PL-PatchSurfer: A Novel Molecular Local Surface-Based Method for Exploring Protein-Ligand Interactions
Abstract
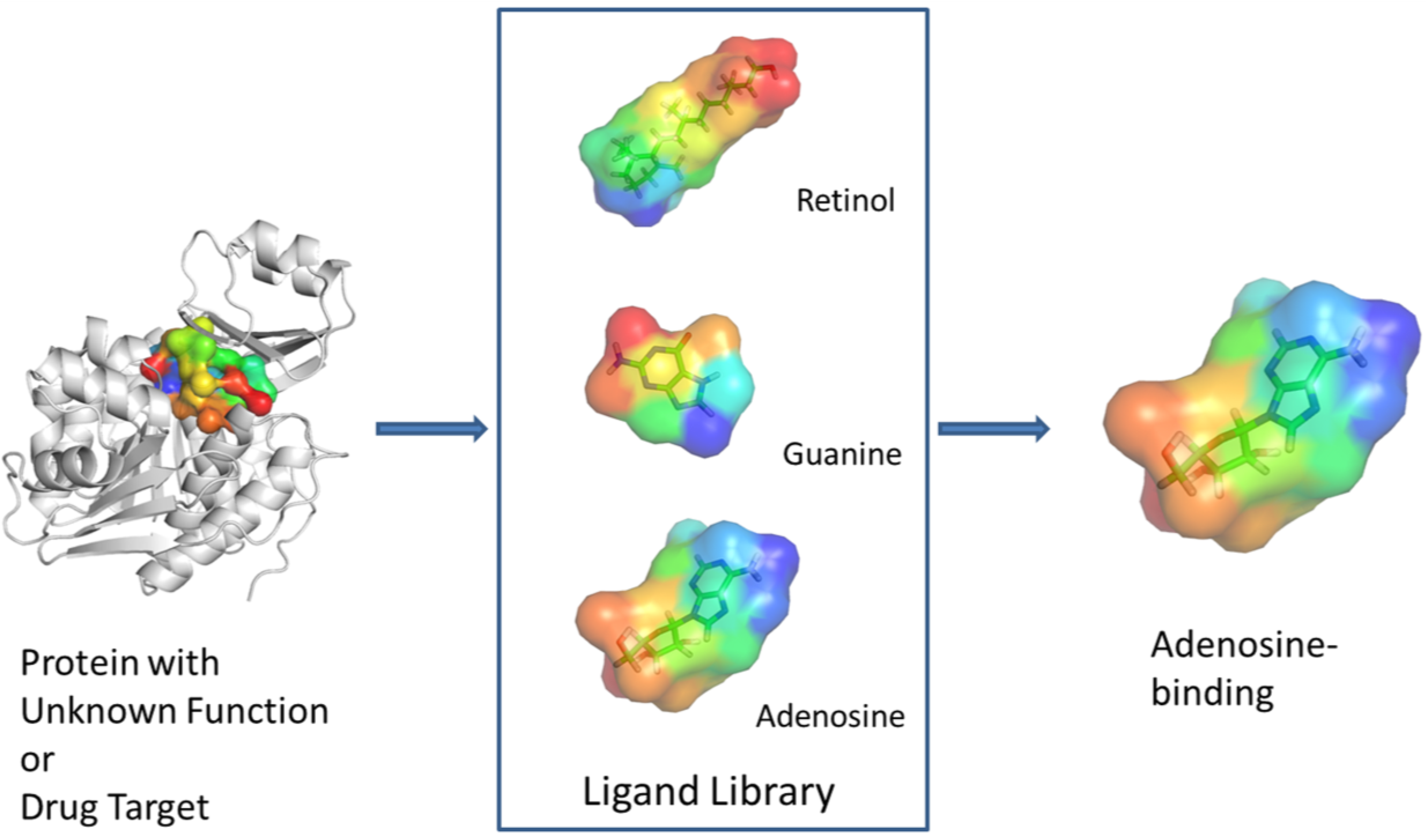
:1. Introduction
2. Results and Discussion
2.1. Binding Ligand Prediction on Huang Data Set
| Ligand Name | # Pockets | # Omega Conformers | Avg # Pocket Patches | Avg # Ligand Patches |
|---|---|---|---|---|
| AND | 12 | 20 | 22.3 | 18.1 |
| BTN | 8 | 20 | 18.6 | 17.7 |
| F6P | 10 | 20 | 19.8 | 16.5 |
| FUC | 8 | 2 | 8.6 | 11.5 |
| GAL | 32 | 3 | 13.9 | 12.7 |
| GUN | 11 | 1 | 14.6 | 10.0 |
| MAN | 15 | 6 | 9.3 | 11.5 |
| MMA | 8 | 10 | 13.0 | 13.4 |
| PIM | 5 | 2 | 14.0 | 12.0 |
| PLM | 24 | 20 | 28.3 | 24.1 |
| RTL | 5 | 20 | 31.2 | 25.9 |
| UMP | 8 | 20 | 22.5 | 19.2 |
| Total | 146 | 144 | - | - |
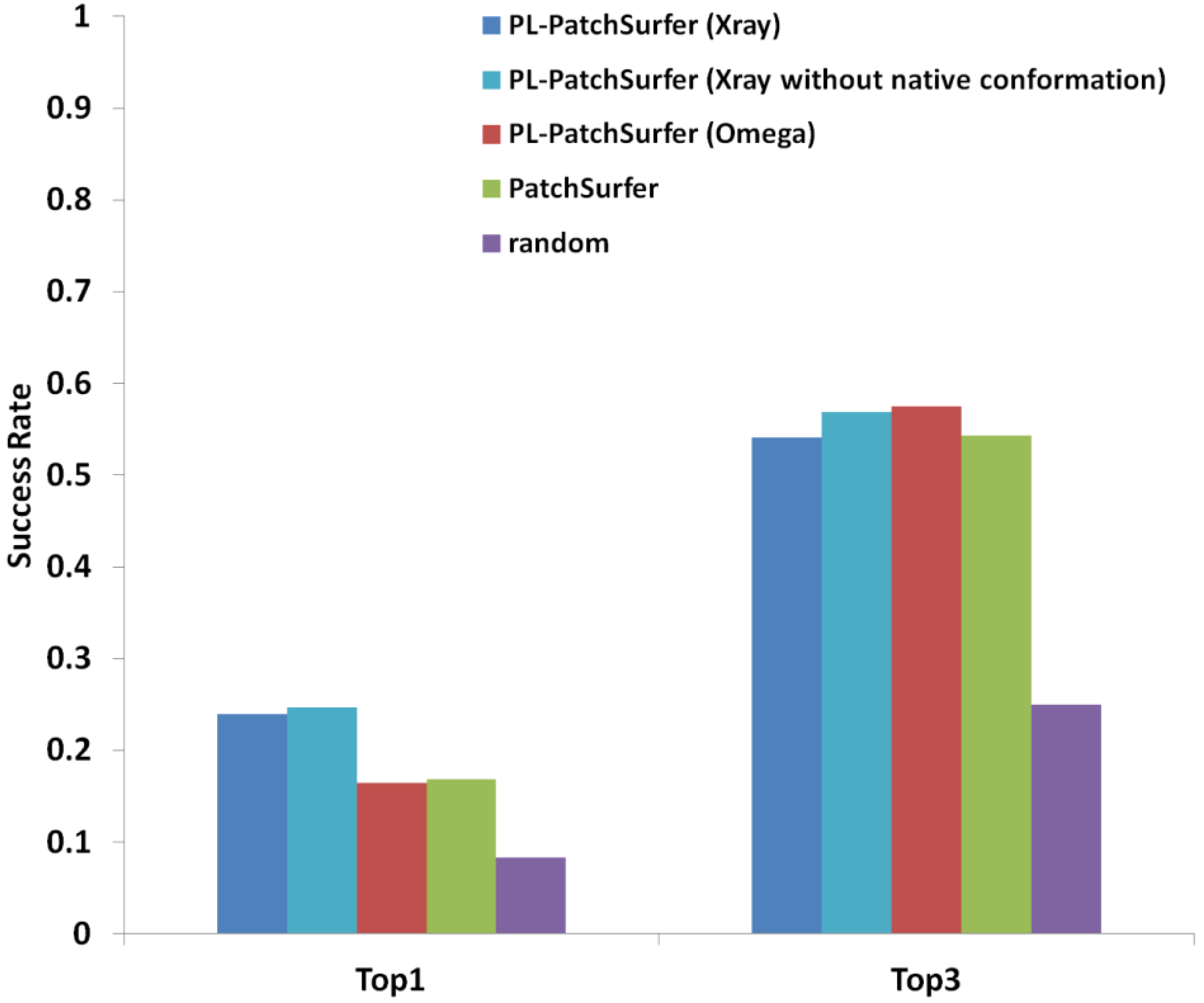
