A Novel Feature Extraction Scheme with Ensemble Coding for Protein–Protein Interaction Prediction
Abstract
:1. Introduction
2. Results and Discussion
2.1. Performance Evaluation







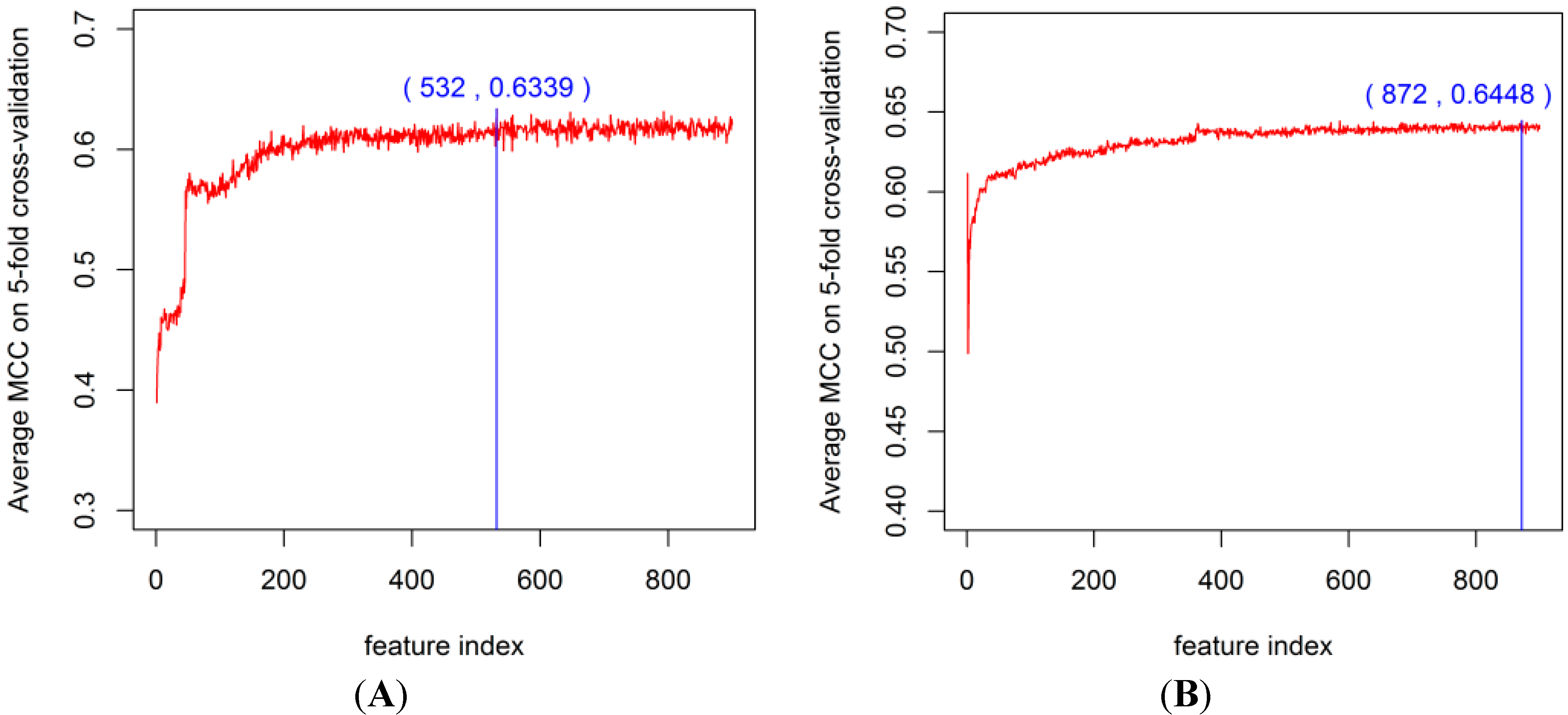
2.2. The DX Result
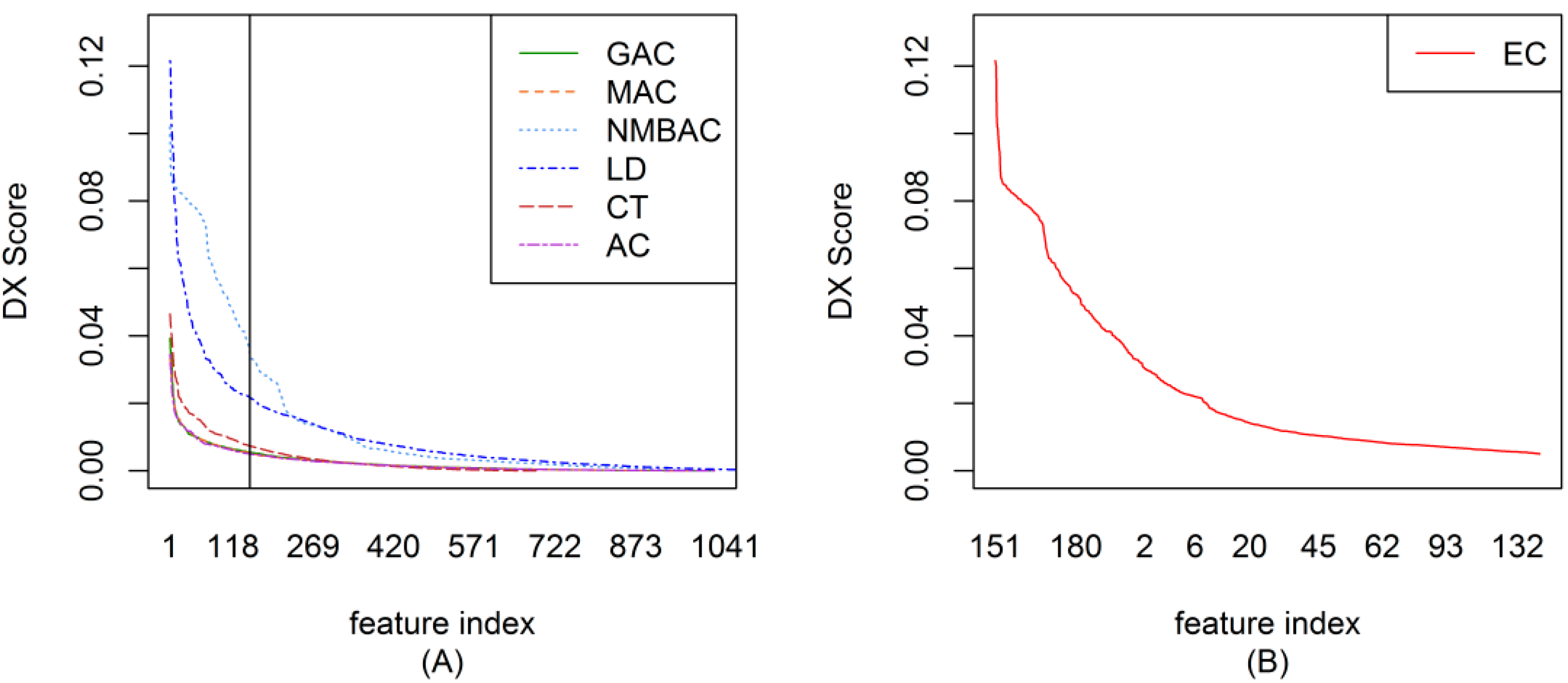
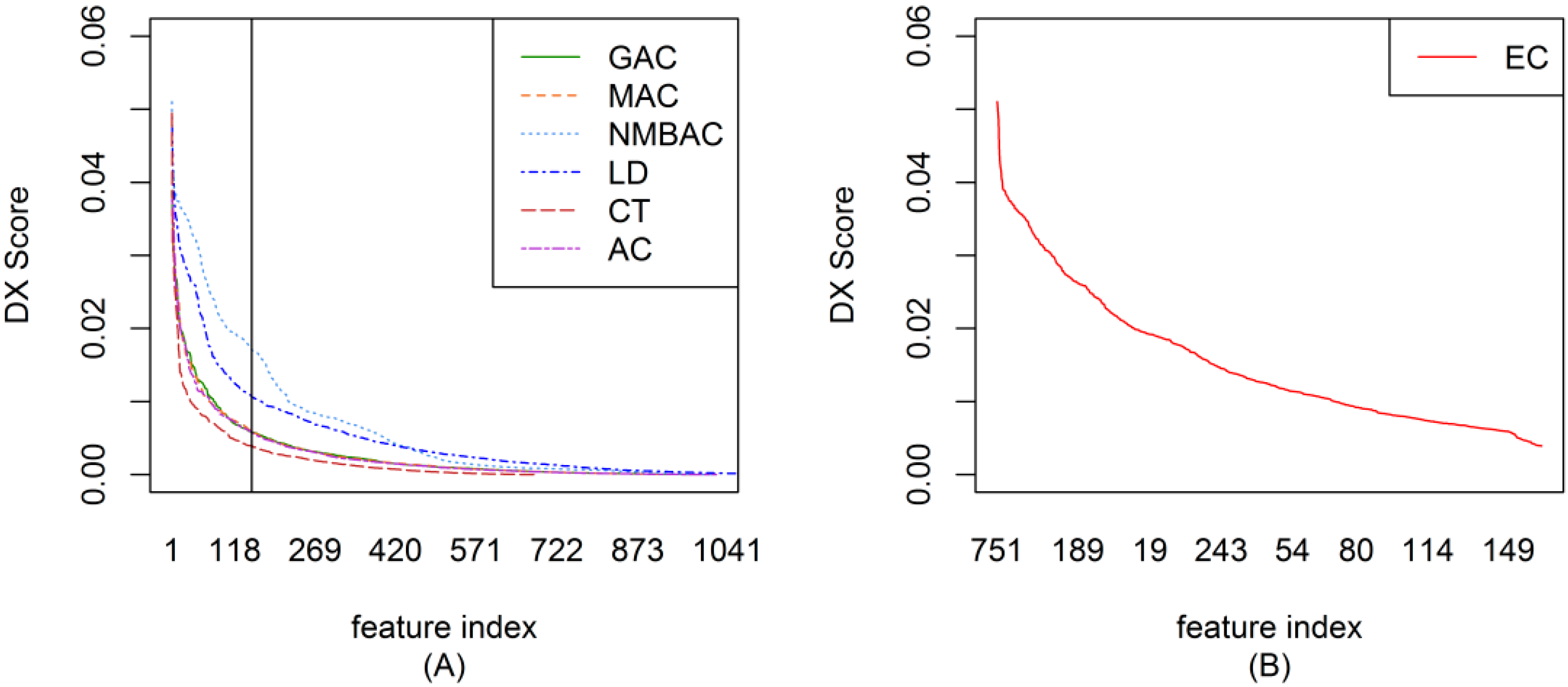
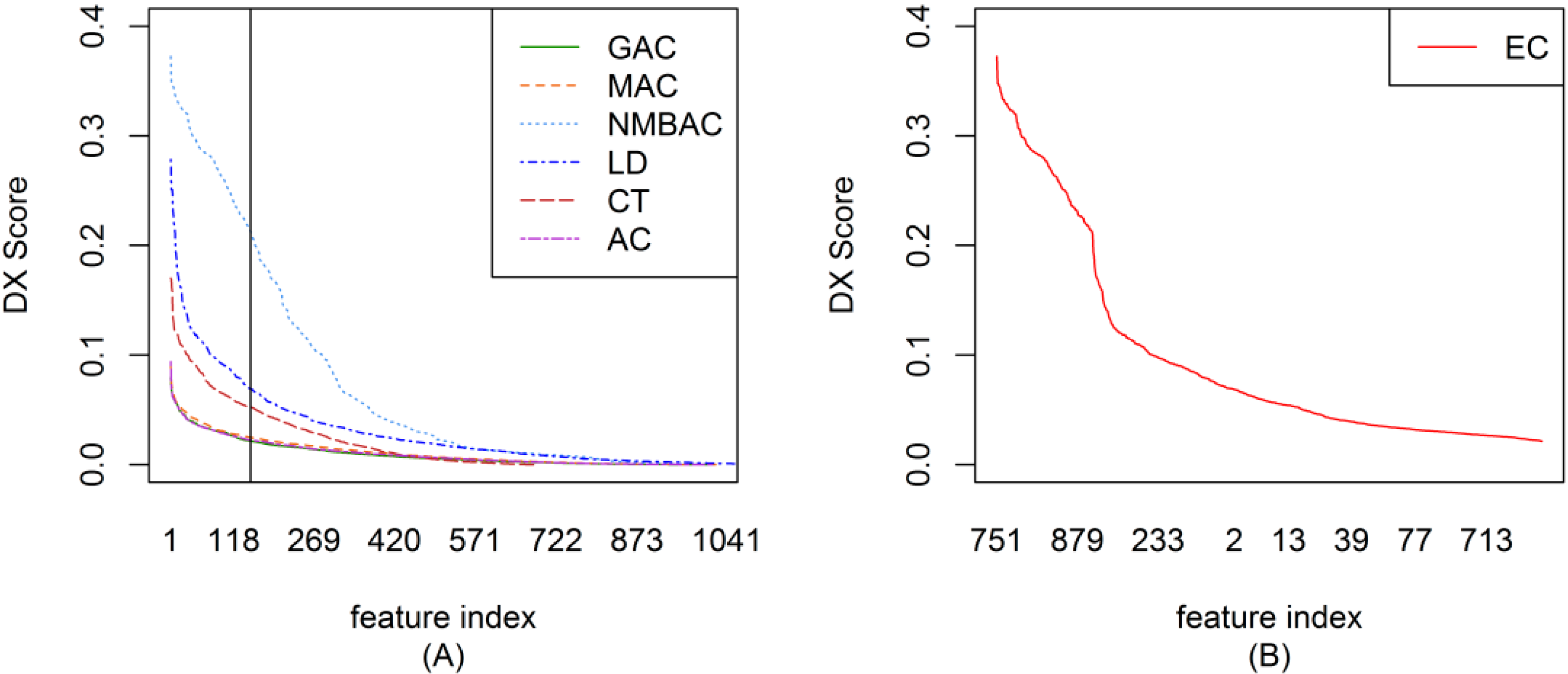
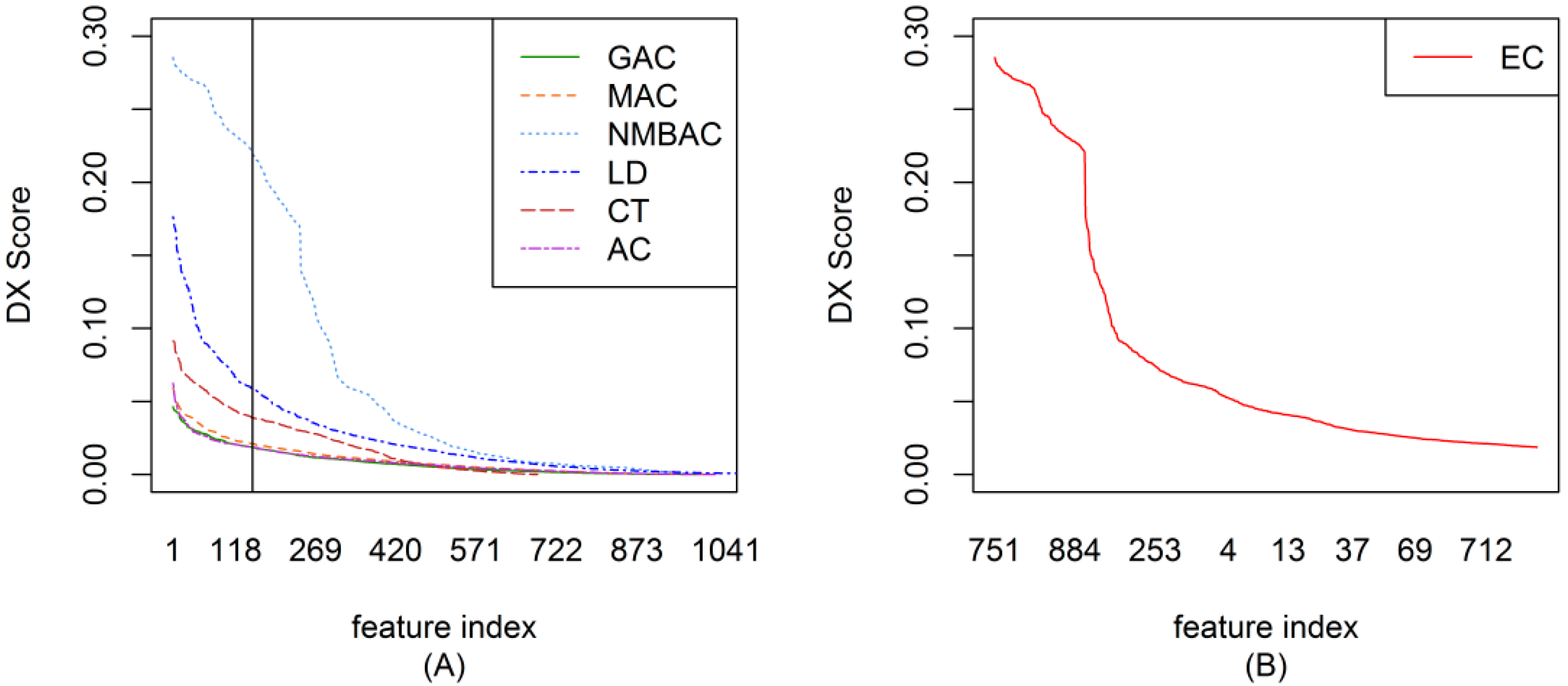
2.3. Feature Importance Evaluation

2.4. Comparison of Prediction Performance by Using Different Methods
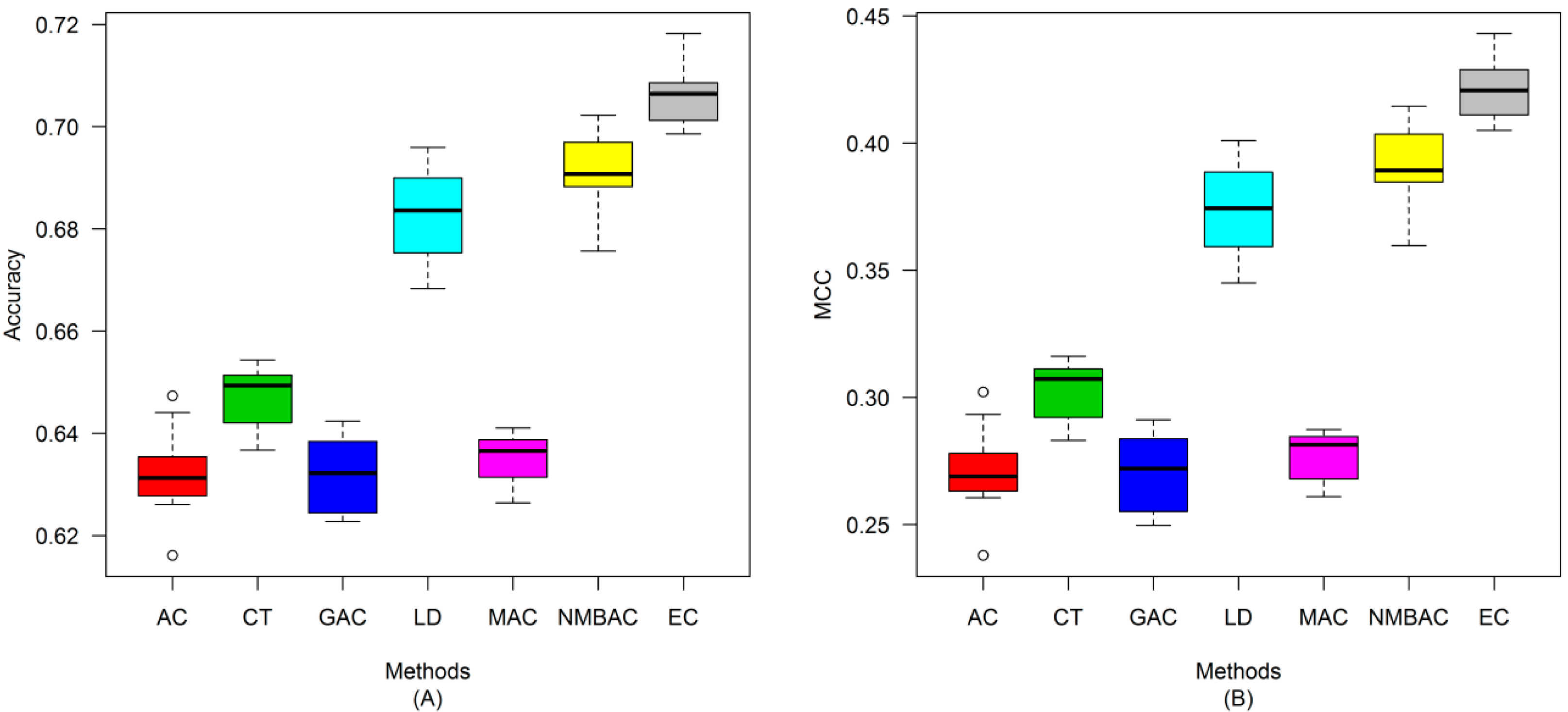
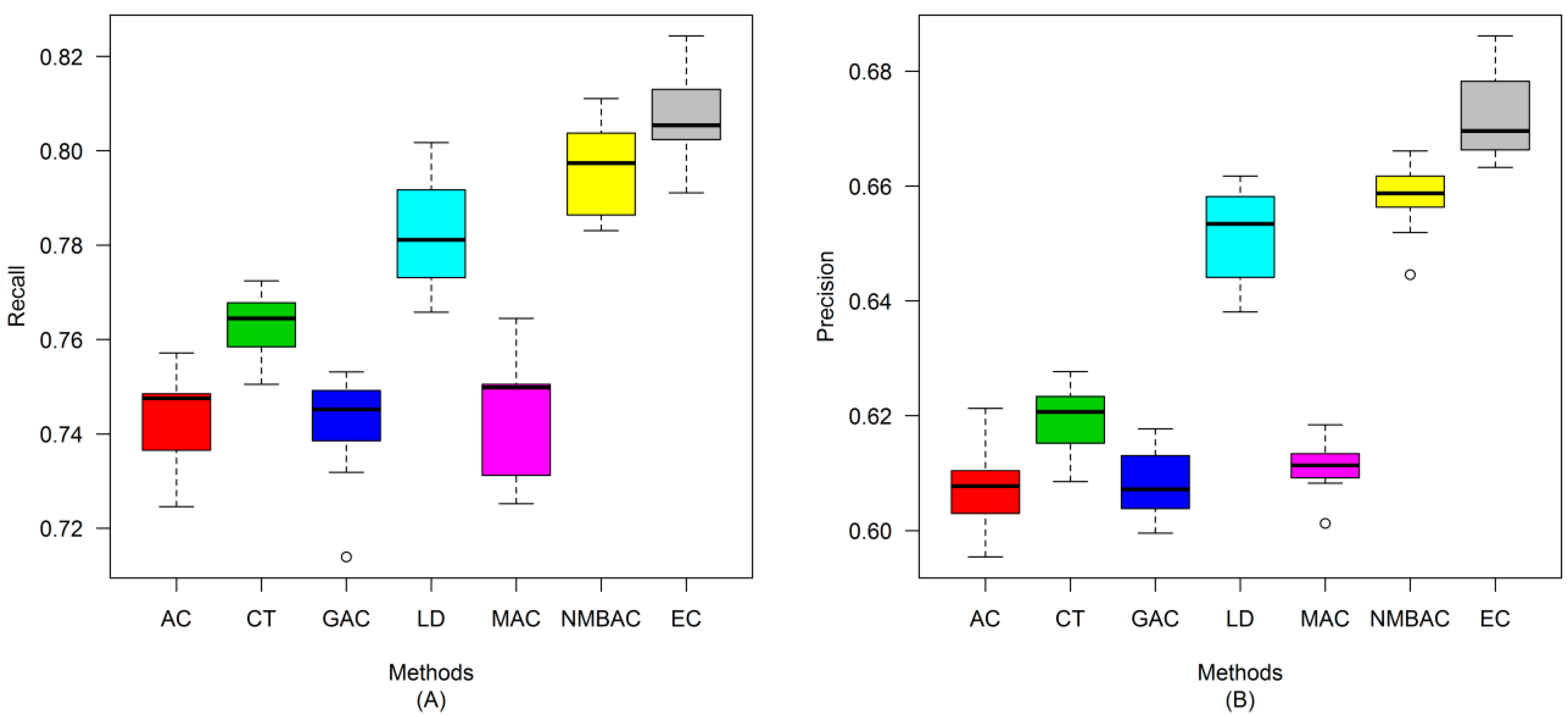
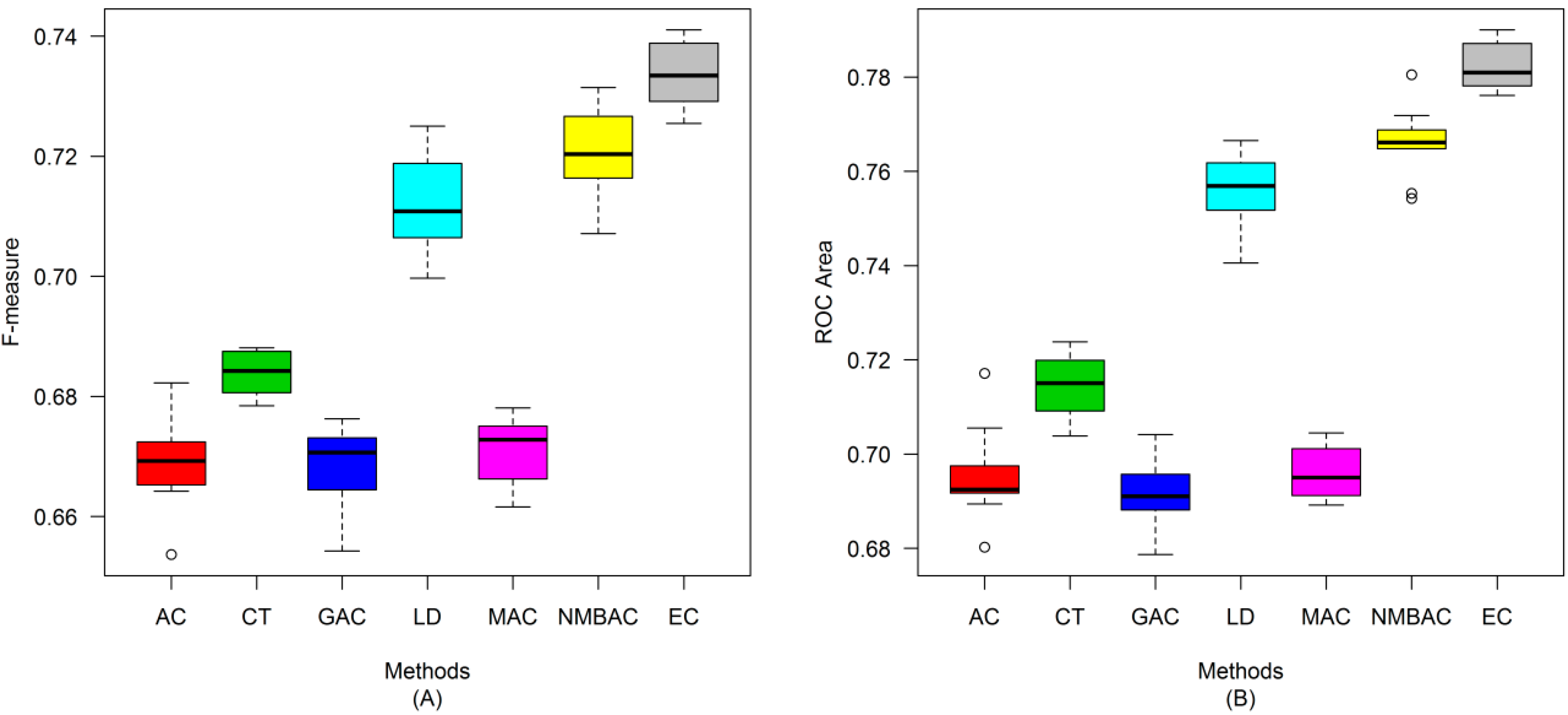
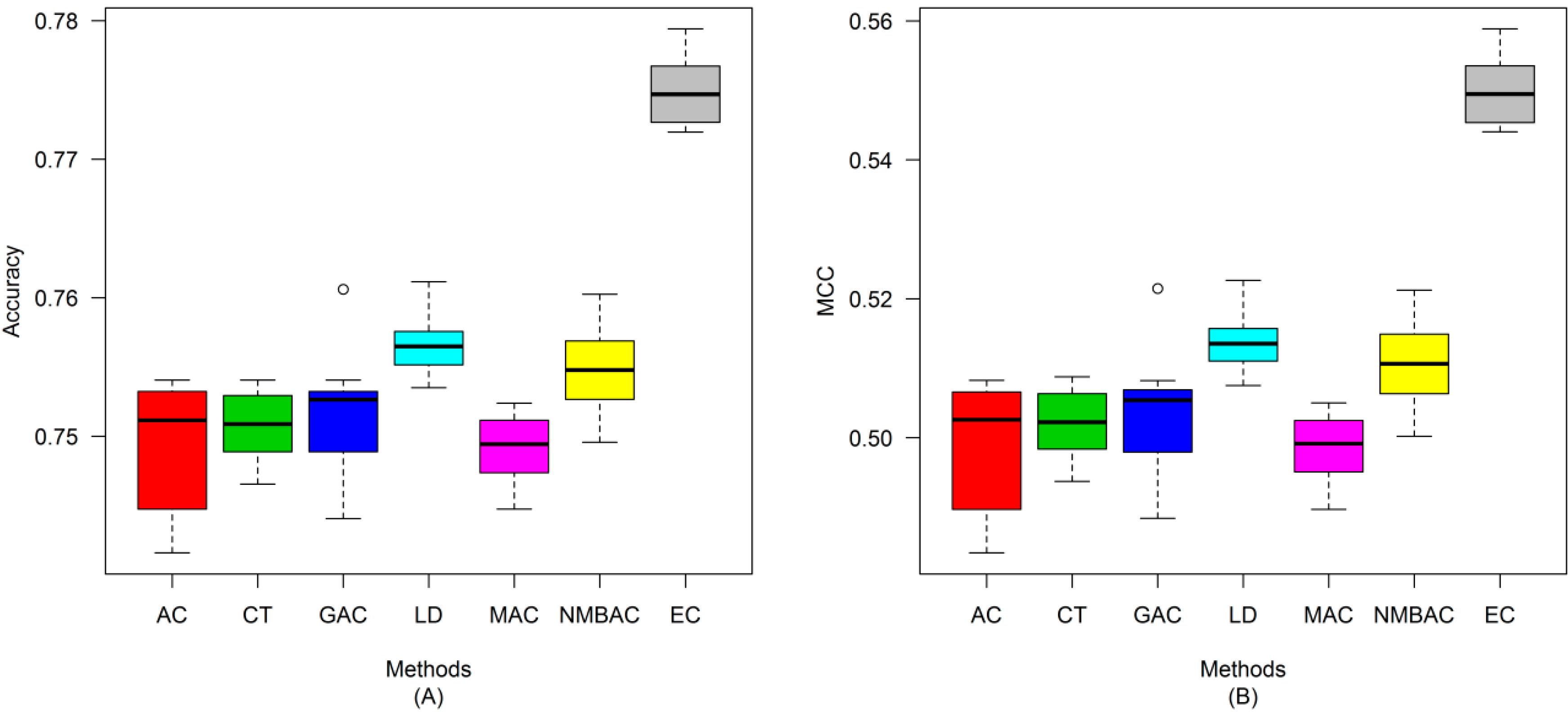
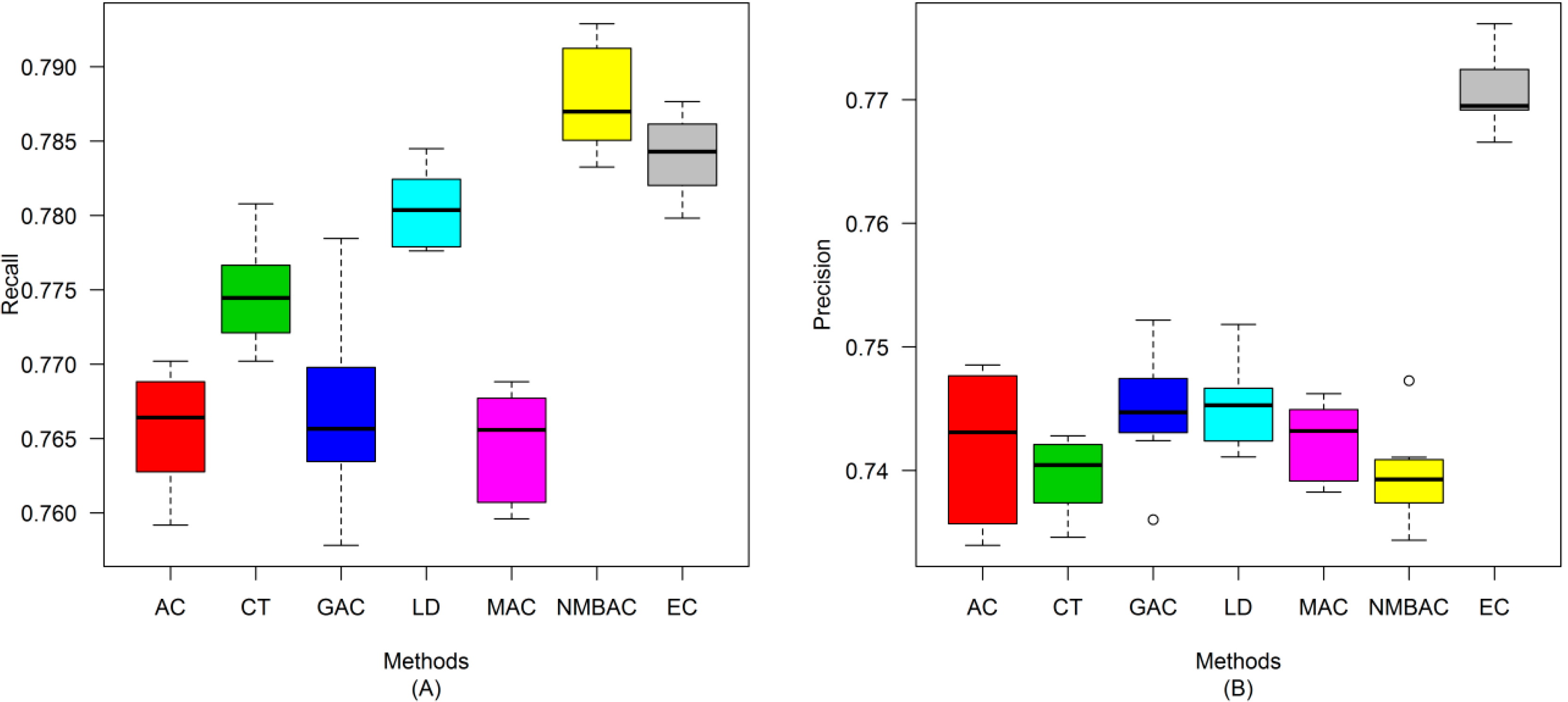

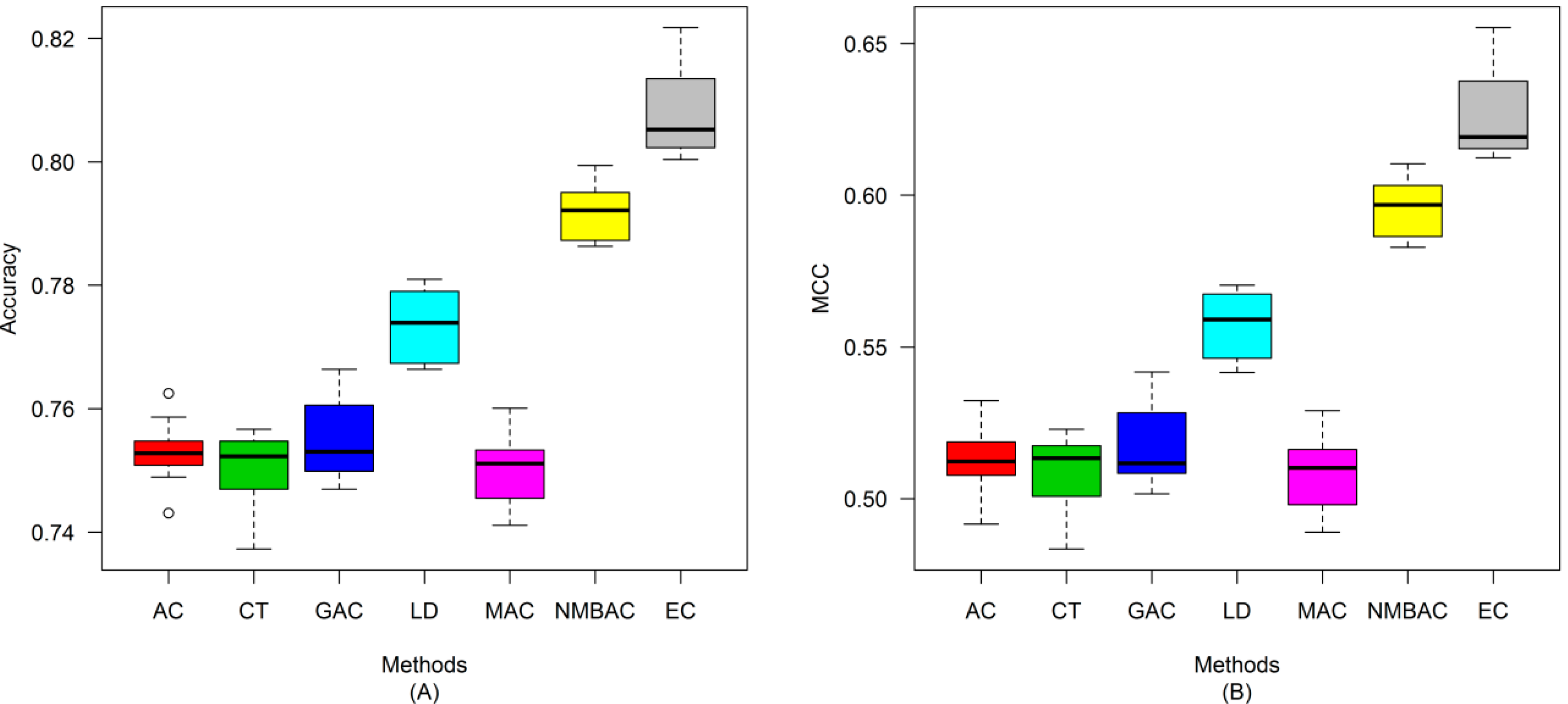
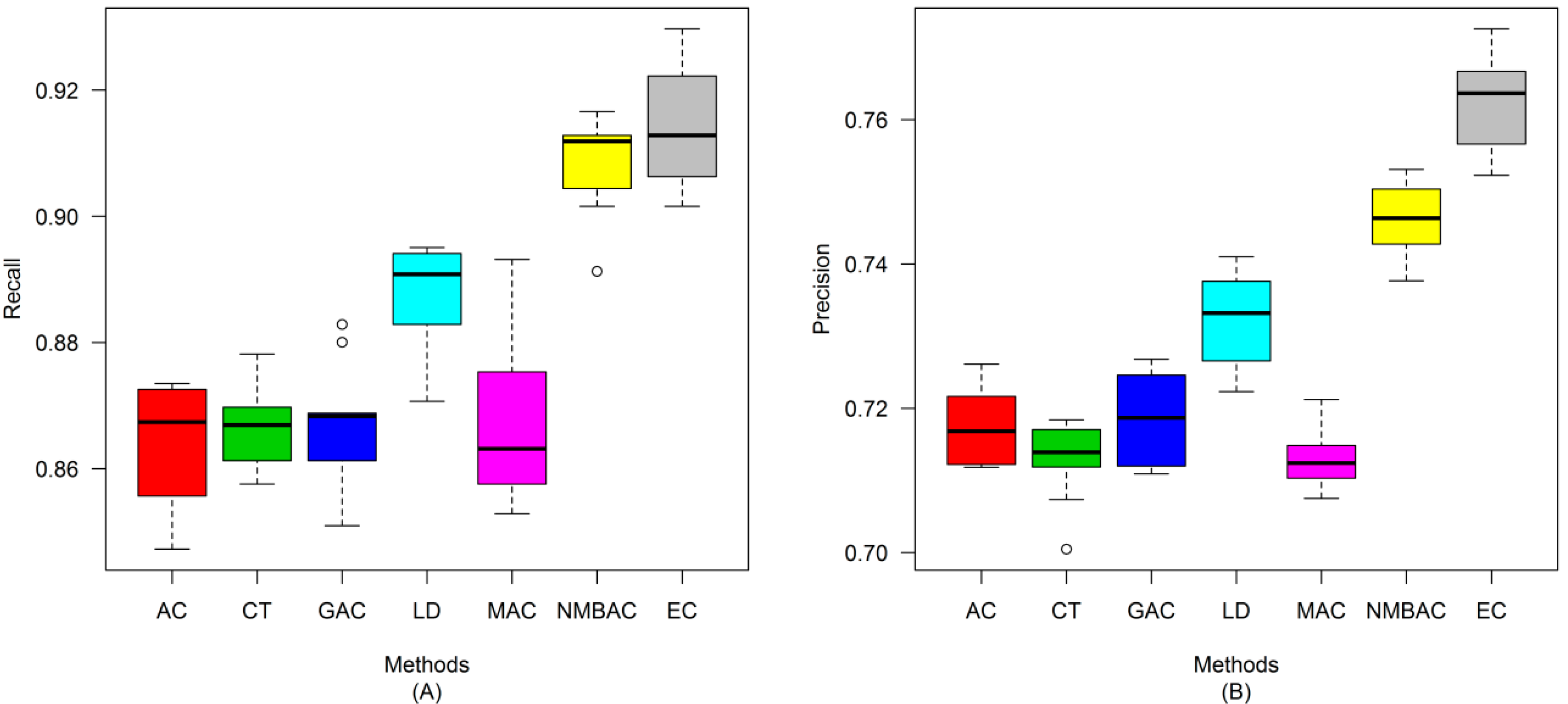
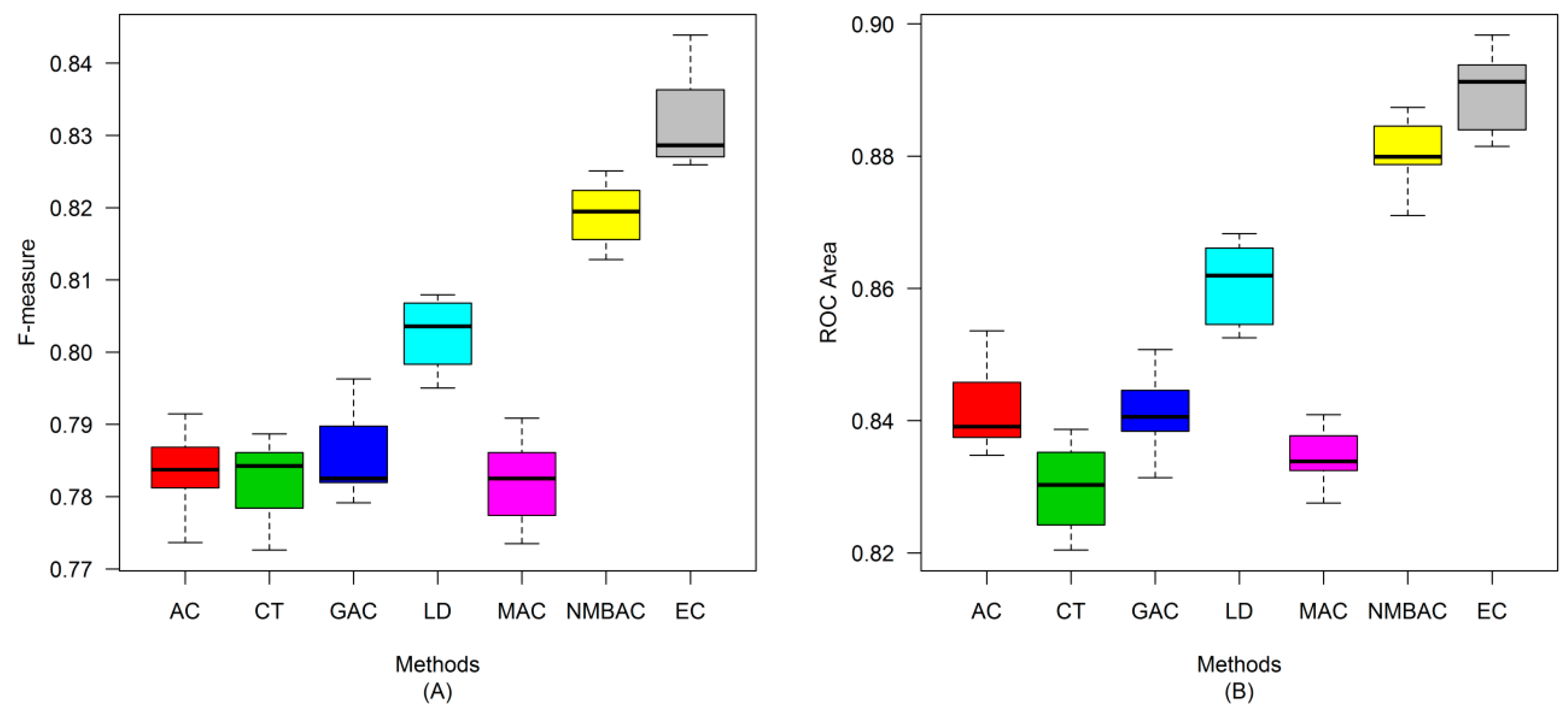


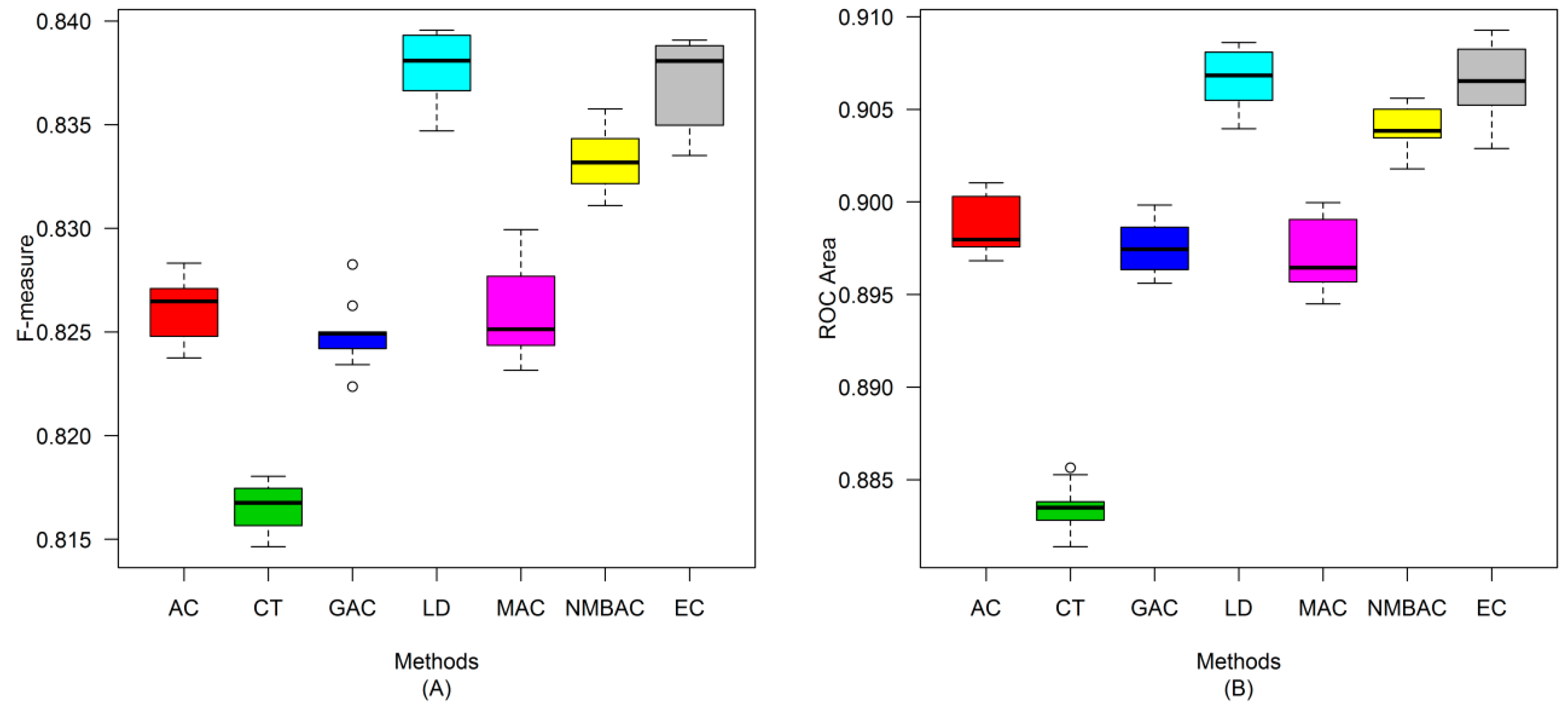
2.5. Comparison with Other Methods on the All Interaction Datasets
| Method | Precision | Recall | F-Measure | Accuracy | MCC | ROC Area |
|---|---|---|---|---|---|---|
| AC-RF | 0.2897 | 0.583 | 0.3871 | 0.5373 | 0.091 | 0.5749 |
| CT-RF | 0.2989 | 0.5645 | 0.3908 | 0.5592 | 0.1058 | 0.5867 |
| LD-RF | 0.3074 | 0.5625 | 0.3975 | 0.5728 | 0.1207 | 0.596 |
| MAC-RF | 0.2894 | 0.5905 | 0.3884 | 0.534 | 0.0916 | 0.576 |
| GAC-RF | 0.2877 | 0.5865 | 0.386 | 0.5325 | 0.0875 | 0.573 |
| NMBAC-RF | 0.31 | 0.5574 | 0.3984 | 0.5783 | 0.1242 | 0.6014 |
| DXEC-RF | 0.3196 | 0.5757 | 0.411 | 0.5866 | 0.1445 | 0.616 |
| Method | Precision | Recall | F-Measure | Accuracy | MCC | ROC Area |
|---|---|---|---|---|---|---|
| AC-RF | 0.3667 | 0.4227 | 0.3927 | 0.6724 | 0.1708 | 0.6222 |
| CT-RF | 0.3664 | 0.4462 | 0.4024 | 0.6679 | 0.1772 | 0.6289 |
