Emerging Regulation and Function of Betatrophin
Abstract
:1. Introduction
2. The Betatrophin Gene
2.1. Identification of Betatrophin
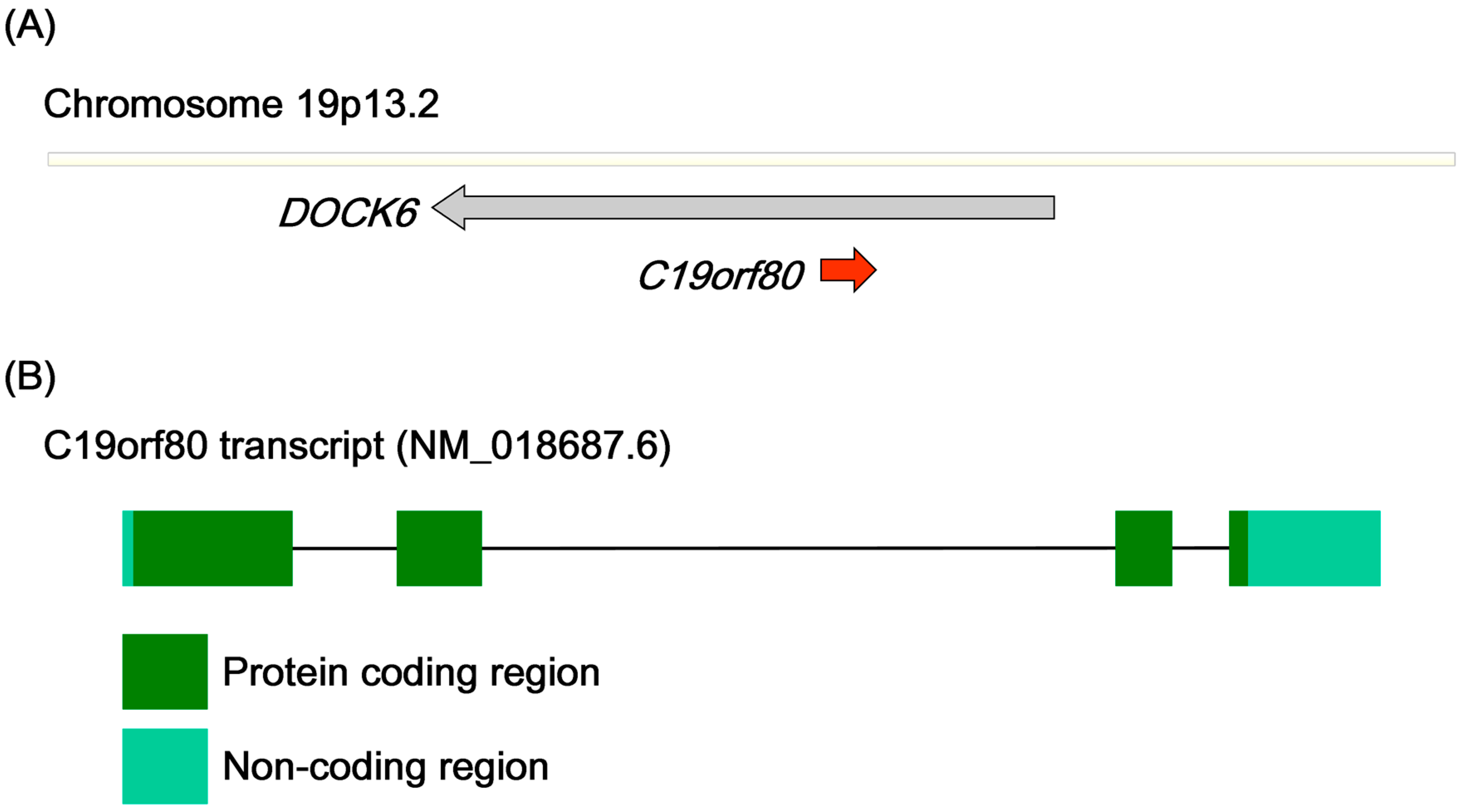

2.2. Gene Structure and Predicted Motifs of Betatrophin
3. Cellular Localization of Betatrophin
3.1. Vesicular Betatrophin
3.2. Secretion of Betatrophin
4. Expression of Betatrophin
| Detection Model | Analysis Factor | Correlation | Detection Method | Reference |
| Age | + | ELISA (EIAAB) | [16] | |
| Obese/overweight | + | ELISA (Phoenix) | [17] | |
| Morbidly obese | NC | ELISA (EIAAB) | [12] | |
| BMI | + | ELISA (Phoenix) | [17] | |
| BMI | − | [17] | ||
| HDL-/LDL-cholesterol | − | [2] | ||
| Type I Diabetes | Type I Diabetes | + | ELISA (EIAAB) | [13] |
| Cholesterol in TID | NC | Sequencing (R59W variant) | [13] | |
| Triacylglycerol in TID | NC | ELISA (EIAAB) | [13] | |
| Type II Diabetes | Type II Diabetes | + | ELISA (EIAAB) ELISA (Phoenix) | [14,16,18] |
| Type II Diabetes | − | ELISA (Cusabio) | [19] | |
| Type II Diabetes | NC | ELISA (EIAAB) | [12] | |
| Glucose in TIID | + | ELISA (Phoenix) | [14] | |
| Glucose in TIID | NC | ELISA (EIAAB) | [12] | |
| Insulin in TIID | + | ELISA (Phoenix) | [14] | |
| Insulin in TIID | NC | ELISA (EIAAB) | [12] | |
| Hemoglobin A1c | + | ELISA (EIAAB) | [16] | |
| BMI in TIID | + | ELISA (Phoenix) | [14] | |
| BMI in TIID | − | ELISA (Cusabio) | [19] | |
| BMI in TIID | NC | ELISA (EIAAB) | [12] | |
| Triacylglycerol in TIID | NC | ELISA (Phoenix) | [14] | |
| Total cholesterol in TIID | + | ELISA (EIAAB) | [12] | |
| Total cholesterol in TIID | NC | ELISA (Phoenix) | [14] | |
| HDL cholesterol in TIID | + | ELISA (Cusabio) | [19] | |
| HDL cholesterol in TIID | NC | ELISA (Phoenix) | [14] | |
| LDL cholesterol in TIID | + | ELISA (EIAAB) | [12] | |
| LDL cholesterol in TIID | NC | ELISA (Phoenix) | [14] | |
| Apolipoprotein B in TIID | + | ELISA (EIAAB) | [12] |
| Regulator | Treatment | Expression/Localization | Model Organism | Reference |
|---|---|---|---|---|
| Positive Regulator | ||||
| Nutrition intake | Caloric intake | Protein in serum | Human | [14] |
| Caloric intake | mRNA | Human adipocytes | [20] | |
| High fat diet | mRNA in liver, BAT and WAT | Mouse | [2,4,9] | |
| Insulin | Insulin | mRNA | Mouse 3T3 and Human adipocytes | [3] |
| Insulin antagonist | S961 | mRNA in liver and WAT | Mouse | [5] |
| Thyroid hormone | Thyroid hormone | mRNA and protein | Human HepG2 cell | [11] |
| Irisin | Humanirisin | mRNA | Mouse 3T3 | [21] |
| Cold stimulation | 4 °C for 4 h | mRNA in BAT | Mouse | [9] |
| Gestation | Gestation | mRNA in liver | Mouse | [5] |
| SREBP1a/SREBP2 | Transgenic mice | mRNA in liver | Mouse | [2] |
| Liver X receptor agonist | T0901317 | mRNA in liver | Mouse | [2] |
| Negative Regulator | ||||
| Fasting | Fasting | mRNA in liver, BAT and WAT | Mouse | [2,4,9] |
| TNFα | TNFα | mRNA | Mouse 3T3 | [3] |
| Lypolysis inducer | db-cAMP, forskolin, Isoproterenol | mRNA | Mouse 3T3 | [3] |
| Analyzed Result | Manipulation of Betatrophin Expression | Model Organism | ||
|---|---|---|---|---|
| Overexpression | Null Mice | Knockdown | ||