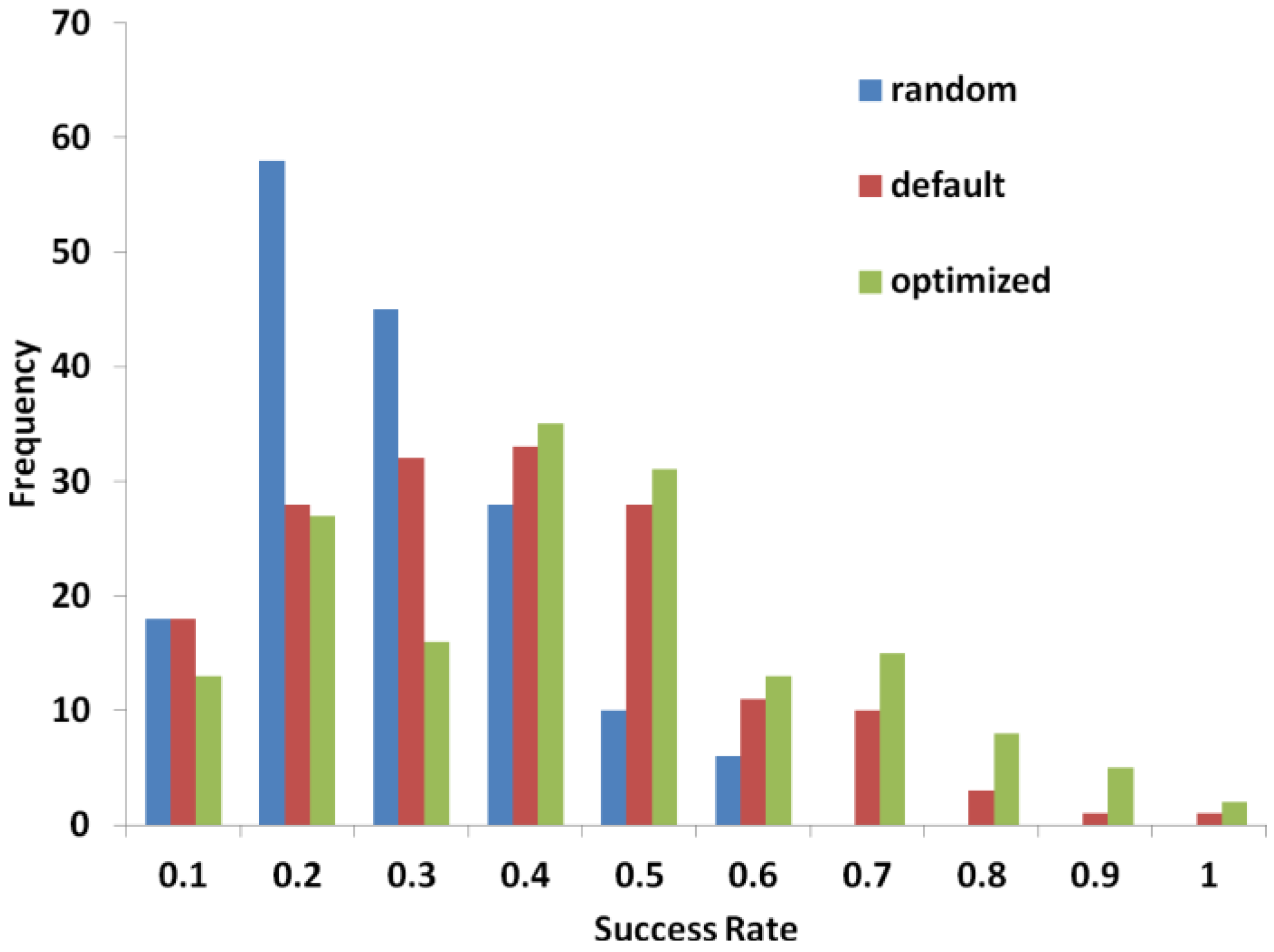
2.2. Optimization of the Search Algorithm on PDBbind Dataset
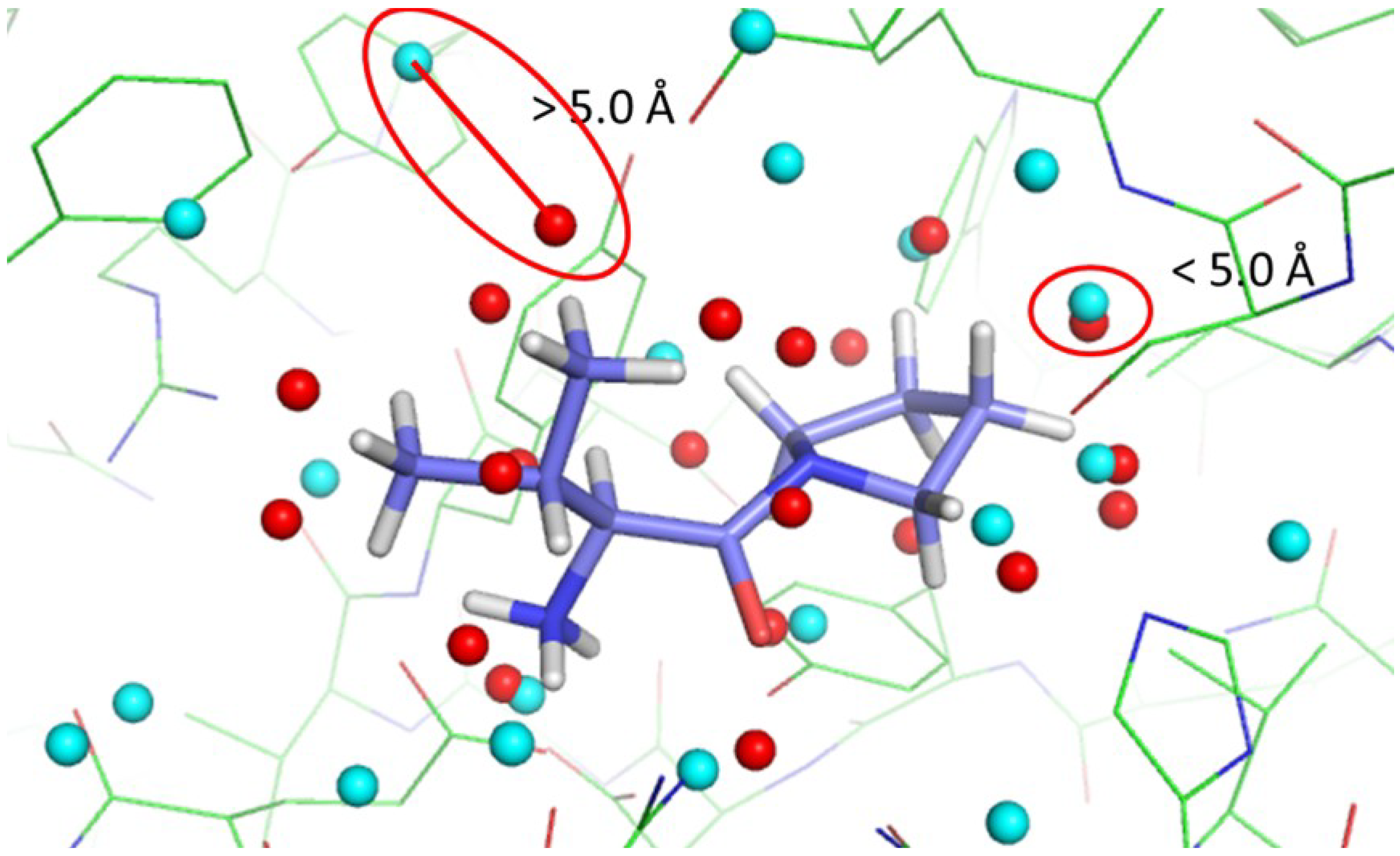
| Setting | 3DZD Difference | Geodesic Distribution | Geodesic Distance | Success Rate | |||
|---|---|---|---|---|---|---|---|
| 3.0 Å | 4.0 Å | 5.0 Å | 6.0 Å | ||||
| Default 1 | 0.32 | 0.48 | 0.2 | 12.3% | 21.6% | 33.1% | 44.0% |
| Optimized | 0.35 | 0.15 | 0.5 | 15.2% | 26.2% | 39.1% | 51.1% |
| Random 2 | - | - | - | 7.6% | 13.4% | 23.7% | 35.8% |
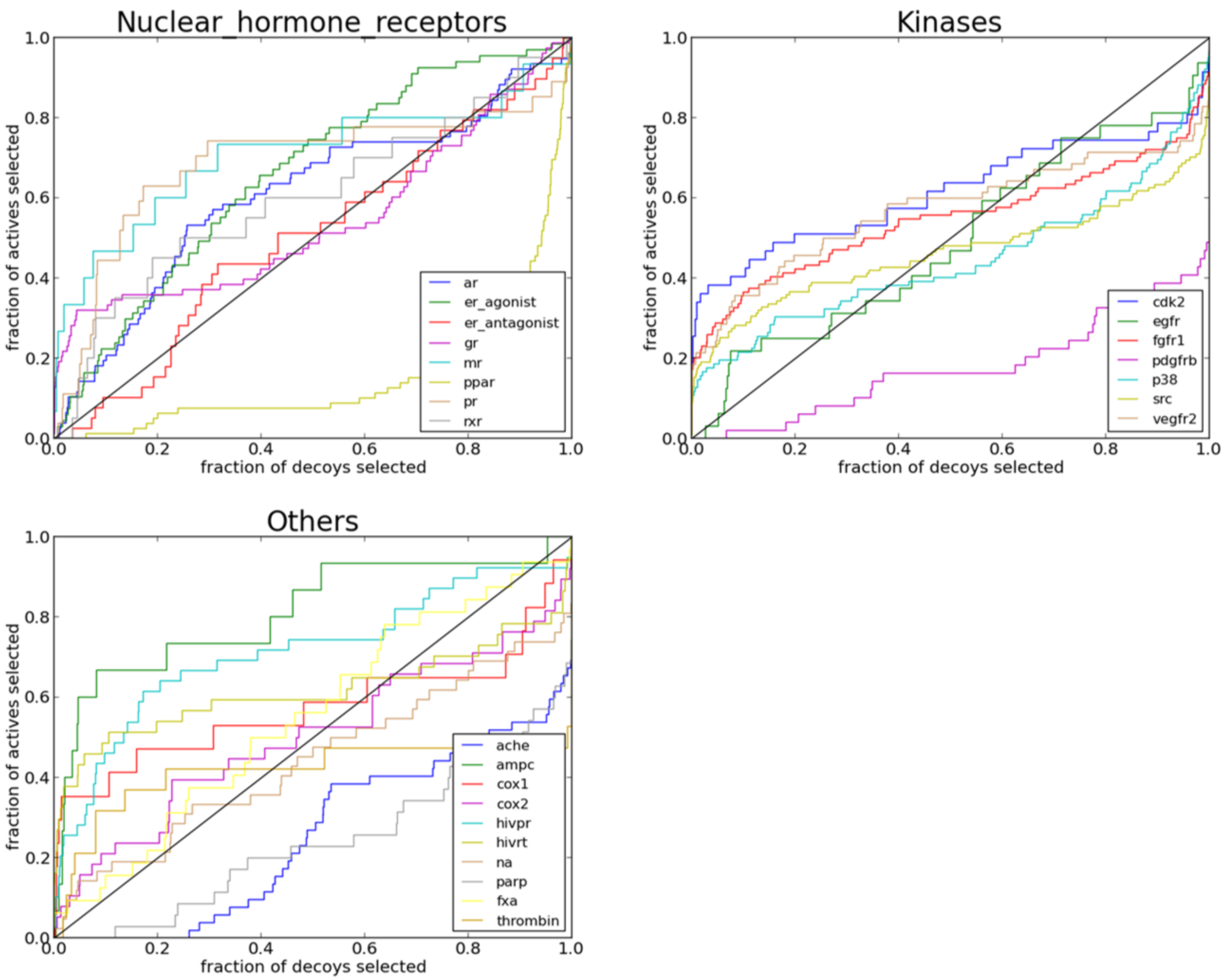
2.3. Performance of PL-PatchSurfer on Directory of Useful Decoys (DUD) Data Set
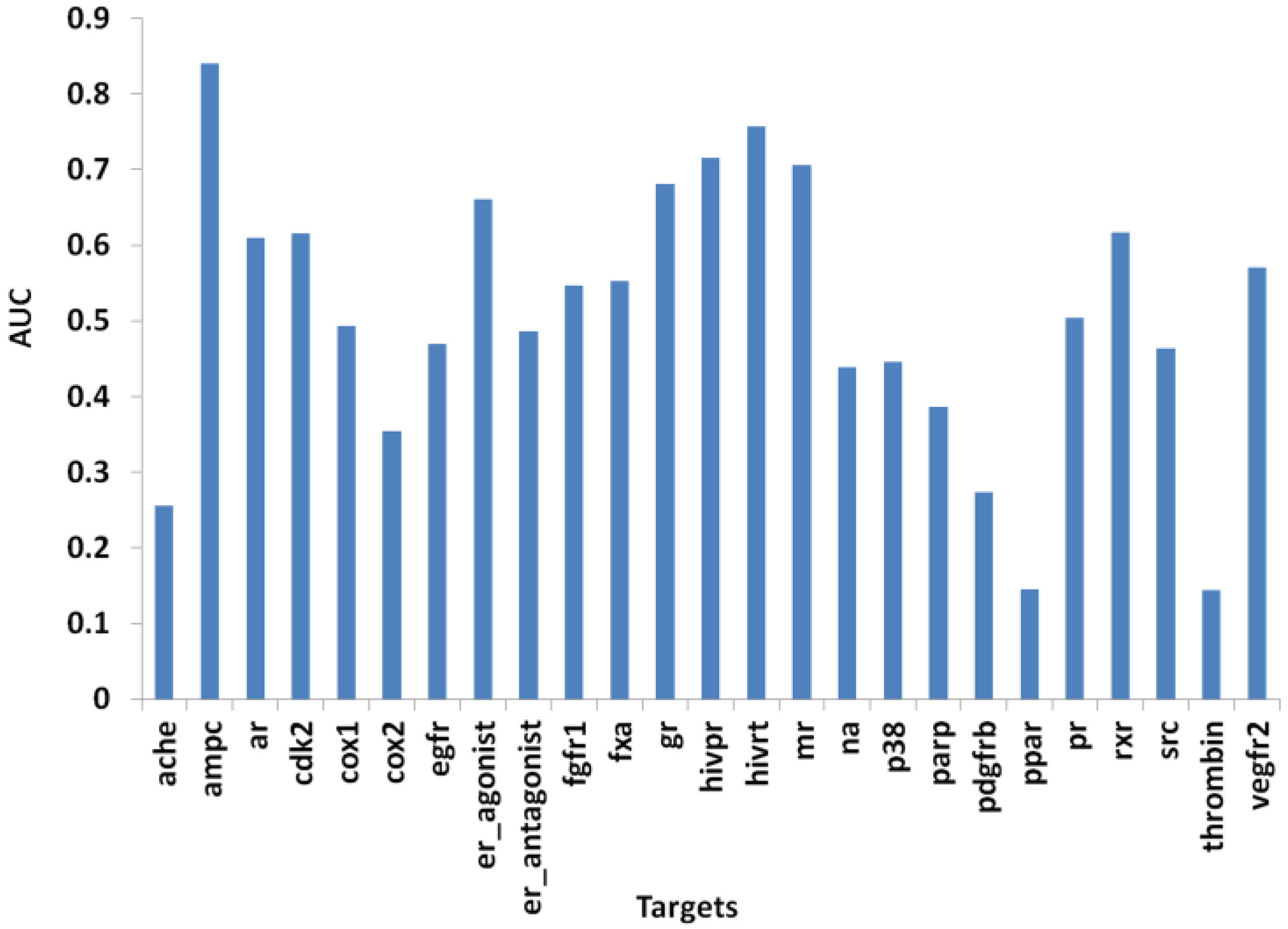
| Protein | PL-PatchSurfer | PharmDock | ||||
|---|---|---|---|---|---|---|
| EF1% | EF10% | EF20% | EF1% | EF10% | EF20% | |
| AChE | 0.00 | 0.00 | 0.00 | 0.00 | 0.19 | 0.14 |