| LD-RF | 0.3611 | 0.4359 | 0.395 | 0.6654 | 0.1679 | 0.6249 |
| MAC-RF | 0.3647 | 0.4279 | 0.3938 | 0.6699 | 0.17 | 0.6264 |
| GAC-RF | 0.3722 | 0.4307 | 0.3993 | 0.6753 | 0.1793 | 0.6272 |
| NMBAC-RF | 0.361 | 0.4427 | 0.3977 | 0.664 | 0.1697 | 0.6293 |
| DXEC-RF | 0.3882 | 0.4257 | 0.4061 | 0.688 | 0.1955 | 0.6467 |
| Method | Precision | Recall | F-Measure | Accuracy | MCC | ROC Area |
|---|---|---|---|---|---|---|
| AC-RF | 0.4148 | 0.7781 | 0.5411 | 0.6798 | 0.367 | 0.7886 |
| CT-RF | 0.4177 | 0.8025 | 0.5494 | 0.6806 | 0.3816 | 0.7971 |
| LD-RF | 0.437 | 0.8128 | 0.5684 | 0.7005 | 0.4112 | 0.8175 |
| MAC-RF | 0.4166 | 0.768 | 0.5402 | 0.6827 | 0.365 | 0.7857 |
| GAC-RF | 0.4092 | 0.7652 | 0.5332 | 0.6749 | 0.3541 | 0.7782 |
| NMBAC-RF | 0.4839 | 0.7528 | 0.5891 | 0.7452 | 0.4382 | 0.8289 |
| DXEC-RF | 0.4711 | 0.8322 | 0.6016 | 0.7326 | 0.4616 | 0.8414 |
| Method | Precision | Recall | F-Measure | Accuracy | MCC | ROC Area |
|---|---|---|---|---|---|---|
| AC-RF | 0.4828 | 0.8528 | 0.6165 | 0.7425 | 0.4851 | 0.8721 |
| CT-RF | 0.4632 | 0.8705 | 0.6047 | 0.7237 | 0.471 | 0.8671 |
| LD-RF | 0.4972 | 0.8802 | 0.6355 | 0.7549 | 0.5153 | 0.8893 |
| MAC-RF | 0.4818 | 0.8549 | 0.6163 | 0.7416 | 0.4851 | 0.8721 |
| GAC-RF | 0.4859 | 0.8431 | 0.6165 | 0.7454 | 0.4838 | 0.8704 |
| NMBAC-RF | 0.4942 | 0.8771 | 0.6322 | 0.7523 | 0.5103 | 0.8846 |
| DXEC-RF | 0.5049 | 0.8809 | 0.6419 | 0.7615 | 0.5242 | 0.8911 |

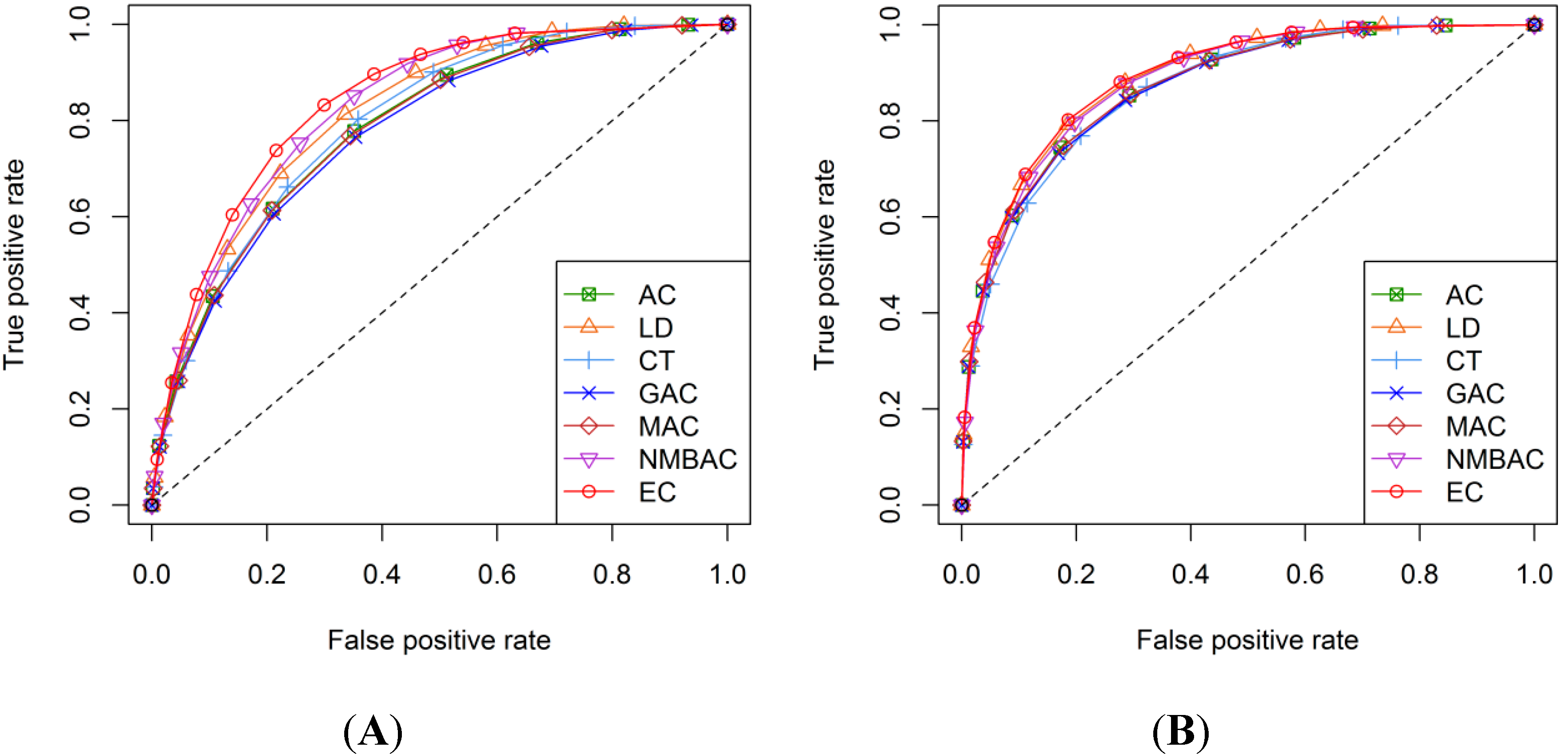
3. Materials and Methods
3.1. Preparation of Datasets
3.2. Molecular Descriptors
3.2.1. AARC (Amino Acid Residue Change) Features
| Dataset | Organism | |||
|---|---|---|---|---|
| Yeast | Human | |||
| Positive | Negative | Positive | Negative | |