| β-Cell proliferation | ↑ [5] | ND | ND | Mouse |
| Insulin production | ↑ [5] | NS | ND | Mouse |
| Blood glucose | ↓ [5] | NS | ND | Mouse |
| TG | Serum TG ↑ [2,4] HepG2 TG ↓ [11] | Serum TG ↓ [3,15] Hepatic TG N.S. [15] | 3T3 Adipocytic TG ↓ [3] HepG2 TG ↑ [11] | Mouse, 3T3 and HepG2 cell |
| ANGPTL3 | Cleavage ↑ [2] ANGPTL3 level ↓ [2] | ANGPTL3 level ↑ [15] | ND | Mouse |
| Autophagy flux | ↑ [11] | ND | ↓ [11] | HepG2 cell |
| Serum LPL activity | ↓ [4] | ↑ [15] | ND | Mouse |
| Mice body weight | ND | ↑ [15] | ND | Mouse |
| Mice fat mass | ND | ↓ [15] | ND | Mouse |
| NEFA | ND | ↓ [15] | ND | Mouse |
| VLDL-TG uptake | ND | In WAT ↓ [15] | ND | Mouse |
4.1. Physiological Expression
4.2. Nutrition-Induced Betatrophin
4.3. Hormonal Regulation of Betatrophin
4.3.1. Insulin
4.3.2. Thyroid Hormone
4.4. Other Factors Modulating Betatrophin Expression
5. Functional Characterization of Betatrophin
5.1. Alterations in Lipid Levels
5.2. Correlation of Betatrophin and ANGPTL3 Functions
5.3. Betatrophin Regulates Autophagic Flux
5.4. Betatrophin Induces Proliferation of Pancreatic Cells
6. Conclusions

Acknowledgments
Author Contributions
Abbreviation
| AFP | α-fetoprotein |
| ANGPTL3 | angiopoietin-like protein 3 |
| ANGPTL8 | angiopoietin-like protein 8 |
| BAT | brown adipose tissue |
| C19orf80 | chromosome 19 open reading frame 80 |
| DOCK6 | dedicator of cytokinesis 6 |
| DOCK7 | dedicator of cytokinesis 7 |
| FAS | fatty acid synthase |
| FNDC5 | fibronectin type III domain-containing 5 |
| HDL | high-density lipoprotein |
| HMGR | 3-hydroxy-3-methylglutaryl-CoA reductase |
| LAMP2 | lysosomal-associated membrane protein 2 |
| LDs | lipid droplets |
| LDLR | low-density lipoprotein receptor |
| MVB | multivesicular body |
| PLIN2 | perilipin2 |
| RIFL | refeeding induced fat and liver |
| T3 | thyroid hormone 3,3',5-triiodo-l-thyronine |
| TG | triglyceride |
| WAT | white adipose tissue |
Conflicts of Interest
References
- Dong, X.Y.; Pang, X.W.; Yu, S.T.; Su, Y.R.; Wang, H.C.; Yin, Y.H.; Wang, Y.D.; Chen, W.F. Identification of genes differentially expressed in human hepatocellular carcinoma by a modified suppression subtractive hybridization method. Int. J. Cancer 2004, 112, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Quagliarini, F.; Wang, Y.; Kozlitina, J.; Grishin, N.V.; Hyde, R.; Boerwinkle, E.; Valenzuela, D.M.; Murphy, A.J.; Cohen, J.C.; Hobbs, H.H. Atypical angiopoietin-like protein that regulates ANGPTL3. Proc. Natl. Acad. Sci. USA 2012, 109, 19751–19756. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Kim, J.Y.; Smas, C.M. Identification of RIFL, a novel adipocyte-enriched insulin target gene with a role in lipid metabolism. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E334–E351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R. Lipasin, a novel nutritionally-regulated liver-enriched factor that regulates serum triglyceride levels. Biochem. Biophys. Res. Commun. 