Proteomic Analysis of Etiolated Juvenile Tetraploid Robinia pseudoacacia Branches during Different Cutting Periods
Abstract
:1. Introduction
2. Results
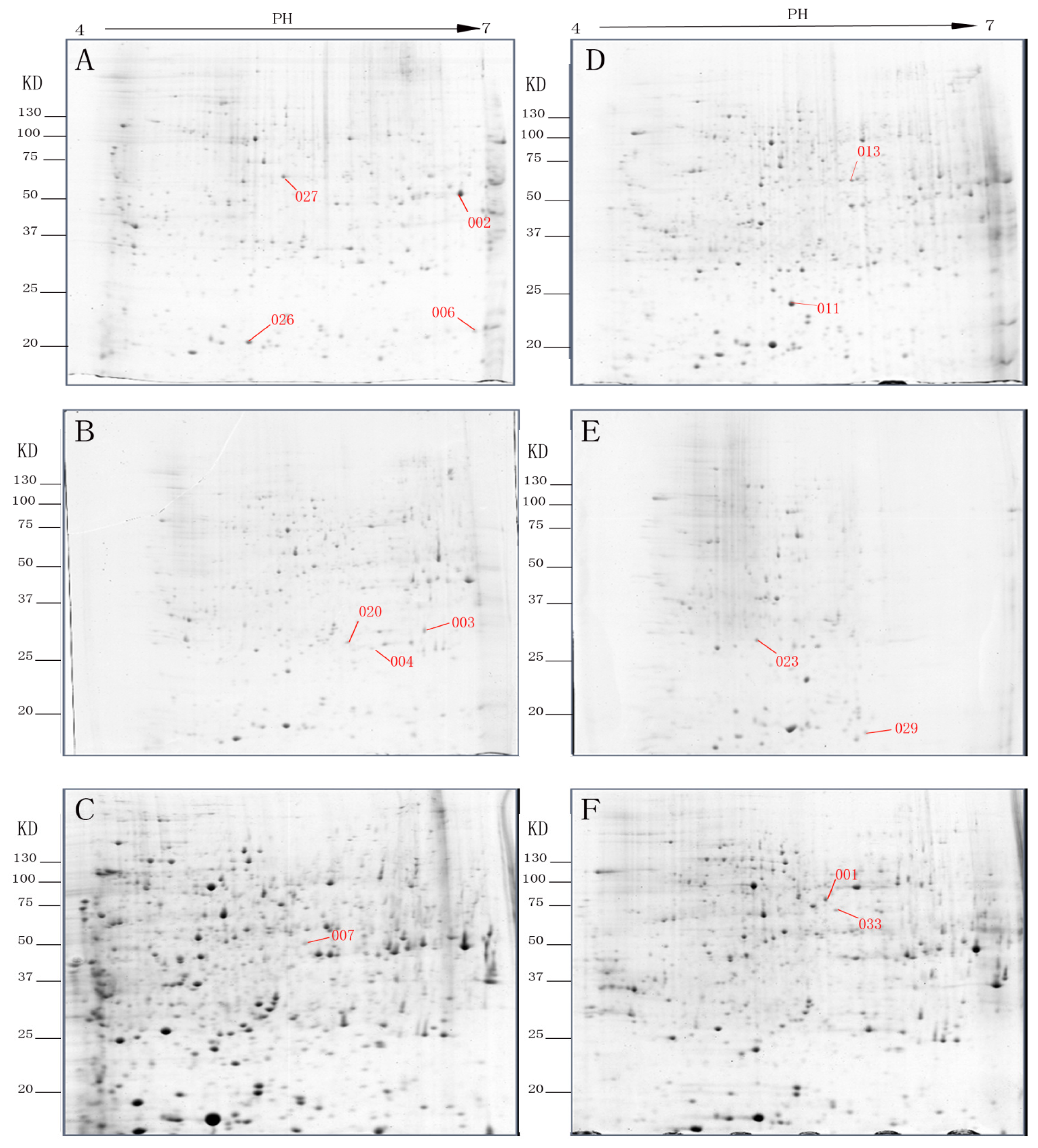
2.1. Comparative Protein Profiles of Non-Etiolated and Etiolated Juvenile Branch Cuttings during Different Rooting Periods
2.2. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight/Time-of-Flight Mass Spectra (MALDI-TOF/TOF-MS) Identification of Etiolation-Responsive Proteins
3. Discussion
3.1. Cell Wall Metabolism and Remodeling
3.2. Proteins Involved in Carbohydrate Metabolism
3.3. Proteins Involved in Metabolism
3.4. Transcription, Translation and Signal Transduction
3.5. Protein Metabolism
4. Experimental Section
4.1. Chemicals and Reagents
4.2. Plant Materials
4.3. Protein Extraction
4.4. Two-Dimensional Electrophoresis (2-DE)
4.5. Image Acquisition and Data Analysis
4.6. Trypsin Digestion
4.7. Protein Identification by MALDI-TOF/TOF-MS
5. Conclusions
Acknowledgments
Conflicts of Interest
- Author ContributionsData analysis and paper writing: Nan Lu. Protein identification: Zhaohe Xu. Two-dimensional electrophoresis: Bingnan Meng. Protein extraction and Sample preparation: Yuhan Sun. Plant materials preparation: Jiangtao Zhang, Shaoming Wang and Yun Li.