| Gold | 1503 | 1503 | 1067 | 992 |
| Silver | 7267 | 7244 | 12680 | 11877 |
| All interactions | 43867 | 131204 | 29714 | 92729 |
3.2.2. The Features of Amino Acid Factors
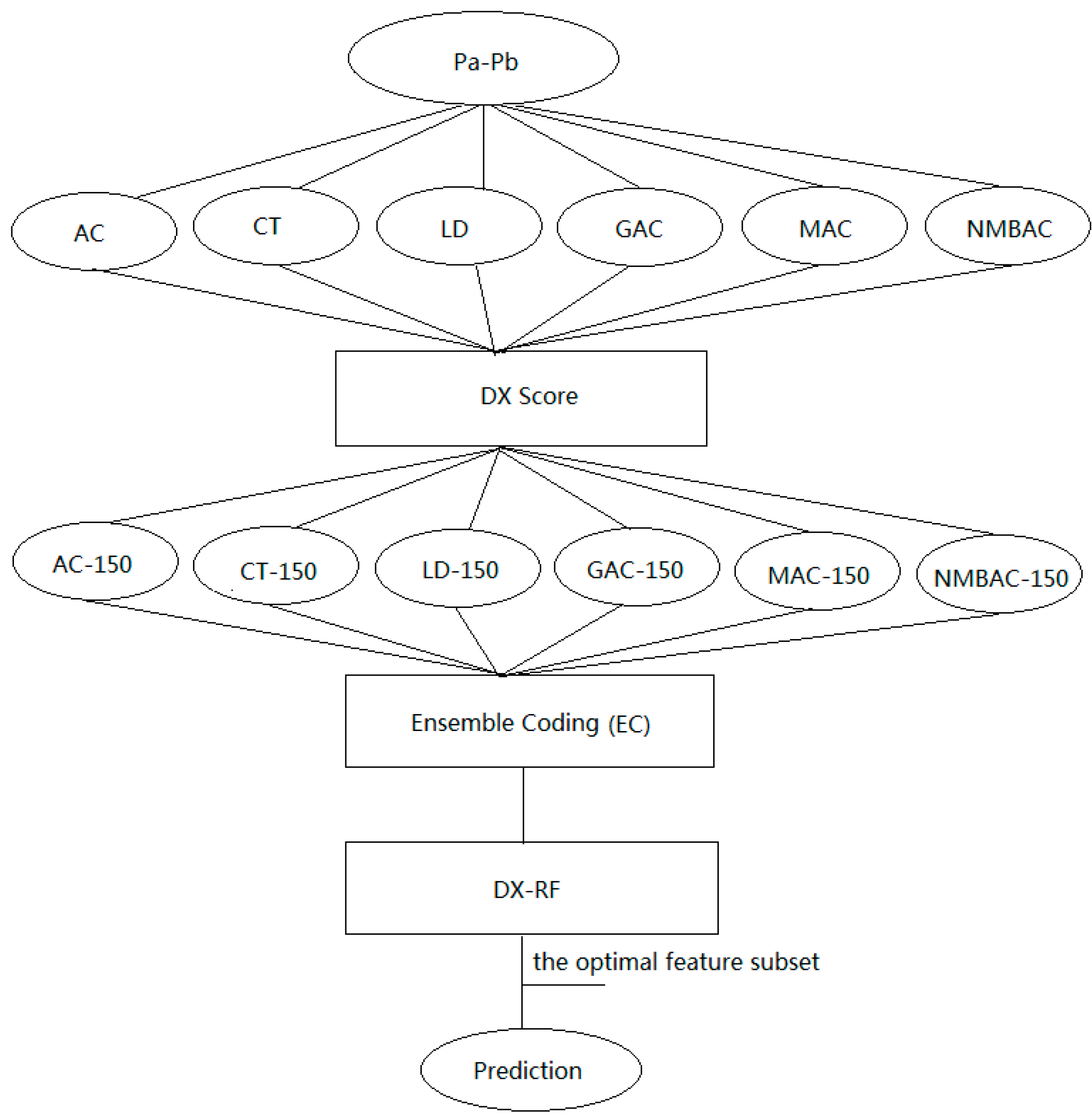
3.3. Feature Selection (DX)

3.4. Ensemble Coding Scheme




4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Braun, P.; Gingras, A.C. History of protein–protein interactions: From egg-white to complex networks. Proteomics 2012, 12, 1478–1498. [Google Scholar] [CrossRef]
- Skrabanek, L.; Saini, H.K.; Bader, G.D.; Enright, A.J. Computational prediction of protein–protein interactions. Mol. Biotechnol. 2008, 38, 1–17. [Google Scholar]
- Jones, S.; Thornton, J.M. Principles of protein–protein interactions. Proc. Natl. Acad. Sci. USA 1996, 93, 13–20. [Google Scholar] [CrossRef]
- Alon, U. Biological networks: The tinkerer as an engineer. Science 2003, 301, 1866–1867. [Google Scholar] [CrossRef]
- Uetz, P.; Giot, L.; Cagney, G.; Mansfield, T.A.; Judson, R.S.; Knight, J.R.; Lockshon, D.; Narayan, V.; Srinivasan, M.; Pochart, P. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 2000, 403, 623–627. [Google Scholar] [CrossRef]
- Ito, T.; Chiba, T.; Ozawa, R.; Yoshida, M.; Hattori, M.; Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 2001, 98, 4569–4574. [Google Scholar] [CrossRef]
- Gavin, A.-C.; Bösche, M.; Krause, R.; Grandi, P.; Marzioch, M.; Bauer, A.; Schultz, J.; Rick, J.M.; Michon, A.-M.; Cruciat, C.-M. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 2002, 415, 141–147. [Google Scholar] [CrossRef]
- Zhu, H.; Bilgin, M.; Bangham, R.; Hall, D.; Casamayor, A.; Bertone, P.; Lan, N.; Jansen, R.; Bidlingmaier, S.; Houfek, T. Global analysis of protein activities using proteome chips. Science 2001, 293, 2101–2105. [Google Scholar] [CrossRef]
