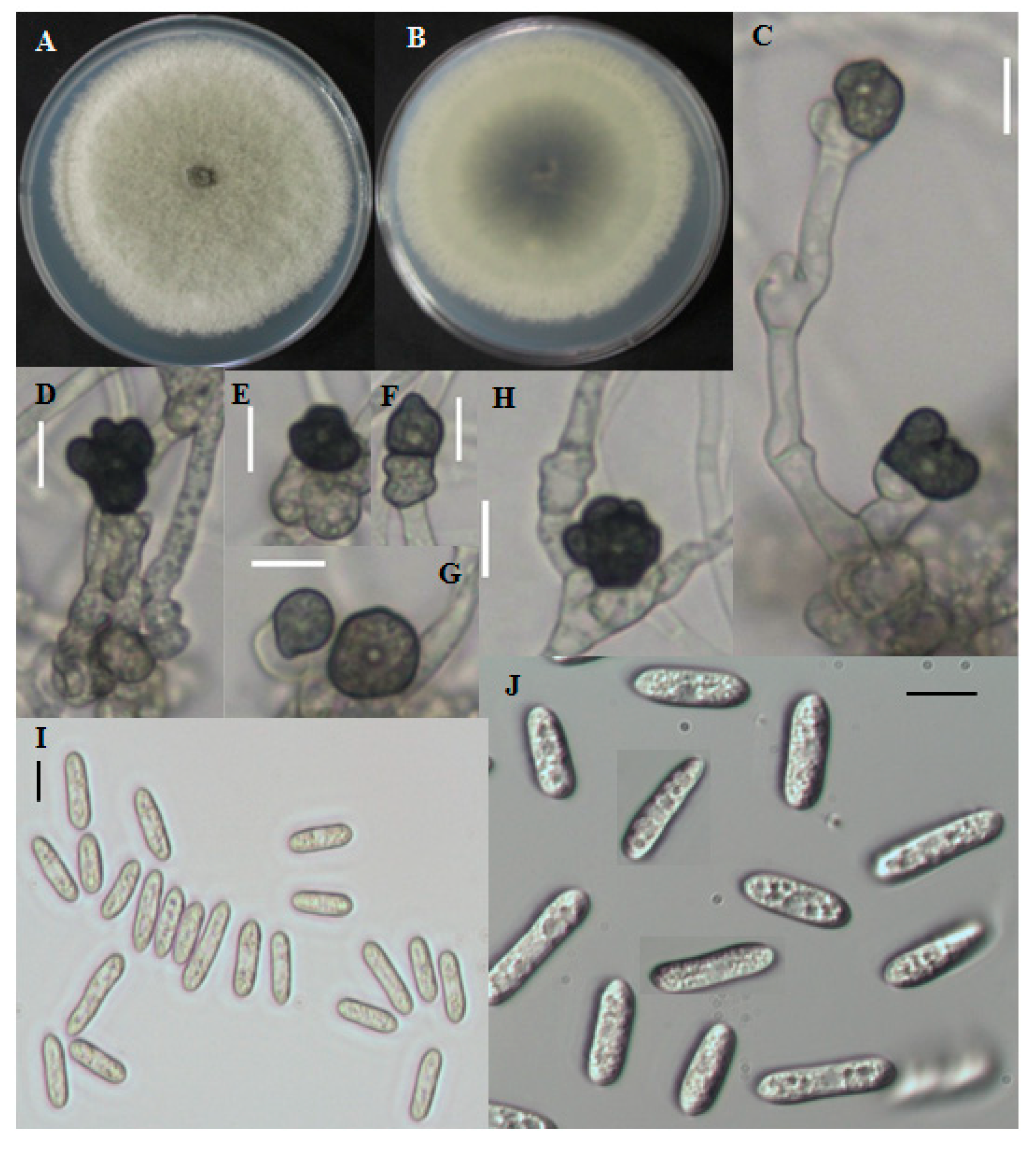
Fast growing colonies on PDA were white at the starting of the growth period. Over time, colonies became gray to dark gray at the center (
Figure 3A,B). The reverse color was grayish to blackish with a white halo. Aerial mycelia were whitish, dense, cottony, and without visible conidial masses. Single-celled conidia were observed after the sporulation period. The cylindrical conidia had obtuse to rounded ends and the size ranged from 8.7–29.5 μm long and 2.8–5.9 μm wide (
Figure 3I,J). The shape of appresoria varied among species. Appresoria were commonly ovoid, sometimes clavate. The sizes of the appresoria were 10.5–14.5 × 6–11 μm (
Figure 3C–H).