2012, 424, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Yi, P.; Park, J.S.; Melton, D.A. Betatrophin: A hormone that controls pancreatic β-cell proliferation. Cell 2013, 153, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Weissglas-Volkov, D.; Aguilar-Salinas, C.A.; Nikkola, E.; Deere, K.A.; Cruz-Bautista, I.; Arellano-Campos, O.; Munoz-Hernandez, L.L.; Gomez-Munguia, L.; Ordonez-Sanchez, M.L.; Reddy, P.M.; et al. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J. Med. Genet. 2013, 50, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Ma, D.; Xu, M. Nested genes in the human genome. Genomics 2005, 86, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Yao, F.; Abou-Samra, A.B.; Zhang, R. Lipasin, thermoregulated in brown fat, is a novel but atypical member of the angiopoietin-like protein family. Biochem. Biophys. Res. Commun. 2013, 430, 1126–1131. [Google Scholar] [CrossRef] [PubMed]
- Clark, H.F.; Gurney, A.L.; Abaya, E.; Baker, K.; Baldwin, D.; Brush, J.; Chen, J.; Chow, B.; Chui, C.; Crowley, C.; et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: A bioinformatics assessment. Genome Res. 2003, 13, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.H.; Ke, P.Y.; Liao, C.J.; Wu, S.M.; Chi, H.C.; Tsai, C.Y.; Chen, C.Y.; Lin, Y.H.; Lin, K.H. Chromosome 19 open reading frame 80 is upregulated by thyroid hormone and modulates autophagy and lipid metabolism. Autophagy 2014, 10, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Fenzl, A.; Itariu, B.K.; Kosi, L.; Fritzer-Szekeres, M.; Kautzky-Willer, A.; Stulnig, T.M.; Kiefer, F.W. Circulating betatrophin correlates with atherogenic lipid profiles but not with glucose and insulin levels in insulin-resistant individuals. Diabetologia 2014, 57, 1204–1208. [Google Scholar] [CrossRef] [PubMed]
- Espes, D.; Lau, J.; Carlsson, P.O. Increased circulating levels of betatrophin in individuals with long-standing type 1 diabetes. Diabetologia 2014, 57, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Berhane, F.; Fite, A.; Seyoum, B.; Abou-Samra, A.B.; Zhang, R. Elevated circulating lipasin/betatrophin in human type 2 diabetes and obesity. Sci. Rep. 2014, 4, 5013. [Google Scholar] [PubMed]
- Wang, Y.; Quagliarini, F.; Gusarova, V.; Gromada, J.; Valenzuela, D.M.; Cohen, J.C.; Hobbs, H.H. Mice lacking ANGPTL8 (betatrophin) manifest disrupted triglyceride metabolism without impaired glucose homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 16109–16114. [Google Scholar] [CrossRef] [PubMed]
- Espes, D.; Martinell, M.; Carlsson, P.O. Increased circulating betatrophin concentrations in patients with type 2 diabetes. Int. J. Endocrinol. 2014, 2014, 323407. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Abou-Samra, A.B.; Zhang, R. An explanation for recent discrepancies in levels of human circulating betatrophin. Diabetologia 2014, 57, 2232–2234. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Sun, W.; Yu, S.; Hong, X.; Qian, W.; Tang, B.; Wang, D.; Yang, L.; Wang, J.; Mao, C.; et al. Increased circulating levels of betatrophin in newly diagnosed type 2 diabetic patients. Diabetes Care 2014, 37, 2718–2722. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Ambrosi, J.; Pascual, E.; Catalan, V.; Rodriguez, A.; Ramirez, B.; Silva, C.; Gil, M.J.; Salvador, J.; Fruhbeck, G. Circulating betatrophin concentrations are decreased in human obesity and type 2 diabetes. J. Clin. Endocrinol. Metab. 2014. doi:org/10.1210/jc.2014-1568. [Google Scholar]