- Shoemaker, B.A.; Panchenko, A.R. Deciphering protein–protein interactions Part I. Experimental techniques and databases. PLoS Comput. Biol. 2007, 3, e42. [Google Scholar] [CrossRef]
- Chepelev, N.; Chepelev, L.; Alamgir, M.; Golshani, A. Large-scale protein–protein interaction detection approaches: Past, present and future. Biotechnol. Biotechnol. Equip. 2008, 22, 513. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Luo, X.; Zhu, W.; Yu, K.; Chen, K.; Li, Y.; Jiang, H. Predicting protein–protein interactions based only on sequences information. Proc. Natl. Acad. Sci. USA 2007, 104, 4337–4341. [Google Scholar]
- Guo, Y.; Yu, L.; Wen, Z.; Li, M. Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acids Res. 2008, 36, 3025–3030. [Google Scholar] [CrossRef]
- Najafabadi, H.S.; Salavati, R. Sequence-based prediction of protein–protein interactions by means of codon usage. Genome Biol. 2008, 9, R87. [Google Scholar] [CrossRef]
- Zhang, Y.-N.; Pan, X.-Y.; Huang, Y.; Shen, H.-B. Adaptive compressive learning for prediction of protein–protein interactions from primary sequence. J. Theor. Biol. 2011, 283, 44–52. [Google Scholar] [CrossRef]
- Liu, C.H.; Li, K.-C.; Yuan, S. Human protein–protein interaction prediction by a novel sequence-based co-evolution method: Co-evolutionary divergence. Bioinformatics 2013, 29, 92–98. [Google Scholar] [CrossRef]
- You, Z.-H.; Lei, Y.-K.; Zhu, L.; Xia, J.; Wang, B. Prediction of protein–protein interactions from amino acid sequences with ensemble extreme learning machines and principal component analysis. BMC Bioinform. 2013, 14, S10. [Google Scholar]