Figure 3.
Morphological features of Colletotrichum fructicola CNU122031. Colony morphology in PDA after 7 days of inoculation at 25 °C (A, obverse; B, Reverse); C–H: appresoria; I,J: Conidia (scale bars C–J = 10 μm).
Figure 3.
Morphological features of Colletotrichum fructicola CNU122031. Colony morphology in PDA after 7 days of inoculation at 25 °C (A, obverse; B, Reverse); C–H: appresoria; I,J: Conidia (scale bars C–J = 10 μm).
2.1.4. Distinguishing Characters
C. fructicola CNU122031 was distinctly separated from the closely related species
C. siamense by colony color, conidial size, and shape.
C. siamense produced a pinkish colony color and the reverse was yellowish to pinkish whereas, the present isolate produced gray to dark gray colony color, and completely matched with the reference species,
C. fructicola strains [
18]. The size of the colony of present isolate was shorter than that of
C. siamense. The CNU122031 isolate produced cylindrical, obtuse conidia but
C. siamense produced fusiform conidia, indicating a clear difference between the species (
Table 2). The fungus,
C. fructicola is a newly isolated species from
L. chinense.
Table 2.
Morphological characters of Colletotrichum species described in this study and closely related reference species.
Table 2.
Morphological characters of Colletotrichum species described in this study and closely related reference species.
| Taxa | Colony | Conidia Shape and Size (μm) | Appresoria Size (μm) | Reference |
|---|
| C. brevisporum | White, mycelium in small tufts, reverse dark in middle | Cylindrical with round ends. 12.2–24.2 × 2.6–6 | 10–16.8 × 5–9.4 | This study |
| C. brevisporum | Aerial mycelia in small tufts, white, sparse with conidial masses, reverse dark green | Cylindrical with round ends, smooth-walled, hyaline. 12–17 × 5–6 | 10.5–14.5 × 8−11 | [13] |
| C. cliviae | White to gray, white at margin, reverse dark brown to greenish black | Cylindrical, straight or slightly curved, obtuse at the ends. 19.5–24.5 × 4.5–7 | 10.5–14.5 × 6−11 | [19] |
