RNA Recognition and Stress Granule Formation by TIA Proteins
Abstract
:1. Introduction
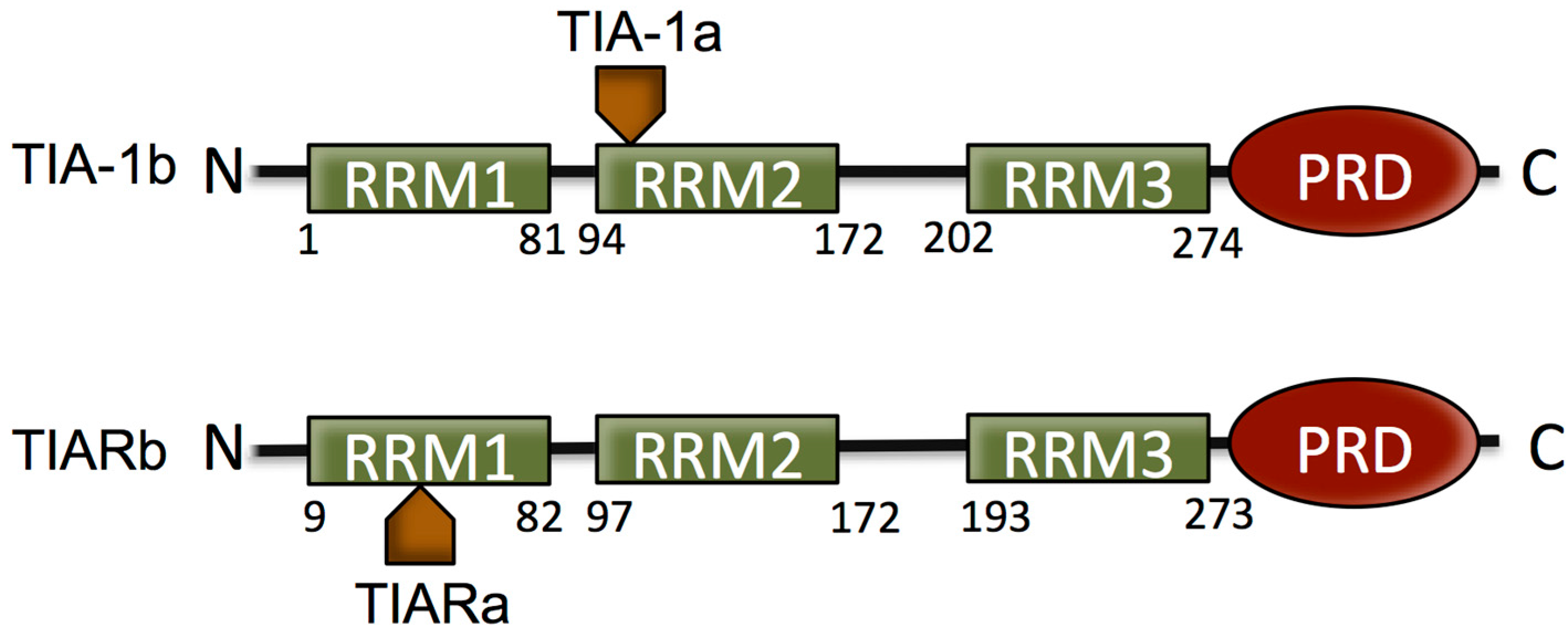
2. The T Cell Restricted Intracellular Antigen (TIA) Proteins
3. Stress Granule Formation in the Cell

4. Specificity of TIA Proteins for Target mRNA
5. Structural Insight into RNA Recognition Motifs of TIA/TIAR

6. TIA Protein Self-Association via the Prion Related Domain
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Buchan, J.R.; Parker, R. Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 2009, 36, 932–941. [Google Scholar]
- Anderson, P.; Kedersha, N. Stress granules. Curr. Biol. 2009, 19, R397–R398. [Google Scholar]
- Scheu, S.; Stetson, D.B.; Reinhardt, R.L.; Leber, J.H.; Mohrs, M.; Locksley, R.M. Activation of the integrated stress response during T helper cell differentiation. Nat. Immunol. 2006, 7, 644–651. [Google Scholar]
- Kim, W.J.; Back, S.H.; Kim, V.; Ryu, I.; Jang, S.K. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 2005, 25, 2450–2462. [Google Scholar]
- Arimoto, K.; Fukuda, H.; Imajoh-Ohmi, S.; Saito, H.; Takekawa, M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008, 10, 1324–1332. [Google Scholar]
- Reineke, L.C.; Lloyd, R.E. Diversion of stress granules and P-bodies during viral infection. Virology 2013, 436, 255–267. [Google Scholar]
- Thedieck, K.; Holzwarth, B.; Prentzell, M.T.; Boehlke, C.; Klasener, K.; Ruf, S.; Sonntag, A.G.; Maerz, L.; Grellscheid, S.N.; Kremmer, E.; et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 2013, 154, 859–874. [Google Scholar]
- Fournier, M.J.; Coudert, L.; Mellaoui, S.; Adjibade, P.; Gareau, C.; Cote, M.F.; Sonenberg, N.; Gaudreault, R.C.; Mazroui, R. Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation. Mol. Cell. Biol. 2013, 33, 2285–2301. [Google Scholar]
- Li, Y.R.; King, O.D.; Shorter, J.; Gitler, A.D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013, 201, 361–372. [Google Scholar]
- Ash, P.E.; Vanderweyde, T.E.; Youmans, K.L.; Apicco, D.J.; Wolozin, B. Pathological stress granules in Alzheimer’s disease. Brain Res. 2014, 1584, 52–58. [Google Scholar]
- Anderson, P.; Kedersha, N. Stress granules: The Tao of RNA triage. Trends Biochem. Sci. 2008, 33, 141–150. [Google Scholar]
- Buchan, J.R. mRNP granules: Assembly, function, and connections with disease. RNA Biol. 2014, 11, 1. [Google Scholar]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar]
- Ohn, T.; Kedersha, N.; Hickman, T.; Tisdale, S.; Anderson, P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 2008, 10, 1224–1231. [Google Scholar]
- Kedersha, N.; Anderson, P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002, 30, 963–969. [Google Scholar]