- Zahiri, J.; Yaghoubi, O.; Mohammad-Noori, M.; Ebrahimpour, R.; Masoudi-Nejad, A. PPIevo: Protein–protein interaction prediction from PSSM based evolutionary information. Genomics 2013, 102, 237–242. [Google Scholar] [CrossRef]
- Shi, M.-G.; Xia, J.-F.; Li, X.-L.; Huang, D.-S. Predicting protein–protein interactions from sequence using correlation coefficient and high-quality interaction dataset. Amino Acids 2010, 38, 891–899. [Google Scholar] [CrossRef]
- Yang, L.; Xia, J.-F.; Gui, J. Prediction of protein–protein interactions from protein sequence using local descriptors. Protein Pept. Lett. 2010, 17, 1085–1090. [Google Scholar] [CrossRef]
- Bock, J.R.; Gough, D.A. Predicting protein–protein interactions from primary structure. Bioinformatics 2001, 17, 455–460. [Google Scholar] [CrossRef]
- Aloy, P.; Russell, R.B. InterPreTS: Protein interaction prediction through tertiary structure. Bioinformatics 2003, 19, 161–162. [Google Scholar] [CrossRef]
- Zhang, Q.C.; Petrey, D.; Deng, L.; Qiang, L.; Shi, Y.; Thu, C.A.; Bisikirska, B.; Lefebvre, C.; Accili, D.; Hunter, T. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature 2012, 490, 556–560. [Google Scholar] [CrossRef]
- Binny Priya, S.; Saha, S.; Anishetty, R.; Anishetty, S. A matrix based algorithm for protein–protein interaction prediction using domain–domain associations. J. Theor. Biol. 2013, 326, 36–42. [Google Scholar] [CrossRef]
- Planas-Iglesias, J.; Bonet, J.; García-García, J.; Marín-López, M.A.; Feliu, E.; Oliva, B. Understanding protein–protein interactions using local structural features. J. Mol. Biol. 2013, 425, 1210–1224. [Google Scholar] [CrossRef]