- Franck, N.; Gummesson, A.; Jernas, M.; Glad, C.; Svensson, P.A.; Guillot, G.; Rudemo, M.; Nystrom, F.H.; Carlsson, L.M.; Olsson, B. Identification of adipocyte genes regulated by caloric intake. J. Clin. Endocrinol. Metab. 2011, 96, E413–E418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, R.; Meng, Y.; Li, S.; Donelan, W.; Zhao, Y.; Qi, L.; Zhang, M.; Wang, X.; Cui, T.; et al. Irisin stimulates browning of white adipocytes through mitogen-activated protein kinase p38 MAP kinase and ERK MAP kinase signaling. Diabetes 2013, 63, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Ojima, H.; Ichikawa, H.; Hirohashi, S.; Kondo, T. Molecular background of α-fetoprotein in liver cancer cells as revealed by global RNA expression analysis. Cancer Sci. 2008, 99, 2402–2409. [Google Scholar] [CrossRef] [PubMed]
- Ammerpohl, O.; Pratschke, J.; Schafmayer, C.; Haake, A.; Faber, W.; von Kampen, O.; Brosch, M.; Sipos, B.; von Schonfels, W.; Balschun, K.; et al. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int. J. Cancer 2011, 130, 1319–1928. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, L.; Brand, C.L.; Hansen, B.F.; Ribel, U.; Shaw, A.C.; Slaaby, R.; Sturis, J. A novel high-affinity peptide antagonist to the insulin receptor. Biochem. Biophys. Res. Commun. 2008, 376, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, L.; Hansen, B.F.; Jensen, P.; Pedersen, T.A.; Vestergaard, K.; Schaffer, L.; Blagoev, B.; Oleksiewicz, M.B.; Kiselyov, V.V.; de Meyts, P. Agonism and antagonism at the insulin receptor. PLoS One 2012, 7, e51972. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Jena, G. S961, an insulin receptor antagonist causes hyperinsulinemia, insulin-resistance and depletion of energy stores in rats. Biochem. Biophys. Res. Commun. 2010, 398, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Karnik, S.K.; Chen, H.; McLean, G.W.; Heit, J.J.; Gu, X.; Zhang, A.Y.; Fontaine, M.; Yen, M.H.; Kim, S.K. Menin controls growth of pancreatic β-cells in pregnant mice and promotes gestational diabetes mellitus. Science 2007, 318, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.R.; Moser, A.H.; Shigenaga, J.K.; Grunfeld, C.; Feingold, K.R. Decreased nuclear hormone receptor expression in the livers of mice in late pregnancy. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E1313–E1320. [Google Scholar] [CrossRef] [PubMed]
- Mazurkiewicz, J.C.; Watts, G.F.; Warburton, F.G.; Slavin, B.M.; Lowy, C.; Koukkou, E. Serum lipids, lipoproteins and apolipoproteins in pregnant non-diabetic patients. J. Clin. Pathol. 1994, 47, 728–731. [Google Scholar] [CrossRef] [PubMed]
- Belo, L.; Caslake, M.; Gaffney, D.; Santos-Silva, A.; Pereira-Leite, L.; Quintanilha, A.; Rebelo, I. Changes in LDL size and HDL concentration in normal and preeclamptic pregnancies. Atherosclerosis 2002, 162, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Sattar, N.; Greer, I.A.; Louden, J.; Lindsay, G.; McConnell, M.; Shepherd, J.; Packard, C.J. Lipoprotein subfraction changes in normal pregnancy: Threshold effect of plasma triglyceride on appearance of small, dense low density lipoprotein. J. Clin. Endocrinol. Metab. 1997, 82, 2483–2491. [Google Scholar] [PubMed]