| C. fructicola | White, becoming gray to dark gray at the centre with age, dark circular margin at the center in reverse | Cylindrical with obtuse to rounded ends. 8.7–29.5 × 2.8–5.9 | 4.1–5.4 × 3−4.9 | This study |
| C. fructicola | White, becoming gray to dark gray at the centre with age, dark circular around the growing margin at the center in reverse | Cylindrical with obtuse to slightly rounded ends, sometimes oblong, hyaline. 9.7–14 × 3–4.3 | 4.7–8.3 × 3.5−5 | [18] |
| C. siamense | White, becoming pale brownish to pinkish, pale yellowish to pinkish colonies in reverse | Fusiform, sometimes with obtuse to slightly rounded ends, sometimes oblong, hyaline. 7–18.3 × 3–4.3 | 4.7–10.7 × 3.3–6.7 | [18] |
2.1.5. CNU122032-Colletotrichum bresvisporum
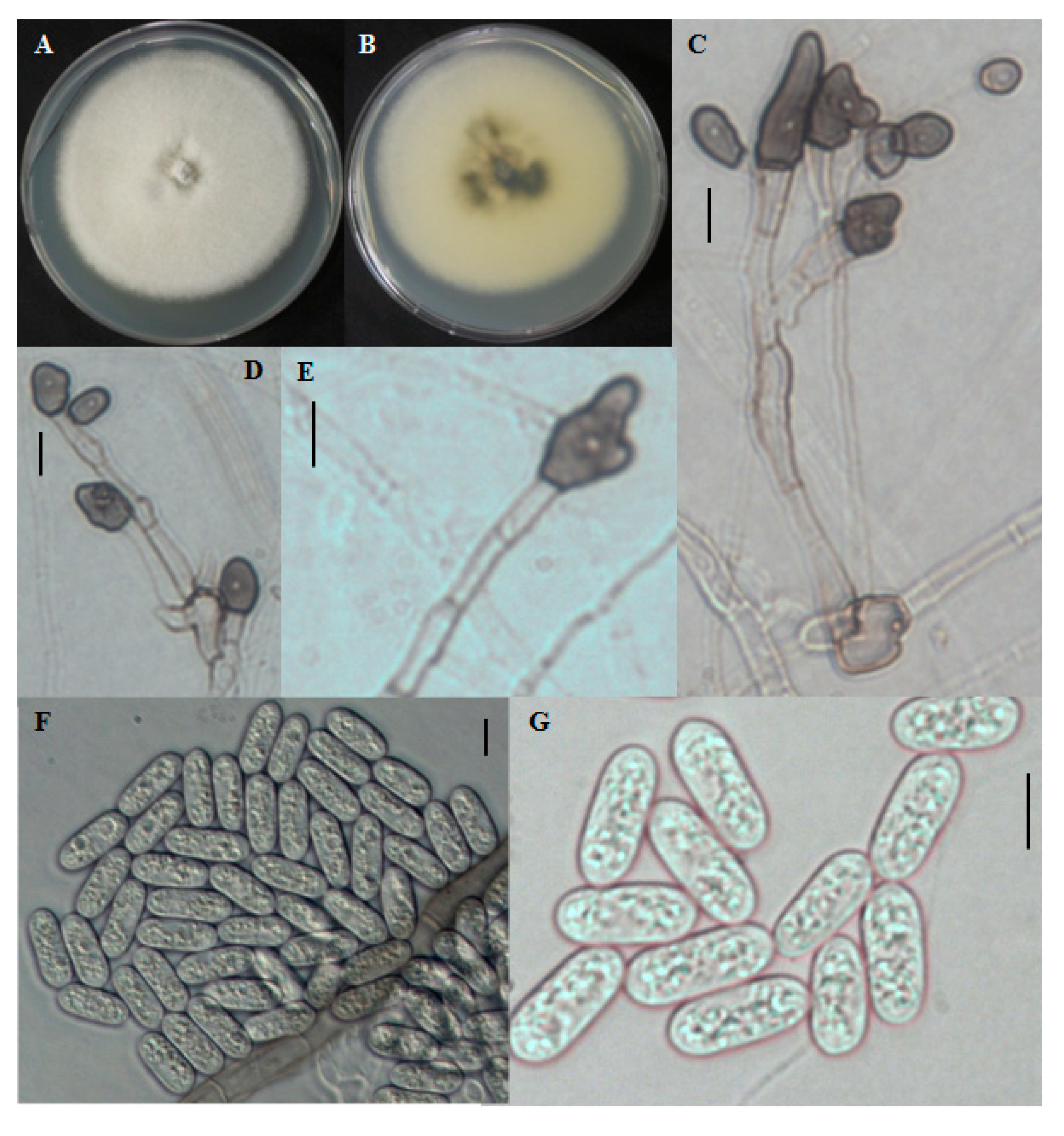
The colony on PDA was white at first, became grayish over time. The mycelium was in small tufts. The reverse was dark blackish at the center and the edge was white to grayish with a white halo (
Figure 4A,B). Aerial mycelia were white in color and dense. Conidial masses were not observed. Conidia produced were single celled. Conidia were cylindrical with rounded ends, smooth-walled, hyaline, and ranged in size from 12.2–24.2 × 2.6–6 μm (
Figure 4F,G). Appresoria were 10–16.8 × 5–9.4 μm in slide cultures, irregular in shape, sometimes ovoid, often becoming complex over time (
Figure 4C–E).
Figure 4.
Morphological features of Colletotrichum brevisporum CNU122032. Colony in PDA after 7 days of inoculation at 25 °C (A, obverse; B, reverse); C–E: appresoria; F,G: Conidia (scale bars C–G = 10 μm).
Figure 4.
Morphological features of Colletotrichum brevisporum CNU122032. Colony in PDA after 7 days of inoculation at 25 °C (A, obverse; B, reverse); C–E: appresoria; F,G: Conidia (scale bars C–G = 10 μm).
2.1.6. Distinguishing Characters
The isolate was identified as C. brevisporum by molecular data analysis. The morphological characteristics supported the molecular identification.