- Ben-Hur, A.; Noble, W.S. Kernel methods for predicting protein–protein interactions. Bioinformatics 2005, 21, i38–i46. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Chang, Z.; DuanMu, H.; Zhang, S.; Li, Z.; Li, Z.; Yu, L.; Li, X. Prediction of human protein–protein interaction by a mixed Bayesian model and its application to exploring underlying cancer-related pathway crosstalk. J. R. Soc. Interface 2011, 8, 555–567. [Google Scholar] [CrossRef]
- Saha, I.; Zubek, J.; Klingstrom, T.; Forsberg, S.; Wikander, J.; Kierczak, M.; Maulik, U.; Plewczynski, D. Ensemble learning prediction of protein–protein interactions using proteins functional annotations. Mol. BioSyst. 2014, 10, 820–830. [Google Scholar] [CrossRef]
- Yang, L.; Tang, X. Protein–protein interactions prediction based on iterative clique extension with gene ontology filtering. Sci. World J. 2014, 2014, 523634. [Google Scholar]
- Souiai, O.; Guerfali, F.; Miled, S.B.; Brun, C.; Benkahla, A. In silico prediction of protein–protein interactions in human macrophages. BMC Res. Notes 2014, 7, 157. [Google Scholar] [CrossRef]
- Sokal, R.R.; Thomson, B.A. Population structure inferred by local spatial autocorrelation: An example from an Amerindian tribal population. Am. J. Phys. Anthropol. 2006, 129, 121–131. [Google Scholar] [CrossRef]
- Xia, J.-F.; Han, K.; Huang, D.-S. Sequence-based prediction of protein–protein interactions by means of rotation forest and autocorrelation descriptor. Protein Pept. Lett. 2010, 17, 137–145. [Google Scholar] [CrossRef]
- Feng, Z.-P.; Zhang, C.-T. Prediction of membrane protein types based on the hydrophobic index of amino acids. J. Protein Chem. 2000, 19, 269–275. [Google Scholar] [CrossRef]
- Lo, S.L.; Cai, C.Z.; Chen, Y.Z.; Chung, M. Effect of training datasets on support vector machine prediction of protein–protein interactions. Proteomics 2005, 5, 876–884. [Google Scholar] [CrossRef]