- Witztum, J.L. Susceptibility of low-density lipoprotein to oxidative modification. Am. J. Med. 1993, 94, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.Y. Multiple mechanisms for regulation of the transcriptional activity of thyroid hormone receptors. Rev. Endocr. Metab. Disord. 2000, 1, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Yen, P.M. Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 2001, 81, 1097–1142. [Google Scholar] [PubMed]
- Hashimoto, K.; Mori, M. Crosstalk of thyroid hormone receptor and liver X receptor in lipid metabolism and beyond. Endocr. J. 2011, 58, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Duntas, L.H. Thyroid disease and lipids. Thyroid 2002, 12, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Mullur, R.; Liu, Y.Y.; Brent, G.A. Thyroid hormone regulation of metabolism. Physiol. Rev. 2014, 94, 355–382. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.J.; Seitz, H.J. Thyroid hormone action on intermediary metabolism. Part II: Lipid metabolism in hypo- and hyperthyroidism. Klin. Wochenschr. 1984, 62, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Blennemann, B.; Leahy, P.; Kim, T.S.; Freake, H.C. Tissue-specific regulation of lipogenic mRNAs by thyroid hormone. Mol. Cell. Endocrinol. 1995, 110, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Matsumoto, S.; Yamada, M.; Satoh, T.; Mori, M. Liver X receptor-α gene expression is positively regulated by thyroid hormone. Endocrinology 2007, 148, 4667–4675. [Google Scholar] [CrossRef] [PubMed]
- Hansson, P.; Valdemarsson, S.; Nilsson-Ehle, P. Experimental hyperthyroidism in man: Effects on plasma lipoproteins, lipoprotein lipase and hepatic lipase. Horm. Metab. Res. 1983, 15, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Ness, G.C.; Pendleton, L.C.; Li, Y.C.; Chiang, J.Y. Effect of thyroid hormone on hepatic cholesterol 7α hydroxylase, LDL receptor, HMG-CoA reductase, farnesyl pyrophosphate synthetase and apolipoprotein A-I mRNA levels in hypophysectomized rats. Biochem. Biophys. Res. Commun. 1990, 172, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Pandak, W.M.; Heuman, D.M.; Redford, K.; Stravitz, R.T.; Chiang, J.Y.; Hylemon, P.B.; Vlahcevic, Z.R. Hormonal regulation of cholesterol 7α-hydroxylase specific activity, mRNA levels, and transcriptional activity in vivo in the rat. J. Lipid Res. 1997, 38, 2483–2491. [Google Scholar] [PubMed]
- Keyes, W.G.; Wilcox, H.G.; Heimberg, M. Formation of the very low density lipoprotein and metabolism of [1–14C]-oleate by perfused livers from rats treated with triiodothyronine or propylthiouracil. Metabolism 1981, 30, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Laker, M.E.; Mayes, P.A. Effect of hyperthyroidism and hypothyroidism on lipid and carbohydrate metabolism of the perfused rat liver. Biochem. J. 1981, 196, 247–255. [Google Scholar] [PubMed]