| AmpC | 11.27 | 6.63 | 3.65 | 0.00 | 0.48 | 0.95 |
| AR | 0.00 | 1.81 | 1.62 | 12.16 | 2.43 | 2.09 |
| CDK2 | 25.26 | 3.81 | 2.34 | 2.00 | 2.40 | 2.20 |
| COX1 | 23.41 | 3.51 | 2.34 | 4.00 | 0.80 | 0.40 |
| COX2 | 2.56 | 1.57 | 1.05 | 4.60 | 1.67 | 1.05 |
| EGFr | 0.00 | 2.18 | 1.24 | 2.25 | 2.16 | 1.94 |
| ER_agonist | 0.00 | 2.23 | 1.64 | 2.99 | 5.37 | 3.21 |
| ER_antagonist | 0.00 | 0.51 | 0.64 | 12.82 | 3.59 | 2.69 |
| FGFr1 | 18.89 | 3.45 | 1.97 | 0.85 | 0.17 | 0.55 |
| FXa | 5.86 | 1.25 | 1.40 | 3.52 | 2.46 | 2.08 |
| GR | 14.89 | 3.33 | 1.79 | 11.54 | 1.92 | 1.35 |
| HIVPR | 15.18 | 5.09 | 3.20 | 24.53 | 7.17 | 4.06 |
| HIVRT | 27.86 | 5.10 | 2.83 | 7.50 | 1.75 | 1.75 |
| MR | 17.97 | 4.66 | 2.66 | 26.67 | 6.00 | 3.67 |
| NA | 2.37 | 1.66 | 0.95 | 4.08 | 1.84 | 1.12 |
| P38 | 12.56 | 2.15 | 1.52 | 1.95 | 1.60 | 1.54 |