- Chen, X.-W.; Liu, M. Prediction of protein–protein interactions using random decision forest framework. Bioinformatics 2005, 21, 4394–4400. [Google Scholar] [CrossRef]
- Jansen, R.; Yu, H.; Greenbaum, D.; Kluger, Y.; Krogan, N.J.; Chung, S.; Emili, A.; Snyder, M.; Greenblatt, J.F.; Gerstein, M. A Bayesian networks approach for predicting protein–protein interactions from genomic data. Science 2003, 302, 449–453. [Google Scholar] [CrossRef]
- Chowdhary, R.; Zhang, J.; Liu, J.S. Bayesian inference of protein–protein interactions from biological literature. Bioinformatics 2009, 25, 1536–1542. [Google Scholar] [CrossRef]
- Keedwell, E.; Narayanan, A. Discovering gene networks with a neural-genetic hybrid. Comput. Biol. Bioinform. 2005, 2, 231–242. [Google Scholar] [CrossRef]
- Hayashida, M.; Kamada, M.; Song, J.; Akutsu, T. Conditional random field approach to prediction of protein–protein interactions using domain information. BMC Syst. Biol. 2011, 5, S8. [Google Scholar] [CrossRef] [Green Version]
- Li, M.-H.; Lin, L.; Wang, X.-L.; Liu, T. Protein–protein interaction site prediction based on conditional random fields. Bioinformatics 2007, 23, 597–604. [Google Scholar] [CrossRef]
- Zweig, M.H.; Campbell, G. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar]
- Li, B.-Q.; Feng, K.-Y.; Chen, L.; Huang, T.; Cai, Y.-D. Prediction of protein–protein interaction sites by random forest algorithm with mRMR and IFS. PLoS One 2012, 7, e43927. [Google Scholar]
- Atchley, W.R.; Zhao, J.; Fernandes, A.D.; Drüke, T. Solving the protein sequence metric problem. Proc. Natl. Acad. Sci. USA 2005, 102, 6395–6400. [Google Scholar] [CrossRef]