- Gupta, B.B.; Chakrabarty, P. Effects of thyroidal, gonadal and adrenal hormones on tissue respiration of streaked frog, Rana limnocharis, at low temperature. Indian J. Exp. Biol. 1990, 28, 23–26. [Google Scholar] [PubMed]
- Muller, M.J.; Seitz, H.J. Thyroid hormone action on intermediary metabolism. Part I: Respiration, thermogenesis and carbohydrate metabolism. Klin. Wochenschr. 1984, 62, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.E. The thermogenic effect of thyroid hormone and its clinical implications. Ann. Intern. Med. 2003, 139, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Weirich, R.T.; Schwartz, H.L.; Oppenheimer, J.H. An analysis of the interrelationship of nuclear and plasma triiodothyronine in the sea lamprey, lake trout, and rat: Evolutionary considerations. Endocrinology 1987, 120, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef] [PubMed]
- Bostrom, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostrom, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Sundin, U.; Mills, I.; Fain, J.N. Thyroid-catecholamine interactions in isolated rat brown adipocytes. Metabolism 1984, 33, 1028–1033. [Google Scholar] [CrossRef] [PubMed]
- Oike, Y.; Akao, M.; Kubota, Y.; Suda, T. Angiopoietin-like proteins: Potential new targets for metabolic syndrome therapy. Trends Mol. Med. 2005, 11, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Abou-Samra, A.B. Emerging roles of lipasin as a critical lipid regulator. Biochem. Biophys. Res. Commun. 2013, 432, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Fugier, C.; Tousaint, J.J.; Prieur, X.; Plateroti, M.; Samarut, J.; Delerive, P. The lipoprotein lipase inhibitor ANGPTL3 is negatively regulated by thyroid hormone. J. Biol. Chem. 2006, 281, 11553–11559. [Google Scholar] [CrossRef] [PubMed]
- Chittur, S.V.; Sangster-Guity, N.; McCormick, P.J. Histone deacetylase inhibitors: A new mode for inhibition of cholesterol metabolism. BMC Genomics 2008, 9, 507. [Google Scholar] [CrossRef] [PubMed]
- Lanford, R.E.; Guerra, B.; Lee, H.; Chavez, D.; Brasky, K.M.; Bigger, C.B. Genomic response to interferon-α in chimpanzees: Implications of rapid down-regulation for hepatitis C kinetics. Hepatology 2006, 43, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Sankaran, S.; Dandekar, S. Hepatitis C virus core protein induces expression of genes regulating immune evasion and anti-apoptosis in hepatocytes. Virology 2006, 354, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Sawada, H.; Takami, K.; Asahi, S. A toxicogenomic approach to drug-induced phospholipidosis: Analysis of its induction mechanism and establishment of a novel in vitro screening system. Toxicol. Sci. 2005, 83, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Czaja, M.J. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology 2011, 140, 1895–1908. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Maiuri, M.C.; Markaki, M.; Megalou, E.; Pasparaki, A.; Palikaras, K.; Criollo, A.; Galluzzi, L.; Malik, S.A.; Vitale, I.; et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010, 1, e10. [Google Scholar] [CrossRef] [PubMed]