| PARP | 0.00 | 0.00 | 0.14 | 36.36 | 4.85 | 3.03 |
| PDGFrb | 0.00 | 0.20 | 0.41 | 0.00 | 0.38 | 0.45 |
| PPARg | 0.00 | 0.00 | 0.06 | 0.00 | 0.00 | 0.43 |
| PR | 3.60 | 2.20 | 2.77 | 3.70 | 2.22 | 1.48 |
| RXR | 0.00 | 2.96 | 2.23 | 0.00 | 1.00 | 2.25 |
| SRC | 15.99 | 2.90 | 1.87 | 0.65 | 2.13 | 1.84 |
| thrombin | 0.00 | 2.08 | 1.57 | 1.54 | 0.92 | 1.15 |
| VEGFr2 | 17.96 | 2.71 | 2.00 | 8.11 | 2.16 | 1.69 |
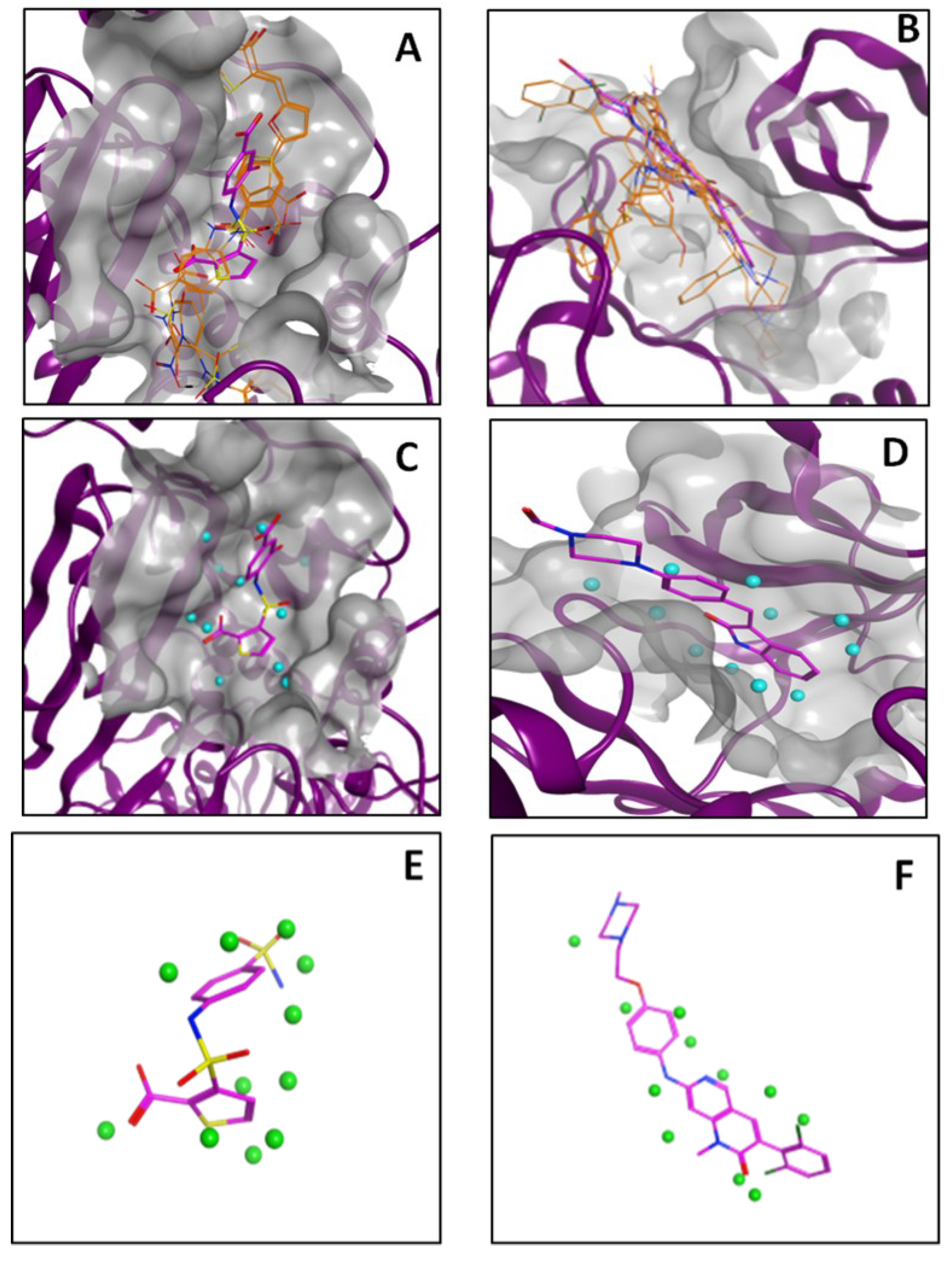
| Protein | 3DZD | Relative Distance | Pocket Size | AUC | AUC before Training |
|---|---|---|---|---|---|
| AChE | 0 | 0.3 | 0.9 | 0.60 | 0.26 |
| PPAR | 0 | 1.0 | 0 | 0.68 | 0.15 |
| COX-2 | 0 | 0 | 1.0 | 0.56 | 0.35 |
| PDGFrb | 0 | 1.0 | 1.0 | 0.42 | 0.27 |
| Thrombin | 0 | 0 | 1.0 | 0.50 | 0.14 |
2.4. Comparison of Computational Time