- Gallouzi, I.E.; Brennan, C.M.; Stenberg, M.G.; Swanson, M.S.; Eversole, A.; Maizels, N.; Steitz, J.A. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 2000, 97, 3073–3078. [Google Scholar]
- Tourriere, H.; Chebli, K.; Zekri, L.; Courselaud, B.; Blanchard, J.M.; Bertrand, E.; Tazi, J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003, 160, 823–831. [Google Scholar]
- Gilks, N.; Kedersha, N.; Ayodele, M.; Shen, L.; Stoecklin, G.; Dember, L.M.; Anderson, P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 2004, 15, 5383–5398. [Google Scholar]
- Goulet, I.; Boisvenue, S.; Mokas, S.; Mazroui, R.; Cote, J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 2008, 17, 3055–3074. [Google Scholar]
- Wolozin, B. Physiological protein aggregation run amuck: Stress granules and the genesis of neurodegenerative disease. Discov. Med. 2014, 17, 47–52. [Google Scholar]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J. Cell Biol. 1999, 147, 1431–1442. [Google Scholar]
- Anderson, P.; Kedersha, N. Visibly stressed: The role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 2002, 7, 213–221. [Google Scholar]
- Piecyk, M.; Wax, S.; Beck, A.R.; Kedersha, N.; Gupta, M.; Maritim, B.; Chen, S.; Gueydan, C.; Kruys, V.; Streuli, M.; et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000, 19, 4154–4163. [Google Scholar]
- Le Guiner, C.; Lejeune, F.; Galiana, D.; Kister, L.; Breathnach, R.; Stevenin, J.; del Gatto-Konczak, F. TIA-1 and TIAR activate splicing of alternative exons with weak 5' splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 2001, 276, 40638–40646. [Google Scholar]
- Beck, A.R.; Medley, Q.G.; O’Brien, S.; Anderson, P.; Streuli, M. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 1996, 24, 3829–3835. [Google Scholar]
- Izquierdo, J.M.; Valcarcel, J. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J. Biol. Chem. 2007, 282, 19410–19417. [Google Scholar]
- Lopez de Silanes, I.; Galban, S.; Martindale, J.L.; Yang, X.; Mazan-Mamczarz, K.; Indig, F.E.; Falco, G.; Zhan, M.; Gorospe, M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell Biol. 2005, 25, 9520–9531. [Google Scholar]
- Srivastava, S.P.; Kumar, K.U.; Kaufman, R.J. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 1998, 273, 2416–2423. [Google Scholar]
- Kedersha, N.; Chen, S.; Gilks, N.; Li, W.; Miller, I.J.; Stahl, J.; Anderson, P. Evidence that ternary complex (eIF2-GTP-tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 2002, 13, 195–210. [Google Scholar]
- Damgaard, C.K.; Lykke-Andersen, J. Translational coregulation of 5' TOP mRNAs by TIA-1 and TIAR. Genes Dev. 25, 2057–2068.