The isolate was closely related to
C. cliviae, but differed in conidial shape and length. Conidia of the present isolate were cylindrical with distinctly round ends, whereas the conidia of
C. cliviae were cylindrical, straight, or curved, and obtuse at the ends. The present isolate did not produce any curved conidia, and not obtuse at the ends (
Table 2). The morphological description of the present isolate completely matched that of
C. brevisporum described by Noireung
et al. 2012 [
13]. This is the first fungal record of
C. brevisporum isolated in Korea.
3. Discussion
Endophytic fungal distributions vary with plant-associated habitats which may affect microbial communities that colonize roots, leaves, stems, branches, fruits, pods, and leaves [
20]. Previous investigations revealed that endophytes were isolated from different plant tissues or organs. In this study, we isolated and characterized endophytic fungi from leaves and fruits of the
L. chinense Mill plant and identified two unreported fungi isolated from the
L. chinense Mill plant.
Endophytic fungal communities associated with various kinds of plants from tropical, sub-tropical, temperate, and Arctic ecosystems were investigated previously and fungal diversity was high [
21]. From the Chinese boxthorn plant we isolated 14 fungi. Among them, 10 morphospecies were selected and sequenced with
ITS gene. Six different genera were observed (
Acremonium,
Colletotrichum,
Cochliobolus,
Fusarium,
Hypocrea and
Nemania) and included common endophytic fungi from different regions around the world. The distribution of dominant species agreed with previous report [
6,
7,
22,
23,
24,
25]. The Dominant fungi described here are commonly associated with disease symptoms in several crop plants. Species of
Acremonium,
Colletotrichum,
Cochliobolus,
Fusarium,
Hypocrea, and
Nemania cause severe disease in many cultivated and non-cultivated plants. The anthracnose of chili pepper caused by
C. acutatum is an example and
Fusarium oxysporum causes disease in the same plant and many other cultivated and non-cultivated plants, and both were found in endophytic association with leaves, roots, pods, and many other plants. They were found in association with leaves and stems of medicinal plant
Tylophora indica in India [
24], and with the orchid species,
Dendrobium loddigesii in China [
6]. We isolated fungi associated with a host plant that did not show any disease symptoms. It is possible that they are latent pathogens.
A number of endophytic fungi consist of sterile mycelia or non-sporulating fungi and consequently cannot be identified by morphological characters. Previously, molecular techniques have been employed successfully for the identification of different endophytic fungal community [
6,
21,
26]. In this study, we also used the molecular strategy to identify fungi specifically utilizing of ITS rDNA gene sequence and phylogenetic analysis.
4. Experimental Section
4.1. Sampling
Chinese boxthorn (
Lycium chinense Mill) plants were selected because of their medicinal applications in Korea. Medicinal plants may live in association with a number of new or unreported microorganisms, especially fungal species. Sampling sites of this study were Cheongyang farmer’s field, South Chungcheong province, Republic of Korea (
Figure 5). Living symptomless leaf and fruit samples were collected and stored in sterile polyethylene bags.
Figure 5.
Chinese boxthorn (Lycium chinense Mill) plant tissues were collected from farmer’s field in Cheongyang locality in Korea.
Figure 5.
Chinese boxthorn (Lycium chinense Mill) plant tissues were collected from farmer’s field in Cheongyang locality in Korea.
4.2. Isolation of Endophytic Fungi
Samples were cleaned under running tap water to remove debris, then air dried and processed within 5 h of collection. Tissues were cut into small (1 cm length and 0.5 cm width) pieces. Then surface sterilized by immersing in 95% ethanol for 1 min, sodium hypochlorite (4% chlorine) for 4 min and 95% ethanol for 30 s, and then samples were washed in sterile water three times to remove the surface sterilization agents. Samples were allowed to dry on paper towel in a laminar air flow chamber. A total of 120 segments were selected and plated (100 segments from leaf samples and 20 segments from fruit samples). Ten segments per petri dish were placed horizontally in potato dextrose agar (PDA, Difco, Franklin Lakes, NJ, USA) and rose bengal chloramphenicol agar (DRBC, Difco, Franklin Lakes, NJ, USA) supplemented with the antibiotic streptomycin sulfate (0.4 mg/mL, SIGMA-ALDRICH, Munich, Germany) to stop bacterial growth. After incubation at 25 °C for 5, 10, and 25 days, individual hyphal tips of the developing fungal colonies were collected and placed onto PDA media and incubated for 5–10 days, and checked for culture purity. Eventually, pure cultures were transferred to PDA slant tubes and eppendorf tubes with 20% glycerol stock solution. Strain numbers were assigned for selected isolates and deposited in the Fungal Herbarium of the Chungnam National University, Daejeon, Korea. Cultures of the isolates were also deposited in the Environmental Microbiology Lab. (EML) Herbarium, Chonnam National University, Gwangju, Korea.