| System | PharmDock | PL-PatchSurfer | AutoDock | Glide |
|---|---|---|---|---|
| 10gs | 585.5 | 1.2 | 43 | 54 |
| 1a30 | 74.0 | 0.5 | 30 | 48 |
| 1bcu | 15.0 | 0.4 | 5 | 22 |
| 1gpk | 22.0 | 0.5 | 7 | 33 |
| 1h23 | 988.9 | 0.8 | 57 | 48 |
| 1lol | 64.6 | 0.7 | 15 | 129 |
| 1loq | 42.6 | 0.7 | 12 | 120 |
| 1mq6 | 348.9 | 0.7 | 34 | 28 |
| 1n2v | 15.6 | 0.4 | 6 | 25 |
| 1q8t | 25.9 | 1.3 | 8 | 31 |
3. Experimental Design
3.1. Definition of the Protein Pocket Surface
3.2. Generation of Ligand Conformations and Computation of the Surface Properties
3.3. Identification of the Surface Patches
3.4. Computation of the 3D Zernike Descriptors on the Surface Patches
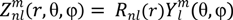
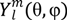 is the spherical harmonics and Rnl(r) is the radial function constructed in a way that
is the spherical harmonics and Rnl(r) is the radial function constructed in a way that 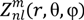 can be converted to polynomials in the Cartesian coordinates,
can be converted to polynomials in the Cartesian coordinates, 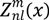 . To obtain the 3DZD of f(x), first 3D Zernike moments are computed:
. To obtain the 3DZD of f(x), first 3D Zernike moments are computed:


3.5. Procedure of Protein–Ligand Patch Comparison
3.5.1. Search Matching Patches between the Pocket and the Ligand
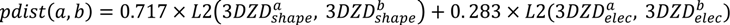
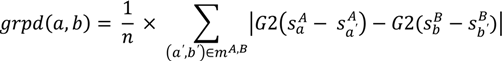
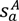 is the coordinate of the center of the patch in pocket A of matched pair a. G2 is the geodesic distance between the centers of the two patches.