- Mazroui, R.; di Marco, S.; Kaufman, R.J.; Gallouzi, I.E. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol. Biol. Cell 2007, 18, 2603–2618. [Google Scholar]
- Dember, L.M.; Kim, N.D.; Liu, K.Q.; Anderson, P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996, 271, 2783–2788. [Google Scholar]
- Barreau, C.; Paillard, L.; Osborne, H.B. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005, 33, 7138–7150. [Google Scholar]
- Caput, D.; Beutler, B.; Hartog, K.; Thayer, R.; Brown-Shimer, S.; Cerami, A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. USA 1986, 83, 1670–1674. [Google Scholar]
- Chen, C.Y.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995, 20, 465–470. [Google Scholar]
- Chen, C.Y.; Xu, N.; Shyu, A.B. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: Different deadenylation kinetics and uncoupling from translation. Mol. Cell Biol. 1995, 15, 5777–5788. [Google Scholar]
- Reyes, R.; Alcalde, J.; Izquierdo, J.M. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 2009, 10, R87. [Google Scholar]
- Kim, H.S.; Kuwano, Y.; Zhan, M.; Pullmann, R., Jr.; Mazan-Mamczarz, K.; Li, H.; Kedersha, N.; Anderson, P.; Wilce, M.C.; Gorospe, M.; et al. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell Biol. 2007, 27, 6806–6817. [Google Scholar]
- Emara, M.M.; Liu, H.; Davis, W.G.; Brinton, M.A. Mutation of mapped TIA-1/TIAR binding sites in the 3' terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J. Virol. 2008, 82, 10657–10670. [Google Scholar]
- Kim, H.S.; Wilce, M.C.J.; Yoga, Y.M.K.; Pendini, N.R.; Gunzburg, M.J.; Cowieson, N.P.; Wilson, G.M.; Williams, B.R.; Gorospe, M.; Wilce, J.A. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res. 2011, 39, 1–14. [Google Scholar]
- Bauer, W.J.; Heath, J.; Jenkins, J.L.; Kielkopf, C.L. Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1. J. Mol. Biol. 2012, 415, 727–740. [Google Scholar]
- Kim, H.S.; Headey, S.J.; Yoga, Y.M.; Scanlon, M.J.; Gorospe, M.; Wilce, M.C.; Wilce, J.A. Distinct binding properties of TIAR RRMs and linker region. RNA Biol. 2013, 10, 579–589. [Google Scholar]
- Suswam, E.A.; Li, Y.Y.; Mahtani, H.; King, P.H. Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 2005, 33, 4507–4518. [Google Scholar]
- Kuwasako, K.; Takahashi, M.; Tochio, N.; Abe, C.; Tsuda, K.; Inoue, M.; Terada, T.; Shirouzu, M.; Kobayashi, N.; Kigawa, T.; et al. Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode. Biochemistry 2008, 47, 6437–6450. [Google Scholar]
- Cruz-Gallardo, I.; Aroca, A.; Gunzburg, M.J.; Sivakumaran, A.; Yoon, J.H.; Angulo, J.; Persson, C.; Gorospe, M.; Karlsson, B.G.; Wilce, J.A.; et al. The binding of TIA-1 to RNA C-rich sequences is driven by its C-terminal RRM domain. RNA Biol. 2014, 11, 6. [Google Scholar]
- Wang, I.; Hennig, J.; Jagtap, P.K.; Sonntag, M.; Valcarcel, J.; Sattler, M. Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1. Nucleic Acids Res. 2014, 42, 5949–5966. [Google Scholar]
- Kumar, A.O.; Swenson, M.C.; Benning, M.M.; Kielkopf, C.L. Structure of the central RNA recognition motif of human TIA-1 at 1.95A resolution. Biochem. Biophys. Res. Commun. 2008, 367, 813–819. [Google Scholar]