References
- Li, Y.; Jiang, J.Z. Research progress of feed tetraploid black locust. Pratacult. Sci 2006, 23, 41–46. [Google Scholar]
- Zhang, G.J.; Li, Y.; He, C.C. Nutrition and growth of leaves at different leaf ages in tetraploidRobinia pseudoacacia. Sci. Silvae Sin 2009, 45, 61–67. [Google Scholar]
- Dong, Z.; Yue, S.M.; Sui, Z.Y. Tetraploid black locust breeding by using root nutrition bags. J. Hebei For. Sci. Technol 2008, 3, 60. [Google Scholar]
- Shang, Z.H. Research on the rapid propagation technology of tetraploidRobinia pseudoacacia. J. Anhui Agric. Sci 2008, 36, 2315–2316. [Google Scholar]
- Meng, B.N.; Peng, Z.D.; Zhang, Z.L.; Xu, H.M.; Li, Y. Research on cuttage of tetraploid black locust (Robinia pseudoacacia L.) hardwood treated by low temperature sand storage and plant growth regulator. Heilongjiang Agric. Sci 2010, 8, 85–88. [Google Scholar]
- Yang, X.F.; Cao, B.H.; Li, S.B.; Ren, Y.H. Studies on the hard stem cutting propagation technique of Robinia pseudoacacia L. Shandong For. Sci. Technol 2007, 2, 50–51. [Google Scholar]
- Frolich, E.F.; Platt, R.G. Use of the etiolation technique in rooting avocado cuttings. Calif. Avocado Soc 1971, 55, 97–109. [Google Scholar]
- Hansen, O.B.; Potter, J.R. Rooting of apple, rhododendron, and mountain laurel cuttings from stock plants etiolated under two temperatures. HortScience 1997, 32, 304–306. [Google Scholar]
- Richards, M.R.; Rupp, L.A. Etiolation improves rooting of bigtooth maple cuttings. HortTechnology 2012, 22, 305–310. [Google Scholar]
- Li, J.H. The Principle and Application of Cutting; Shanghai Science and Technology Press: Shanghai, China, 1987; pp. 10–12. [Google Scholar]
- Lu, N.; Meng, B.N.; Sun, Y.H.; Li, Y.; Wang, S.M.; Guo, Z.M.; Wang, Q.L.; Xu, H.M. Dynamic of physiology and biochemistry during etiolated shoots cutting of tetraploidRobinia pseudoacacia. J. Northeast. For. Univ 2013, 11, 5–9. [Google Scholar]
- Konishi, H.; Yamane, H.; Maeshima, M.; Komatsu, S. Characterization of fructose-bisphosphate aldolase regulatedby gibberellin in roots of rice seedling. Plant Mol. Biol 2004, 56, 839–848. [Google Scholar]
- Sorin, C.; Negroni, L.; Balliau, T.; Corti, H.; Jacquemot, M.P.; Davanture, M.; Sandberg, G.; Zivy, M.; Bellini, C. Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol 2006, 140, 349–364. [Google Scholar]
- Wang, W.; Scali, M.; Vignani, R.; Spadafora, A.; Sensi, E.; Mazzuca, S.; Cresti, M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 2003, 24, 2369–2375. [Google Scholar]
- Ringli, C.; Keller, B.; Ryser, U. Glycine-rich proteins as structural components of plant cell walls. Cell. Mol. Life Sci 2001, 58, 1430–1441. [Google Scholar]
- Sagi, G.; Katz, A.; Guenoune-Gelba, D.; Epel, B.L. Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell 2005, 17, 1788–1800. [Google Scholar]
- Silk, W.K. Steady form from changing cells. Int. J. Plant Sci 1992, 153, S49–S58. [Google Scholar]
- Beemster, G.T.; Baskin, T.I. Analysis of cell division and elongation underlying the developmental acceleration of root growth inArabidopsis thaliana. Am. Soc. Plant Biol 1998, 116, 1515–1526. [Google Scholar]