- Andreo, U.; Guo, L.; Chirieac, D.V.; Tuyama, A.C.; Montenont, E.; Brodsky, J.L.; Fisher, E.A. Insulin-stimulated degradation of apolipoprotein B100: Roles of class II phosphatidylinositol-3-kinase and autophagy. PLoS One 2013, 8, e57590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.; Bassik, M.C.; Moresi, V.; Sun, K.; Wei, Y.; Zou, Z.; An, Z.; Loh, J.; Fisher, J.; Sun, Q.; et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012, 481, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Irrcher, I.; Adhihetty, P.J.; Sheehan, T.; Joseph, A.M.; Hood, D.A. PPARγ coactivator-1α expression during thyroid hormone- and contractile activity-induced mitochondrial adaptations. Am. J. Physiol. Cell Physiol. 2003, 284, C1669–C1677. [Google Scholar] [CrossRef] [PubMed]
- Leone, T.C.; Lehman, J.J.; Finck, B.N.; Schaeffer, P.J.; Wende, A.R.; Boudina, S.; Courtois, M.; Wozniak, D.F.; Sambandam, N.; Bernal-Mizrachi, C.; et al. PGC-1α deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005, 3, e101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Xu, D. Cold stress accentuates pressure overload-induced cardiac hypertrophy and contractile dysfunction: Role of TRPV1/AMPK-mediated autophagy. Biochem. Biophys. Res. Commun. 2013, 442, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.Y.; Chen, S.S. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Investig. 2011, 121, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Morissette, G.; Moreau, E.; C.-Gaudreault, R.; Marceau, F. Massive cell vacuolization induced by organic amines such as procainamide. J. Pharmacol. Exp. Ther. 2004, 310, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Virgin, H.W.; Levine, B. Autophagy genes in immunity. Nat. Immunol. 2009, 10, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.J.; Wang, Z.V.; Battiprolu, P.K.; Jiang, N.; Morales, C.R.; Kong, Y.; Rothermel, B.A.; Gillette, T.G.; Hill, J.A. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc. Natl. Acad. Sci. USA 2011, 108, 4123–4128. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Le Lay, J.; Yu, M.; Naji, A.; Kaestner, K.H. Elevated mouse hepatic betatrophin expression does not increase human β-cell replication in the transplant setting. Diabetes 2014, 63, 1283–1288. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.F. Betatrophin versus bitter-trophin and the elephant in the room: Time for a new normal in β-cell regeneration research. Diabetes 2014, 63, 1198–1199. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, Y.-H.; Yeh, Y.-H.; Chen, W.-J.; Lin, K.-H. Emerging Regulation and Function of Betatrophin. Int. J. Mol. Sci. 2014, 15, 23640-23657. https://doi.org/10.3390/ijms151223640
Tseng Y-H, Yeh Y-H, Chen W-J, Lin K-H. Emerging Regulation and Function of Betatrophin. International Journal of Molecular Sciences. 2014; 15(12):23640-23657. https://doi.org/10.3390/ijms151223640
Chicago/Turabian StyleTseng, Yi-Hsin, Yung-Hsin Yeh, Wei-Jan Chen, and Kwang-Huei Lin. 2014. "Emerging Regulation and Function of Betatrophin" International Journal of Molecular Sciences 15, no. 12: 23640-23657. https://doi.org/10.3390/ijms151223640