- Auweter, S.D.; Oberstrass, F.C.; Allain, F.H. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res. 2006, 34, 4943–4959. [Google Scholar]
- Cruz-Gallardo, I.; Aroca, A.; Persson, C.; Karlsson, B.G.; Diaz-Moreno, I. RNA binding of T-cell intracellular antigen-1 (TIA-1) C-terminal RNA recognition motif is modified by pH conditions. J. Biol. Chem. 2013, 288, 25986–25994. [Google Scholar]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar]
- Jarrett, J.T.; Lansbury, P.T., Jr. Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer’s disease and scrapie? Cell 1993, 73, 1055–1058. [Google Scholar]
- Bandyopadhyay, U.; Cuervo, A.M. Chaperone-mediated autophagy in aging and neurodegeneration: Lessons from α-synuclein. Exp. Gerontol. 2007, 42, 120–128. [Google Scholar]
- Bucciantini, M.; Nosi, D.; Forzan, M.; Russo, E.; Calamai, M.; Pieri, L.; Formigli, L.; Quercioli, F.; Soria, S.; Pavone, F.; et al. Toxic effects of amyloid fibrils on cell membranes: The importance of ganglioside GM1. FASEB J. 2012, 26, 818–831. [Google Scholar]
- Furukawa, Y.; Kaneko, K.; Matsumoto, G.; Kurosawa, M.; Nukina, N. Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J. Neurosci. 2009, 29, 5153–5162. [Google Scholar]
- Kato, M.; Han, T.W.; Xie, S.; Shi, K.; Du, X.; Wu, L.C.; Mirzaei, H.; Goldsmith, E.J.; Longgood, J.; Pei, J.; et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149, 753–767. [Google Scholar]
- Wolozin, B. Regulated protein aggregation: Stress granules and neurodegeneration. Mol. Neurodegener. 2012, 7, 56. [Google Scholar]
- Vanderweyde, T.; Yu, H.; Varnum, M.; Liu-Yesucevitz, L.; Citro, A.; Ikezu, T.; Duff, K.; Wolozin, B. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 2012, 32, 8270–8283. [Google Scholar]
- Liu-Yesucevitz, L.; Bilgutay, A.; Zhang, Y.J.; Vanderweyde, T.; Citro, A.; Mehta, T.; Zaarur, N.; McKee, A.; Bowser, R.; Sherman, M.; et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: Analysis of cultured cells and pathological brain tissue. PLoS One 2010, 5, e13250. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waris, S.; Wilce, M.C.J.; Wilce, J.A. RNA Recognition and Stress Granule Formation by TIA Proteins. Int. J. Mol. Sci. 2014, 15, 23377-23388. https://doi.org/10.3390/ijms151223377
Waris S, Wilce MCJ, Wilce JA. RNA Recognition and Stress Granule Formation by TIA Proteins. International Journal of Molecular Sciences. 2014; 15(12):23377-23388. https://doi.org/10.3390/ijms151223377
Chicago/Turabian StyleWaris, Saboora, Matthew Charles James Wilce, and Jacqueline Anne Wilce. 2014. "RNA Recognition and Stress Granule Formation by TIA Proteins" International Journal of Molecular Sciences 15, no. 12: 23377-23388. https://doi.org/10.3390/ijms151223377
APA StyleWaris, S., Wilce, M. C. J., & Wilce, J. A. (2014). RNA Recognition and Stress Granule Formation by TIA Proteins. International Journal of Molecular Sciences, 15(12), 23377-23388. https://doi.org/10.3390/ijms151223377