- Beemster, G.T.; Baskin, T.I. Stunted plant 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root ofArabidopsis. Plant Physiol 2000, 12, 1718–1727. [Google Scholar]
- Showalter, A.M. Structure and function of plant cell wall proteins. Plant Cell 1993, 5, 9–23. [Google Scholar]
- Darvill, A.; McNeil, M.; Albersheim, P. The primary cell walls of flowering plants. In The Biochemistry of Plants; Academic Press: New York, NY, USA, 1980; Volume 9, p. 91. [Google Scholar]
- Hayashi, T. Xyloglucans in the primary cell wall. Ann. Rev. Plant Biol 1989, 40, 139–168. [Google Scholar]
- Brummell, D.A.; Camirand, A.; Maclachlan, G. Differential distribution of xyloglucan glycosyl transferases in pea Golgi dictyosome and secretory vesicles. J. Cell Sci 1990, 96, 705–710. [Google Scholar]
- Driouich, A.; Faye, L.; Staehelin, L.A. The plant Golgi apparatus: A factory for complex polysaccharides and glycoproteins. Trends Biochem. Sci 1993, 18, 210–214. [Google Scholar]
- Staehelin, L.; Moore, I. The plant Golgi apparatus structure, functional organization and trafficking mechanisms. Annu. Rev. Plant Physiol 1995, 46, 261–288. [Google Scholar]
- Dhugga, K.S.; Tiwari, S.C.; Ray, P.M. A reversibly gly-cosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: Purification, gene cloning, and trans-Golgi localization. Proc. Natl. Acad. Sci. USA 1997, 94, 7679–7684. [Google Scholar]
- Sherrier, D.J.; VandenBosch, K.A. Secretion of cell wall polysaccharides in Vicia root hairs. Plant J 1994, 5, 185–195. [Google Scholar]
- Ludwig-Müller, J.; Vertocnik, A.; Town, C.D. Analysis of indole-3-butyric acid-induced adventitious root formation on Arabidopsis stem segments. J. Exp. Bot 2005, 56, 2095–2105. [Google Scholar]
- Brinker, M.; Zyl, L.V.; Liu, W.B.; Craig, D.; Sederoff, R.R.; Clapham, D.H.; Arnold, S.V. Microarray analyses of gene expression during adventitious root development inPinus contorta. Plant Physiol 2004, 135, 1526–1539. [Google Scholar]
- Qin, Q.P.; Zhang, S.L.; Chen, J.W.; Wu, J.Y.; Chen, K.S. The relationship of fructokinase and sugar accumulation during fruit development in satsuma mandarin. J. Plant Physiol. Mol. Biol 2004, 30, 435–440. [Google Scholar]
- Pego, J.V.; Smeekens, S. Plant fructokinases: A sweet family get-together. Trends Plant Sci 2000, 5, 531–536. [Google Scholar]
- Williams, L.E.; Lemoine, R.; Sauer, N. Sugar transporters in higher plants a diversity of roles and complex regulation. Trends Plant Sci 2000, 5, 283–290. [Google Scholar]
- Fumio, T.; Kumi, S.N.; Kazutaka, K.; Mitsuo, S.; Hitoshi, S. Sugar-induced adventitious roots in Arabidopsis seedlings. J. Plant Res 2003, 116, 83–91. [Google Scholar]
- Forsthoefel, N.R.; Cushman, M.; Cushman, J.C. Posttranscriptional and posttranslational control of enolase expression in the facultative Crassulacean acid metabolism plant Mesembryanthemum crystallinum L. Plant Physiol 1995, 108, 1185–1195. [Google Scholar]
- Wasaki, J.; Yonetan, R.; Kuroda, S. Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ 2003, 26, 1515–1523. [Google Scholar]