4.3. Genomic DNA Extraction, PCR and Sequencing
Genomic DNA was extracted from 10 pure culture isolates using the method of Park
et al. [
27]. Primers ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3') were used for the amplification of the fungal rDNA internal transcribed spacers (ITS) regions of all isolates.
PCR amplification was carried out [
28] in an i-Cycler (BIO-RAD, Hercules, CA, USA) for 30 cycles of 94 °C for 1 min denaturing, 55 °C for 1 min annealing and 72 °C for 1.30 min extension. Initial denaturing at 94 °C was extended to 5 min and the final extension was for 10 min at 72 °C. For the specific identification of the
Colletotrichum species, two additional genes were used for PCR amplification: Actin (Forward ACT-512F & reverse ACT-783R) and Glyceraldehyde-3-phosphate dehydrogenase (Forward GDF & reverse GDR) (
Table 3). The PCR conditions for these genes are showing in
Table 3. All PCR products were purified using the Wizard PCR prep kit (Promega, Madison, WI, USA). Purified double-stranded PCR fragments were directly sequenced with BigDye terminator cycle sequencing kits (Applied Biosystems, Foster City, CA, USA) following the manufacturer instructions. The gel electrophoresis and data collection were performed on an ABI prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Table 3.
Primers used in this study, with sequences and sources.
Table 3.
Primers used in this study, with sequences and sources.
| Gene | Product Name | Primer | Direction | Sequence (5'–3') | Reference |
|---|
| ACT | Actin | ACT-512F | Forward | ATGTGCAAGGCCGGTTTCGC | [29] |
| ACT-783R | Reverse | TACGAGTCCTTCTGGCCCAT | [29] |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | GDF | Forward | GCCGTCAACGACCCCTTCATTGA | [30] |
| GDR | Reverse | GGGTGGAGTCGTACTTGAGCATGT | [30] |
| ITS | Internal transcribed spacer | ITS-1F | Forward | CTTGGTCATTTAGAGGAAGTAA | [28] |
| ITS-4 | Reverse | TCCTCCGCTTATTGATATGC | [28] |
4.4. Phylogenetic Analysis
The rDNA
ITS, Actin, and GAPDH gene sequences were compared by BLAST search with sequence available in the GenBank database. Sequences generated from materials in this study and retrieved from GenBank (
Table 4) were initially aligned using CLUSTAL X [
31], and the alignment was refined manually using PHYDIT program version 3.2 [
32]. A neighbor-joining tree for the ITS sequences was constructed with Kimura’s 2-parameter distance model [
33] using the PHYLIP 3.57c package [
34]. Maximum parsimony trees were constructed for the combined datasets of
ITS, Actin, and GAPDH gene sequences using MEGA5 program. Bootstrap analysis using 1000 replications was performed to assess the relative stability of the branches.
Table 4.
Isolates used in this study for molecular data analysis.
Table 4.
Isolates used in this study for molecular data analysis.