is the coordinate of the center of the patch in pocket A of matched pair a. G2 is the geodesic distance between the centers of the two patches.3.5.2. Score the Overall Fit of the Ligand into the Pocket
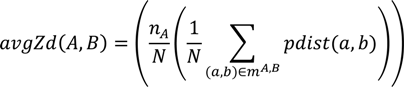

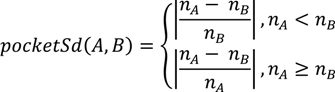
3.6. Data Set and Evaluation Methods
3.6.1. Huang Dataset
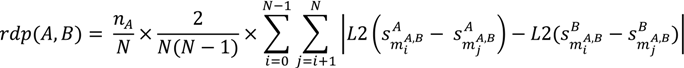
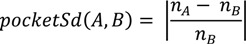
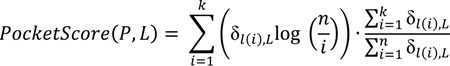
3.6.2. PDBbind Dataset


3.6.3. DUD Set
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The protein data bank. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Hoffmann, B.; Zaslavskiy, M.; Vert, J.-P.; Stoven, V. A new protein binding pocket similarity measure based on comparison of clouds of atoms in 3D: Application to ligand prediction. BMC Bioinform. 2010, 11, 99. [Google Scholar]
- Kinoshita, K.; Nakamura, H. Identification of the ligand binding sites on the molecular surface of proteins. Protein Sci. 2005, 14, 711–718. [Google Scholar]
- Shulman-Peleg, A.; Nussinov, R.; Wolfson, H.J. Siteengines: Recognition and comparison of binding sites and protein–protein interfaces. Nucleic Acids Res. 2005, 33, W337–W341. [Google Scholar]
- Schmitt, S.; Kuhn, D.; Klebe, G. A new method to detect related function among proteins independent of sequence and fold homology. J. Mol. Biol. 2002, 323, 387–406. [Google Scholar]
- Najmanovich, R.; Kurbatova, N.; Thornton, J. Detection of 3D atomic similarities and their use in the discrimination of small molecule protein–binding sites. Bioinformatics 2008, 24, i105–i111. [Google Scholar]
- Sael, L.; Kihara, D. Binding ligand prediction for proteins using partial matching of local surface patches. Int. J. Mol. Sci. 2010, 11, 5009–5026. [Google Scholar]
- Sael, L.; Kihara, D. Detecting local ligand-binding site similarity in nonhomologous proteins by surface patch comparison. Proteins 2012, 80, 1177–1195. [Google Scholar]
- Brylinski, M.; Skolnick, J. A threading-based method (findsite) for ligand-binding site prediction and functional annotation. Proc. Natl. Acad. Sci. USA 2008, 105, 129–134. [Google Scholar]
- Porter, C.T.; Bartlett, G.J.; Thornton, J.M. The catalytic site atlas: A resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004, 32, D129–D133. [Google Scholar]
- Arakaki, A.K.; Zhang, Y.; Skolnick, J. Large-scale assessment of the utility of low-resolution protein structures for biochemical function assignment. Bioinformatics 2004, 20, 1087–1096. [Google Scholar]
- Mestres, J.; Knegtel, R.M.A. Similarity versus docking in 3D virtual screening. Perspect. Drug Discov. Des. 2000, 20, 191–207. [Google Scholar] [CrossRef]
- Willett, P.; Barnard, J.M.; Downs, G.M. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998, 38, 983–996. [Google Scholar]
- Cheng, A.; Diller, D.J.; Dixon, S.L.; Egan, W.J.; Lauri, G.; Merz, K.M. Computation of the physio-chemical properties and data mining of large molecular collections. J. Comput. Chem. 2002, 23, 172–183. [Google Scholar]
- Rarey, M.; Dixon, J.S. Feature trees: A new molecular similarity measure based on tree matching. J. Comput. Aided Mol. Des. 1998, 12, 471–490. [Google Scholar]
- Srinivasan, J.; Castellino, A.; Bradley, E.K.; Eksterowicz, J.E.; Grootenhuis, P.D.J.; Putta, S.; Stanton, R.V. Evaluation of a novel shape-based computational filter for lead evolution: Application to thrombin inhibitors. J. Med. Chem. 2002, 45, 2494–2500. [Google Scholar]
- Wild, D.J.; Willett, P. Similarity searching in files of three-dimensional chemical structures. Alignment of molecular electrostatic potential fields with a genetic algorithm. J. Chem. Inf. Comput. Sci. 1996, 36, 159–167. [Google Scholar] [CrossRef]
- Sheridan, R.P.; Kearsley, S.K. Why do we need so many chemical similarity search methods? Drug Discov. Today 2002, 7, 903–911. [Google Scholar] [CrossRef]
- Martin, Y. Distance comparisons (disco): A new strategy for examining 3D structure-activity relationships. In Classical and Three-Dimensional Qsar in Agrochemistry; Hansch, C., Fujita, T., Eds.; American Chemical Society: Washington, DC, USA, 1995; pp. 318–329. [Google Scholar]
- Lill, M. Multi-dimensional qsar in drug discovery. Drug Discov. Today 2007, 12, 1013–1017. [Google Scholar]
- Dixon, S.L.; Smondyrev, A.M.; Knoll, E.H.; Rao, S.N.; Shaw, D.E.; Friesner, R.A. Phase: A new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J. Comput. Aided Mol. Des. 2006, 20, 647–671. [Google Scholar] [CrossRef]
- Sousa, S.F.; Fernandes, P.A.; Ramos, M.J. Protein–ligand docking: Current status and future challenges. Proteins 2006, 65, 15–26. [Google Scholar]
- Cavasotto, C.N.; Orry, A.J. Ligand docking and structure-based virtual screening in drug discovery. Curr. Top. Med. Chem. 2007, 7, 1006–1014. [Google Scholar]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar]
- Goodford, P.J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985, 28, 849–857. [Google Scholar]
- Goodsell, D.S. Computational docking of biomolecular complexes with autodock. CSH Protoc. 2009. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Ewing, T.; Makino, S.; Skillman, A.; Kuntz, I. Dock 4.0: Search strategies for automated molecular docking of flexible molecule databases. J. Comput. Aided Mol. Des. 2001, 15, 411–428. [Google Scholar] [CrossRef]
- Hu, B.; Lill, M.A. Pharmdock: A pharmacophore-based docking program. J. Cheminform. 2014, 6, 14. [Google Scholar]
- Norel, R.; Lin, S.L.; Wolfson, H.J.; Nussinov, R. Shape complementarity at protein–protein interfaces. Biopolymers 1994, 34, 933–940. [Google Scholar]
- Norel, R.; Petrey, D.; Wolfson, H.J.; Nussinov, R. Examination of shape complementarity in docking of unbound proteins. Proteins: Struct. Funct. Bioinf. 1999, 36, 307–317. [Google Scholar] [CrossRef]
- Chen, R.; Weng, Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins: Struct. Funct. Bioinf. 2002, 47, 281–294. [Google Scholar] [CrossRef]
- Venkatraman, V.; Yang, Y.D.; Sael, L.; Kihara, D. Protein–protein docking using region-based 3D zernike descriptors. BMC Bioinform. 2009, 10, 407. [Google Scholar]
- Sael, L.; Kihara, D. Constructing patch-based ligand-binding pocket database for predicting function of proteins. BMC Bioinform. 2012, 13, S7. [Google Scholar]
- Chikhi, R.; Sael, L.; Kihara, D. Real-time ligand binding pocket database search using local surface descriptors. Proteins: Struct. Funct. Bioinf. 2010, 78, 2007–2028. [Google Scholar] [CrossRef]
- Kihara, D.; Sael, L.; Chikhi, R.; Esquivel-Rodriguez, J. Molecular surface representation using 3D zernike descriptors for protein shape comparison and docking. Curr. Protein Pept. Sci. 2011, 12, 520–530. [Google Scholar]
- Wang, R.; Fang, X.; Lu, Y.; Wang, S. The pdbbind database: Collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J. Med. Chem. 2004, 47, 2977–2980. [Google Scholar]
- Wang, R.; Fang, X.; Lu, Y.; Yang, C.Y.; Wang, S. The pdbbind database: Methodologies and updates. J. Med. Chem. 2005, 48, 4111–4119. [Google Scholar]
- Huang, B.; Schroeder, M. Ligsitecsc: Predicting ligand binding sites using the connolly surface and degree of conservation. BMC Struct. Biol. 2006, 6, 19. [Google Scholar]
- Kahraman, A.; Morris, R.J.; Laskowski, R.A.; Favia, A.D.; Thornton, J.M. On the diversity of physicochemical environments experienced by identical ligands in binding pockets of unrelated proteins. Proteins 2010, 78, 1120–1136. [Google Scholar]
- Kinoshita, K.; Murakami, Y.; Nakamura, H. Ef-seek: Prediction of the functional sites of proteins by searching for similar electrostatic potential and molecular surface shape. Nucleic Acids Res. 2007, 35, W398–W402. [Google Scholar]
- Omega: Version 2.2.0. Available online: http://www.eyesopen.com (accessed on 25 August 2014).