- Han, H.; Zhang, S.G.; Sun, X.M. A review on the molecular mechanism of plants rooting modulated by auxin. Afr. J. Biotechnol 2009, 8, 348–353. [Google Scholar]
- Ghannoum, O.; Evans, J.R.; Chow, W.S.; Andrews, T.J.; Conroy, J.P.; Caemmerer, S.V. Faster Rubisco is the key to superiornitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiol 2005, 137, 638–650. [Google Scholar]
- Medrano, H.; Parry, M.A.J.; Socias, X.; Lawlor, D.W. Long term water stress inactivates RuBisCO in submediterranean clover. Ann. Appl. Biol 1997, 131, 491–501. [Google Scholar]
- Wei, J.H.; Song, Y.R. Recent advances in study of lignin biosynthesis and manipulation. Acta Bot. Sin 2001, 43, 771–779. [Google Scholar]
- Kajita, S.; Katayama, Y.; Omori, S. Alterations in the biosynthesis of lignin in transgenic plants with chimeric genes for 4-coumarate: Coenzyme A ligase. Plant Cell Physiol 1996, 37, 957–965. [Google Scholar]
- Kajita, S.; Hishiyama, S.; Tomimura, Y.; Omori, S. Structural characterization of modified lignin in transgenic tobacco plants in which the activity of 4-coumarate: Coenzyme A ligase is depressed. Plant Physiol 1997, 11, 871–879. [Google Scholar]
- Lee, D.; Meyer, K.; Chapple, C.; Douglas, C.J. Antisense suppression of 4-coumarate: Coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell Online 1997, 9, 1985–1998. [Google Scholar]
- Cho, H.Y.; Chang, C.Y.; Huang, L.C.; Tsai, J.B.; Liu, Z.H. Indole-3-butyric acid suppresses the activity of peroxidase while inducing adventitious roots inCinnamomum kanehirae. Bot. Stud 2011, 52, 153–160. [Google Scholar]
- Steele, M.I.; Lorenz, D.; Hatter, K.; Park, A.; Sokatch, J.R. Characterization of the mmsAB operon of Pseudomonas aeruginosa PAO encoding ethylmalonate-semialdehyde dehydrogenase and 3-hydroxyisobutyrate dehydrogenase. J. Biol. Chem 1992, 267, 13585–13592. [Google Scholar]
- Kirch, H.H.; Bartels, D.; Wei, Y.; Schnable, P.S.; Wood, A.J. The ALDH gene superfamily ofArabidopsis. Trends Plant Sci 2004, 9, 371–377. [Google Scholar]
- Kirch, H.H.; Nair, A.; Bartels, D. Novel ABA-and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantagineum andArabidopsis thaliana. Plant J 2001, 28, 555–567. [Google Scholar]
- Gao, C.; Han, B. Evolutionary and expression study of the aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene 2009, 431, 86–94. [Google Scholar]
- Chen, X.; Zeng, Q.; Wood, A.J. The stress-responsive Tortula ruralis gene ALDH21A1 describes a novel eukaryotic aldehyde dehydrogenase protein family. J. Plant Physiol 2002, 159, 677–684. [Google Scholar]
- Vogel, J.; Hubsehman, T.; Borner, T.; Hess, W.R. Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: Support for MatK as an essential splice factor. J. Mol. Biol 1997, 270, 179–187. [Google Scholar]
- Jia, J.; Zhang, L.G. mRNA differential display and Est sequence analysis of aborted bud and normal bud in Radishi (Raphanus sativus). J. Nucl. Agric. Sci 2008, 22, 426–431. [Google Scholar]
- Westermann, P.; Nygard, O. The spatial arrangement of the complex between eukaryotic initiation factor eIF-3 and 40S ribosomal sub-unit. Biochim. Biophys. Acta 1983, 741, 103–108. [Google Scholar]