- Kawashima, S.; Kanehisa, M. AAindex: Amino acid index database. Nucleic Acids Res. 2000, 28, 374–374. [Google Scholar] [CrossRef]
- Wang, J.T.-L.; Ma, Q.; Shasha, D.; Wu, C.H. New techniques for extracting features from protein sequences. IBM Syst. J. 2001, 40, 426–441. [Google Scholar]
- Solovyev, V.V.; Makarova, K.S. A novel method of protein sequence classification based on oligopeptide frequency analysis and its application to search for functional sites and to domain localization. Comput. Appl. Biosci. 1993, 9, 17–24. [Google Scholar]
- Horne, D. Prediction of protein helix content from an autocorrelation analysis of sequence hydrophobicities. Biopolymers 1988, 27, 451–477. [Google Scholar] [CrossRef]
- Li, Z.; Lin, H.; Han, L.; Jiang, L.; Chen, X.; Chen, Y. PROFEAT: A web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res. 2006, 34, W32. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newslett. 2009, 11, 10–18. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Du, X.; Cheng, J.; Zheng, T.; Duan, Z.; Qian, F. A Novel Feature Extraction Scheme with Ensemble Coding for Protein–Protein Interaction Prediction. Int. J. Mol. Sci. 2014, 15, 12731-12749. https://doi.org/10.3390/ijms150712731
Du X, Cheng J, Zheng T, Duan Z, Qian F. A Novel Feature Extraction Scheme with Ensemble Coding for Protein–Protein Interaction Prediction. International Journal of Molecular Sciences. 2014; 15(7):12731-12749. https://doi.org/10.3390/ijms150712731
Chicago/Turabian StyleDu, Xiuquan, Jiaxing Cheng, Tingting Zheng, Zheng Duan, and Fulan Qian. 2014. "A Novel Feature Extraction Scheme with Ensemble Coding for Protein–Protein Interaction Prediction" International Journal of Molecular Sciences 15, no. 7: 12731-12749. https://doi.org/10.3390/ijms150712731