| Species | Isolate | Host | Origin | Accession No. |
|---|
| ITS | gpd | ACT |
|---|
| C. fructicola | CNU122031 | Lycium chinense | Korea | KJ651254 | KJ651266 | KJ651264 |
| C. brevisporum | CNU122032 | Lycium chinense | Korea | KJ651255 | KJ651267 | KJ651265 |
| C. alatae | C1276.6 | Discorea alata | India | JX010191 | JX010011 | JX009470 |
| C. aotearoa | C77 | Vitex lucens | NZ b | JX010221 | JX010023 | JX009500 |
| C. asianum | CBS 573.97 | Mangifera indica | Brazil | KC566732 | KC566586 | KC566878 |
| C. brevisporum | GZAAS5 | Citrus sp. | China | JQ247623 | JQ247599 | JQ247647 |
| C. brevisporum | LC0600 | Neoregalia sp. | Thailand | JN050238 | JN050227 | JN050216 |
| C. brevisporum | LC0870 | Pandanas pygmaeus | Thailand | JN050239 | JN050228 | JN050217 |
| C. cliviae | CSSS1 | Clivia miniata | China | GQ485607 | JX546611 | GU085861 |
| C. dracaenophilum | CBS 121453 | Dracaena sanderiana | Bulgaria | EU003533 | NA a | NA a |
| C. fructicola | C1316.21 | Theobroma cacao | Panama | JX010173 | JX009992 | JX009581 |
| C. fructicola | CBS 111.14 | NA a | Brazil | KC566785 | KC566639 | KC566931 |
| C. fructicola | CBS 272.51 | NA a | Brazil | KC566783 | KC566637 | KC566929 |
| C. fructicola | C1315.3 | Coffea arabica | Thailand | JX010165 | JX010033 | JX009501 |
| C. fructicola | C1216.5 | Persea americana | Australia | JX010166 | JX009946 | JX009529 |
| C. gloeosporioides | CBS 273.51 | Citrus limon | Italy | JX010148 | JX010054 | JX009558 |
| C. gloeosporioides | CBS 100471 | NA | Brazil | KC566719 | KC566573 | KC566865 |
| C. musae | CBS 116870 | Musa sp. | USA | JX010146 | JX010050 | JQ005840 |
| C. nupharicola | C1275.26 | Nuphar lutea | USA | JX010188 | JX010031 | JX009582 |
| C. siamense | C1316.5 | Hymenocallis americana | China | JX010278 | JX010019 | JX009441 |
| C. thailandicum | MFUCC1101 | Hibiscus rosa-sinensis | Thailand | JN050242 | JN050231 | JN050220 |
| C. theobromicola | C1316.13 | T. cacao | Panama | JX010294 | JX010006 | JX009444 |
| C. tropicale | CBS 124946 | T. cacao | Panama | KC566806 | KC566660 | KC566952 |
| Glomerella glycines | ATCC 62257 | Glycine max | USA | KC110794 | KC110812 | KC110830 |
4.5. Morphological Observation
The isolates CNU122031 and CNU122032 were used for morphological description. Colony characteristics (color, size, and texture) were determined after 7 days at 25 °C in the dark grown on PDA plates. Conidial morphology was examined in standard conditions [
13]. Plates were maintained in a chamber without humidity control at 25 °C and held under a fluorescent light/dark cycle of 12/12 h. Conidia were mounted in lactophenol picric acid solution (SIGMA-ALDRICH, Munich, Germany) and measured using a light microscope (OLYMPUS BX50, Tokyo, Japan) and an Artray Artcam 300MI digital camera system (Artray Co., Ltd., Tokyo, Japan). Randomly selected conidia were counted for the morphological description. A slide culture technique [
35] was used for the production of appresoria and their size and shape were examined.
5. Conclusions
One of the main objectives of the present study was to find unreported fungal species in Korea. The ITS sequence-based identification showed that CNU122031 and CNU122032 were consistent with C. fructicola and C. brevisporum, respectively, both of which were previously unrecorded from Chinese borxthorn plant and C. brevisporum is new record in Korea. Further study was needed to clarify the identification. A multi-locus molecular study was carried out with Actin and GAPDH gene sequences. The combined dataset produced a clear molecular identification for these two fungi. Each fungus was isolated as endophytic fungi previously. C. brevisporum was reported as an endophytic new fungus from Thailand. The distinguishing characteristics of the present fungi and their reference species and related species was described. We conclude that Chinese boxthorn in Korea is the host species of unrecorded fungi and plays a potentially important role in understanding microbial diversity in Korea.
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ0089712013): Study on the organic cultivation techniques utilizing ecological method of Cnidium officinale Makino, Schisandrae chinense Ballon, and Lycium chinense Miller”, Rural Development Administration, Republic of Korea.
Author Contributions
N.C.P. was in charge of the whole project design, experiments, and writing; T.H.R. and H.R.K. performed sample & data collection; Y.K.K. provided technical and administrative support; H.B.L. made critical revisions and analyzed morphological data; N.C.P., J.H.L., and K.S.S. were in charge of the DNA extraction, PCR, and molecular phylogenetic data analysis; S.H.Y. and Y.N.Y. guided the conception, managed funding and supervised the study.