- Cheng, J.; Li, K.; Lu, Y.Y.; Wang, L.; Liu, Y. Bioinformatics analysis of human hepatitis C virus core protein-binding protein 6 gene and protein. World Chin. J. Digestol 2003, 11, 378–384. [Google Scholar]
- Chen, X.; Li, W.Z.; Shao, Y.; Zeng, Q.C. Progress in RNA-binding proteins in animals and plants. Biotechnol. Bull 2007, 3, 9–15. [Google Scholar]
- Walter, P.; Lingappa, V.R. Mechanism of protein translocation across the endoplasmic reticulum membrane. Ann. Rev. Cell Biol 1986, 2, 499–516. [Google Scholar]
- Wolin, S.L.; Waiter, P. Signal recognition particle mediates a transient elongation arrest of preprolaetin in reticulocyte lysate. J. Cell Biol 1989, 109, 2617–2622. [Google Scholar]
- Meyer, E.I.; Krause, E.; Dobberstein, B. Secretory protein translocation across membranes: The role of the “docking protein”. Nature 1982, 297, 647–650. [Google Scholar]
- Rǒmisch, K.; Webb, J.; Lingelbach, K.; Gausepohl, H.; Dobberstein, B. The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J. Cell Biol 1990, 111, 1793–1802. [Google Scholar]
- Li, Q.B.; Haskell, D.W.; Guy, C.L. Coordinate and noncoordinate expression of the stress 70 family and other molecular chaperones at high and low temperature in spinach and tomato. Plant Mol. Biol 1999, 39, 21–34. [Google Scholar]
- Collins, J.F.; Pawloski, D.C.; Davis, M.G.; Ball, N.; Dorn, G.W.; Walsh, R.A. The role of the cytoskeleton in left ventricular pressure overload hypertrophy and failure. J. Mol. Cell. Cardiol 1996, 28, 1435–1443. [Google Scholar]
- Reed, R.C.; Brady, S.R.; Muday, G.K. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 1998, 118, 1369–1378. [Google Scholar]
| Protein ID | Accession No. | Description | Score | SC (%) | Theoretical Mr (kDa)/pI | Observed Mr (kDa)/pI |
|---|---|---|---|---|---|---|
| 001 | gi|116061758 | Putative RNA-binding protein | 92 | 28 | 72.4/8.51 | 78.3/5.26 |
| 002 | gi|168009419 | Methylmalonate-semialdehyde dehydrogenase (MM-ALDH) | 74 | 17 | 61.5/8.51 | 57.1/6.33 |
| 003 | gi|108864006 | Signal recognition particle 54 kDa protein, chloroplast precursor, putative, expressed [Oryza sativa (japonica cultivar-group)] | 80 | 19 | 53.3/9.37 | 38.8/6.14 |
| 004 | gi|60650116 | Actin [Pyrus communis] | 106 | 28 | 38.4/5.47 | 28.6/5.91 |
| 005 | gi|242080859 | Hypothetical protein SORBIDRAFT_07g005770 [Sorghum bicolor] | 82 | 21 | 71.8/6.17 | 65.8/5.43 |
| 006 | gi|121761863 | Ribosomal protein S4 [Plagiomnium cf. tezukae Wyatt 1808] | 83 | 12 | 23.5/10.39 | 23.88/6.61 |
| 007 | gi|42521309 | Enolase [Glycine max] | 87 | 15 | 47.7/5.31 | 52.2/5.05 |
| 008 | gi|152143640 | Chloroplast photosynthetic water oxidation complex 33 kDa subunit precursor [Morus nigra] | 78 | 16 | 28.2/5.48 | 35.2/5.84 |
| 009 | gi|242080859 | Hypothetical protein SORBIDRAFT_07g005770 [Sorghum bicolor] | 82 | 21 | 71.8/6.17 | 75.4/5.88 |
| 010 | gi|270306046 | Unnamed protein product [Vitis vinifera] | 98 | 16 | 43.2/9.36 | 50.3/5.43 |
| 011 | gi|67079128 | Ribulose-1,5-bisphosphatecarboxylase/oxygenase large subunit [Chasmanthium latifolium] | 90 | 14 | 25.2/5.82 | 24.5/5.52 |
| 012 | gi|255559120 | Cytosolic purine 5-nucleotidase, putative [Ricinus communis] | 82 | 12 | 62.7/6.67 | 72.1/6.22 |