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with omega: Algorithm and validation using high quality structures from the protein databank and cambridge structural database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar]
- Hawkins, P.C.D.; Nicholls, A. Conformer generation with omega: Learning from the dataset and the analysis of failures. J. Chem. Inf. Model. 2012, 52, 2919–2936. [Google Scholar]
- Rarey, M.; Kramer, B.; Lengauer, T. Time-efficient docking of flexible ligands into active sites of proteins. In Proceedings of the 3rd International Conference on Intelligent Systems for Molecular Biology, Cambridge, UK, 16–19 July 1995; pp. 300–308.
- Abagyan, R.; Totrov, M.; Kuznetsov, D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994, 15, 488–506. [Google Scholar]
- Jain, A. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003, 46, 499–511. [Google Scholar]
- Joseph-McCarthy, D.; Thomas, B.E.; Belmarsh, M.; Moustakas, D.; Alvarez, J.C. Pharmacophore-based molecular docking to account for ligand flexibility. Proteins 2003, 51, 172–188. [Google Scholar]
- Huang, N.; Shoichet, B.K.; Irwin, J.J. Benchmarking sets for molecular docking. J. Med. Chem. 2006, 49, 6789–6801. [Google Scholar]
- Cross, J.B.; Thompson, D.C.; Rai, B.K.; Baber, J.C.; Fan, K.Y.; Hu, Y.; Humblet, C. Comparison of several molecular docking programs: Pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2009, 49, 1455–1474. [Google Scholar]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar]
- Canterakis, N. 3D zernike moments and zernike affine invariants for 3D image analysis and recognition. In Proceedings of the 11th Scandinavian Conference on Image Analysis, Kangerlusssuaq, Greenland, 7–11 June 1999; SCIA: Kangerlusssuaq, Greenland, 1999. [Google Scholar]
- Novotni, M.; Klein, R. 3D zernike descriptors for content based shape retrieval. In Proceedings of the Eighth ACM Symposium on Solid Modeling and Applications, Seattle, WA, USA, 16–20 June 2003; ACM: New York, NY, USA, 2003; pp. 216–225. [Google Scholar]
- Demange, G.; Gale, D.; Sotomayor, M. Multi-item auctions. J. Polit. Econ. 1986, 94, 863–872. [Google Scholar]
- Gehlhaar, D.K.; Verkhivker, G.M.; Rejto, P.A.; Sherman, C.J.; Fogel, D.B.; Fogel, L.J.; Freer, S.T. Molecular recognition of the inhibitor ag-1343 by HIV-1 protease: Conformationally flexible docking by evolutionary programming. Chem. Biol. 1995, 2, 317–324. [Google Scholar]
- Hu, B.; Lill, M.A. Exploring the potential of protein-based pharmacophore models in ligand pose prediction and ranking. J. Chem. Inf. Model. 2013, 53, 1179–1190. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hu, B.; Zhu, X.; Monroe, L.; Bures, M.G.; Kihara, D. PL-PatchSurfer: A Novel Molecular Local Surface-Based Method for Exploring Protein-Ligand Interactions. Int. J. Mol. Sci. 2014, 15, 15122-15145. https://doi.org/10.3390/ijms150915122
Hu B, Zhu X, Monroe L, Bures MG, Kihara D. PL-PatchSurfer: A Novel Molecular Local Surface-Based Method for Exploring Protein-Ligand Interactions. International Journal of Molecular Sciences. 2014; 15(9):15122-15145. https://doi.org/10.3390/ijms150915122
Chicago/Turabian StyleHu, Bingjie, Xiaolei Zhu, Lyman Monroe, Mark G. Bures, and Daisuke Kihara. 2014. "PL-PatchSurfer: A Novel Molecular Local Surface-Based Method for Exploring Protein-Ligand Interactions" International Journal of Molecular Sciences 15, no. 9: 15122-15145. https://doi.org/10.3390/ijms150915122
APA StyleHu, B., Zhu, X., Monroe, L., Bures, M. G., & Kihara, D. (2014). PL-PatchSurfer: A Novel Molecular Local Surface-Based Method for Exploring Protein-Ligand Interactions. International Journal of Molecular Sciences, 15(9), 15122-15145. https://doi.org/10.3390/ijms150915122