Conflict of Interest
The authors declare no conflict of interest.
References
- Fisher, P.J.; Petrini, O. Location of fungal endophytes in tissues of Suaeda fruiticosa: apreliminary study. Trans. Br. Mycol. Soc. 1987, 89, 246–249. [Google Scholar] [CrossRef]
- Spurr, H.W.; Welty, R.E. Characterization of endophytic fungi in healthy leaves of Nicotiana spp. Phytopathology 1975, 65, 417–422. [Google Scholar] [CrossRef]
- Fisher, P.J.; Petrini, O.; Lappin, S.H.M. The distribution of some fungal and bacterial endophytes in maize (Zea mays L.). New Phytol. 1992, 122, 299–305. [Google Scholar] [CrossRef]
- Larran, S.; Perello, A.; Simon, M.R.; Moreno, V. Isolation and analysis of endophytic microorganisms in wheat (Triticum aestivum L.) leaves. World J. Microbiol. Biotechnol. 2002, 18, 683–686. [Google Scholar]
- Paul, N.C.; Kim, W.K.; Woo, S.K.; Park, M.S.; Yu, S.H. Fungal endophytes in roots of Aralia species and their antifungal activity. Plant Pathol. J. 2007, 23, 287–294. [Google Scholar] [CrossRef]
- Chen, X.M.; Dong, H.L.; Hu, K.X.; Sun, Z.R.; Chen, J.; Guo, S.X. Diversity and antimicrobial and plant-growth-promoting activities of endophytic fungi in Dendrobium loddigesii Rolfe. J. Plant Growth Regul. 2010, 29, 328–337. [Google Scholar] [CrossRef]
- Margherita, D.; Salvatore, F.; Matteo, C. Endophytic fungi occurring in fennel, lettuce, chicory and celery-commercial crops in Southern Italy. Mycol. Res. 2008, 112, 100–107. [Google Scholar]
- Baltruschat, H.; Fodor, J.; Harrach, B.D.; Niemczyk, E.; Barna, B.; Gullner, G.; Janeczko, A.; Kogel, K.H.; Schafer, P.; Schwarczinger, I.; et al. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 2008, 180, 500–510. [Google Scholar]
- Tanaka, A.; Christensen, M.J.; Takemoto, D.; Park, P.; Scott, B. Reactive oxygen species play a role in regulating a fungus—Perennial ryegrass mutualistic interaction. Plant Cell 2006, 18, 1052–1066. [Google Scholar] [CrossRef]
- Tudzynski, B.; Sharon, A. Biosynthesis, biological role and application of fungal phyto-hormones. In Osiewacz of Pestalotiopsis isolated as endophytes from medicinal plants. Fungal Div. 2002, 24, 37–54. [Google Scholar]
- Potterat, O. Goji (Lycium barbarum and L. chinense): Phytochemistry, pharmacology and safety in the perspective of traditional uses and recent popularity. Planta Med. 2010, 76, 7–19. [Google Scholar] [CrossRef]
- Ballarin, S.M.; Lopez-Mataz, M.A.; Saenz, D.A.; Perez-Cinto, N.; Carnes, J. Anaphylaxis associated with the ingestion of Goji Berries (Lycium barbarum). J. Investig. Allergol. Clin. Immunol. 2011, 21, 567–570. [Google Scholar]
- Noireung, P.; Phoulivong, S.; Fang, L.; Cai, L.; Eric, H.C.M.; Ekachai, C.; Jones, E.B.G.; Ali, H.B.; Hyde, D.K. Novel species of Colletotrichum revealed by morphology and molecular analysis. Cryptogam. Mycol. 2012, 33, 347–362. [Google Scholar] [CrossRef]
- Cai, L.; Udayanga, D.; Manamgoda, D.S.; Maharachchikumbura, S.S.N.; Mckenzie, E.H.C.; Guo, L.D.; Liu, X.X.; Bahkali, A.H.; Hyde, K.D. The need to carry out re-inventory of plant pathogens. Trop. Plant Pathol. 2011, 36, 205–213. [Google Scholar]
- Hyde, K.D.; Cai, L.; Mckenzie, E.H.C.; Yang, Y.L.; Zhang, J.Z.; Prihastuti, H. Colletotrichum: A catalogue of confusion. Fungal Div. 2009, 39, 1–17. [Google Scholar]