| 013 | gi|166156335 | Maturase K [Protea neriifolia] | 83 | 17 | 60.0/9.51 | 64.2/5.83 |
| 014 | gi|147814811 | Hypothetical protein [Vitis vinifera] | 104 | 25 | 78.8/6.23 | 85.7/6.54 |
| 015 | gi|242081717 | Hypothetical protein SORBIDRAFT_07g022905 [Sorghum bicolor] | 73 | 8 | 21.0/5.73 | 19.2/6.47 |
| 016 | gi|168044879 | Predicted protein [Physcomitrella patens subsp. patens] | 74 | 9 | 38.8/9.45 | 40.8/5.43 |
| 017 | gi|13928452 | 14-3-3 Protein [Vigna angularis] | 108 | 21 | 29.2/4.66 | 35.2/5.21 |
| 018 | gi|255559120 | Cytosolic purine 5-nucleotidase, putative [Ricinus communis] | 82 | 12 | 62.7/6.67 | 72.6/6.08 |
| 019 | gi|224141801 | Predicted protein [Populus trichocarpa] | 84 | 18 | 60.9/7.04 | 68.2/5.92 |
| 020 | gi|224055984 | Actin 1 [Populus trichocarpa] | 130 | 26 | 41.7/5.31 | 33.7/5.82 |
| 021 | gi|225448323 | Predicted: hypothetical protein [Vitis vinifera] | 126 | 28 | 41.6/5.31 | 35.2/6.05 |
| 022 | gi|125563066 | Hypothetical protein OsI_30711 [Oryza sativa Indica Group] | 78 | 25 | 88.7/9.13 | 85.5/5.67 |
| 023 | gi|162463414 | Golgi associated protein se-wap4 [Zea mays] | 88 | 18 | 41.2/5.75 | 32.8/5.30 |
| 024 | gi|255554359 | Conserved hypothetical protein [Ricinus communis] | 74 | 14 | 60.7/6.12 | 68.4/5.76 |
| 025 | gi|225437076 | Predicted: hypothetical protein isoform [Vitis vinifera] | 82 | 16 | 63.9/7.18 | 71.4/6.37 |
| 026 | gi|194466127 | Fructokinase [Arachis hypogaea] | 76 | 11 | 20.1/5.07 | 20.6/5.27 |
| 027 | gi|425194 | Heat shock protein [Spinacia oleracea] | 112 | 25 | 70.8/5.15 | 68.6/5.50 |
| 028 | gi|212276328 | Hypothetical protein LOC100191878 [Zea mays] | 82 | 20 | 59.5/9.72 | 55.3/6.18 |
| 029 | gi|224174082 | 4-Coumarate-coa ligase [Populus trichocarpa] | 86 | 14 | 16.6/9.03 | 12.3/5.78 |
| 030 | gi|255618262 | Conserved hypothetical protein [Ricinus communis] | 82 | 12 | 20.3/11.86 | 22.3/6.16 |
| 031 | gi|116055419 | Unnamed protein product [Ostreococcus tauri] | 89 | 22 | 52.5/9.06 | 50.7/5.96 |
| 032 | gi|115486767 | Os11g0701800 | 87 | 11 | 33.9/9.33 | 35.1/6.21 |
| 033 | gi|79325139 | Glycine-rich protein [Arabidopsis thaliana] | 77 | 16 | 60.5/5.28 | 72.2/5.33 |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lu, N.; Xu, Z.; Meng, B.; Sun, Y.; Zhang, J.; Wang, S.; Li, Y. Proteomic Analysis of Etiolated Juvenile Tetraploid Robinia pseudoacacia Branches during Different Cutting Periods. Int. J. Mol. Sci. 2014, 15, 6674-6688. https://doi.org/10.3390/ijms15046674
Lu N, Xu Z, Meng B, Sun Y, Zhang J, Wang S, Li Y. Proteomic Analysis of Etiolated Juvenile Tetraploid Robinia pseudoacacia Branches during Different Cutting Periods. International Journal of Molecular Sciences. 2014; 15(4):6674-6688. https://doi.org/10.3390/ijms15046674
Chicago/Turabian StyleLu, Nan, Zhaohe Xu, Bingnan Meng, Yuhan Sun, Jiangtao Zhang, Shaoming Wang, and Yun Li. 2014. "Proteomic Analysis of Etiolated Juvenile Tetraploid Robinia pseudoacacia Branches during Different Cutting Periods" International Journal of Molecular Sciences 15, no. 4: 6674-6688. https://doi.org/10.3390/ijms15046674