- Phoulivong, S.; Cai, L.; Chen, H.; Mckenzie, E.H.C.; Abd-Elsalam, K.; Chukeatirote, E.; Hyde, K.D. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Div. 2010, 44, 33–43. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D.; Taylor, P.W.J.; Weir, B.; Waller, J.; Abang, M.M.; Zhang, J.Z.; Yang, Y.L.; Phoulivong, S.; Liu, Z.Y.; et al. A polyphasic approach for studying Colletotrichum. Fung. Div. 2009, 39, 183–204. [Google Scholar]
- Prihastuti, H.; Cai, L.; Chen, H.; Mckenzie, E.H.C.; Hude, K.D. Characterization of Colletotrhichum species with coffee barriers in northern Thailand. Fungal Div. 2009, 39, 89–109. [Google Scholar]
- Yang, Y.L.; Liu, Z.Y.; Cai, L.; Hyde, K.D.; Yu, Z.N.; Mckenzie, E.H.C. Colletotrichum anthracnose of Amaryllidaceae. Fungal Div. 2009, 39, 123–149. [Google Scholar]
- Rubini, M.R.; Silva-Ribeiro, R.T.; Pomella, A.W.V.; Maki, C.S.; Araujo, W.L.; Santos, D.R.; Azevedo, J.L. Diversity of endophytic fungal community of cacao (Theobroma cacao L.) and biological control of Crinipellis perniciosa, causal agent of witches broom disease. Int. J. Biol. Sci. 2005, 1, 24–33. [Google Scholar]
- Rosa, L.H.; Vieira, M.L.A.; Santiago, I.F.; Rosa, C.A. Endophytic fungi community associated with the dicotyledonous plant Colobanthus quitensis (Kunth) Bartl. (Caryophyllaceae) in Antarctica. FEMS Microbiol. Ecol. 2010, 73, 178–189. [Google Scholar]
- Flor, N.R.; Roberto, A.S.; Zolia, N.G.; Luis, B.F. Diversity of endophytic fungi of Taxus globosa (Mexican yew). Fungal Div. 2010, 47, 65–74. [Google Scholar]
- Huang, W.Y.; Cai, Y.Z.; Surveswaran, S.; Hyde, K.D.; Corke, H.; Sun, M. Molecular phylogenetic identification of endophytic fungi isolated from three Artemisia species. Fungal Div. 2009, 36, 69–88. [Google Scholar]
- Kumar, S.; Kaushik, N.; Ebel, R.E.; Ebel, R.; Proksch, P. Isolation, characterization, and bioactivity of endophytic fungi of Tylophora indica. World J. Microbiol. Biotechnol. 2011, 27, 571–577. [Google Scholar] [CrossRef]
- Sanchez, M.S.; Bills, G.F.; Zabalgogeazcoa, I. Diversity and structure of the fungal endophytic assemblages from two sympatric coastal grasses. Fungal Div. 2008, 33, 87–100. [Google Scholar]
- Sette, L.D.; Passarini, M.R.Z.; Delarmelina, C.; Salati, F.; Duarte, M.C.T. Molecular characterization and antimicrobial activity of endophytic fungi from coffee plants. World J. Microbiol. Biotechnol. 2006, 22, 1185–1195. [Google Scholar] [CrossRef]
- Park, M.S.; Seo, G.S.; Bae, K.S.; Yu, S.H. Characterization of Trichoderma spp. Associated with green mold oyster mushroom by PCR-RFLP and sequence analysis of ITS regions of rDNA. Plant Pathol. J. 2005, 21, 229–236. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. PCR Protocols: A Guide to the Methods and Applications; Innes, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; p. 315. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Templeton, M.D.; Rikkerink, E.H.; Solon, S.L.; Crowhurst, R.N. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene 1992, 122, 225–230. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. ClustalX: Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 487–4878. [Google Scholar]
- Chun, J. Computer-Assisted Classification and Identification of Actinomycetes. Ph.D. Thesis, University of Newcastle, New Castle Upon Tyne, UK, July 1995. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequence. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits of phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Johnston, P.R.; Jones, D. Relationship among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia 1997, 89, 420–430. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).